TransFLP — A Method to Genetically Modify Vibrio cholerae Based on Natural Transformation and FLP-recombination
Summary
A quick method to modify the genome of V. cholerae is described. These modifications include the deletion of single genes, gene clusters and genomic islands as well as the integration of short sequences (e.g. promoter elements or affinity-tag sequences). The method is based on the natural transformation and FLP-recombination.
Abstract
Several methods are available to manipulate bacterial chromosomes1-3. Most of these protocols rely on the insertion of conditionally replicative plasmids (e.g. harboring pir-dependent or temperature-sensitive replicons1,2). These plasmids are integrated into bacterial chromosomes based on homology-mediated recombination. Such insertional mutants are often directly used in experimental settings. Alternatively, selection for plasmid excision followed by its loss can be performed, which for Gram-negative bacteria often relies on the counter-selectable levan sucrase enzyme encoded by the sacB gene4. The excision can either restore the pre-insertion genotype or result in an exchange between the chromosome and the plasmid-encoded copy of the modified gene. A disadvantage of this technique is that it is time-consuming. The plasmid has to be cloned first; it requires horizontal transfer into V. cholerae (most notably by mating with an E. coli donor strain) or artificial transformation of the latter; and the excision of the plasmid is random and can either restore the initial genotype or create the desired modification if no positive selection is exerted. Here, we present a method for rapid manipulation of the V. cholerae chromosome(s)5 (Figure 1). This TransFLP method is based on the recently discovered chitin-mediated induction of natural competence in this organism6 and other representative of the genus Vibrio such as V. fischeri7. Natural competence allows the uptake of free DNA including PCR-generated DNA fragments. Once taken up, the DNA recombines with the chromosome given the presence of a minimum of 250-500 bp of flanking homologous region8. Including a selection marker in-between these flanking regions allows easy detection of frequently occurring transformants.
This method can be used for different genetic manipulations of V. cholerae and potentially also other naturally competent bacteria. We provide three novel examples on what can be accomplished by this method in addition to our previously published study on single gene deletions and the addition of affinity-tag sequences5. Several optimization steps concerning the initial protocol of chitin-induced natural transformation6 are incorporated in this TransFLP protocol. These include among others the replacement of crab shell fragments by commercially available chitin flakes8, the donation of PCR-derived DNA as transforming material9, and the addition of FLP-recombination target sites (FRT)5. FRT sites allow site-directed excision of the selection marker mediated by the Flp recombinase10.
Protocol
The TransFLP method (Figure 1) is exemplified by three different approaches: I) the deletion of two adjacent genes encoding virulence determinants of V. cholerae (e.g., ΔctxAB); II) the removal of a pathogenicity island (e.g., ΔVPI-1); and III) the integration of a T7 RNA polymerase-dependent promoter sequence upstream of a gene-of-interest, which subsequently allows its artificial expression in the respective strain background (e.g., T7-tfoX) (Figure 2).
1. Preparation of Chitin Flakes
- Weight 50-80 mg of chitin flakes in a standard 1.5 ml plastic tube. This can be prepared in bulk. Chitin flakes are commercially available from Sigma (cat. number C9213).
- Autoclave the flakes keeping the lids of the tubes open.
- Immediately close the lids after the autoclave has cooled down.
- Store autoclaved chitin flakes at room temperature.
2. Optional: Artificial Transformation of V. cholerae with Flp Recombinase-encoding Plasmid
- Prepare electrocompetent V. cholerae cells using standard methods11. Store aliquots of competent cells at -80 °C.
- Add 1-2 μl of a regular mini-preparation of plasmid pBR-flp5 to the electrocompetent V. cholerae cells and transfer the mixture into an electroporation cuvette (0.2 cm gap width).
- Apply a pulse at 1.6 kV.
- Add 0.9 ml SOC medium, gently transfer cell to a standard 14 ml tube. Incubate non-moving for 2.5 to 3 hr at 30 °C.
- Plate 100 μl and 300 μl on LB plates containing 100 μg/ml ampicillin and incubate at 30 °C overnight.
- Purify single clone and store as glycerol stock for future TransFLP experiments.
3. Oligonucleotide Design and Polymerase Chain Reaction
- At least six oligonucleotides are required to amplify the DNA region(s) of interest as well as the FRT-flanked antibiotic cassette by PCR (Figure 3). It is recommended to also include a pair of oligonucleotides priming outside the inserted PCR fragment. This allows checking for integration and correct excision of the FRT-flanked antibiotic resistance cassette (e.g. depicted as ‘chk-up’ and ‘chk-down’ primers in Figures 3 to 5).
- Design the oligonucleotides #2, #3, #4, and #5 with care. They should be able to sufficiently anneal (e.g. ~28 bp) to the template DNA. The template DNA is genomic DNA (gDNA) for primers #2 and #5 and FRT-Kan-FRT-containing plasmid DNA such as pROD1712 and pBR-FRT-KAN-FRT (this study) for primers #3 and #4 (Figure 3A).
- Design the primers #2 to #5 allowing extensive base-pairing at their 5′-end between #2 / #3 and #4 / #5, respectively (Figure 3A inset). Two rounds of PCR follow.
- Perform three independent PCR reactions as a 1st round of PCR. The first one will amplify the upstream region of the gene / DNA region-of-interest with the aid of oligonucleotides #1 and #2. The PCR should result in a fragment of at least 500 bp in length to allow homologous recombination within the cells. Shorter fragments (~250 bp) can be sufficient8 but are not recommended.
- In parallel perform the second PCR, which amplifies the FRT-flanked antibiotic cassette. For the examples provided here we mainly used aph (kanR). Use oligonucleotides #3 and #4 for this reaction (Figure 3A).
- Concomitantly, amplify the downstream DNA region by PCR (third sample). The PCR fragment should also be at least 500 bp in length. Perform the PCR with gDNA as template and oligonucleotides #5 and #6 (Figure 3A).
- After purification of all three PCR fragments, perform the 2nd round of PCR. Use an equal mixture of all three fragments obtained in the first round as template. The amplification is catalyzed by oligonucleotides #1 and #6. The resulting PCR fragment (Figure 3B) serves as transforming DNA in the natural transformation experiment (Figure 1).
4. Chitin-induced Natural Transformation
- Grow V. cholerae cells aerobically in rich medium at 30 °C until they reach an optical density at 600 nm of approximately 0.5.
- Harvest the bacteria by centrifugation. Wash pellet once in defined artificial medium (DASW6) before resuspending the cells in 2 volumes of DASW. Add 1 ml of culture to 50-80 mg of sterile chitin flakes (explained under point 1). If bacteria harbor plasmid pBR-flp (optional point 2), add ampicillin (50 μg/ml) to the culture. Incubate at 30 °C for 16-24 hr without movement.
- Add ≥ 200 ng of the 2nd-round PCR-derived fragment (explained under point 3). Mix carefully without extensively detaching the bacteria from the chitin surfaces. Incubate at 30 °C for 24 hr without movement.
- Vortex the culture extensively for ≥ 30 sec. Spread 100-300 μl on selective LB medium plates (e.g. kanamycin or gentamicin-containing LB plates for examples provided here). Use double-selective plates if the bacteria harbor the plasmid pBR-flp by adding ampicillin concomitantly to the other antibiotics. Incubate plates at 30 °C for 16-24 hr or until colonies are visible.
- Isolate single transformants from selective plates. Such transformants have replaced the original chromosomal locus by the PCR fragment due to a double crossover event. These strains can be directly used for further experiments in case the removal of the antibiotic cassette is not necessary.
5. Removal of Selective Cassette(s) by Flp-recombination
- Artificially transform your transformants with plasmid pBR-flp as described under point 2 if not done before the natural transformation assay.
- Grow the bacteria on LB agar plates containing ampicillin at 37 °C for 16-24 hr. Options: change the temperature in between for 2-3 hr to 40 °C as expression of flp from plasmid pBR-flp is de-repressed at higher temperatures5,10; transfer bacteria to fresh plates after approx. 8 hr of incubation.
- Test for antibiotic-sensitivity (demonstrated here for kanamycin; Figure 1) by restreaking the clones in parallel on antibiotic-containing and antibiotic-free agar plates. Isolate single sensitive colonies. Freeze ampicillin-resistant clone as glycerol stock if you intend to further modify the strain with other deletion / insertion using TransFLP.
6. Plasmid Curing
- Grow culture overnight under aerobic condition and at 30 °C. Use rich medium without the addition of any antibiotic.
- Optional: Grow fresh culture for 3-6 hr by diluting the overnight culture 1:100 in fresh antibiotic-free medium.
- Plate or streak dilution(s) of culture on plain LB agar plate(s) and incubate at 30 °C for 8-16 hr (until colonies are visible).
- Test for ampicillin-sensitivity of the clones by restreaking the clones in parallel on antibiotic-containing and antibiotic-free agar plates (Figure 1). Freeze ampicillin-sensitive strain as glycerol stock.
Representative Results
Representative results of the three examples are shown in Figures 4 to 6. The first approach aimed at deleting the neighboring genes ctxA and ctxB. Together they encode the major virulence factor of V. cholerae, cholera toxin. We designed oligonucleotides for the TransFLP method as described above and according to Figure 3. The parental strain, the intermediate strain harboring the FRT-flanked antibiotic resistance cassette at the original DNA locus of ctxAB and the final strain deleted for ctxAB and freed of the resistance cassette were all tested for their genotype (Figure 4). The method of choice was whole-cell PCR. We used primers that do not anneal to the transforming PCR fragment but only to chromosomal DNA (Figure 3). This allowed us to verify the site-specific exchange of the PCR fragment with the homologous region of the chromosome (Figure 4A). The PCR fragments were separated by standard agarose gel electrophoresis to determine their size (Figure 4B). This strategy can be used to check strain intermediates and to confirm the final strain construct(s).
For the second example, the removal of a whole genomic island, we chose the Vibrio pathogenicity island 1 (VPI-1) as target13. The standard TransFLP procedure described above was unsuccessful in this case. It was known from previous studies that such a large DNA region can be exchanged by a similar sized DNA region using chitin-induced natural transformation9. We therefore hypothesized that the size difference between the region-to-be-deleted and the rather short transforming PCR fragment (< 3.5 kb) might be the limiting factor. We therefore decided to use the TransFLP method in two steps: we first integrated the aph gene (kanR) preceded by a single FRT site at the start of VPI-1 (illustrated in Figure 5A). We then performed a second round of natural transformation, which allowed us to insert an additional antibiotic resistance cassette (gmR, conferring gentamicin resistance) followed by a second FRT site at the end of the pathogenicity island (Figure 5A). The respective double-resistant strain was electroporated to insert plasmid pBR-flp and the TransFLP method was continued as described above. The resulting strain lacked the whole VPI-1 as verified by PCR of purified genomic DNA (Figure 5B).
For the third example we integrated a T7 RNA polymerase-dependent promoter sequence upstream of a gene-of-interest, in our case the gene encoding the major regulator of natural transformation tfoX6. This was done using the TransFLP method with a slight modification from the standard protocol: we included the T7 RNA dependent promoter consensus sequence according to Ref.14 as overhangs into the oligonucleotides #4 and #5. With this strategy the T7 RNA polymerase-dependent promoter sequence was kept on the chromosome even after the antibiotic resistance cassette was excised (Figure 6). We integrated the construct into V. cholerae strain ADVCH-T7POL, which harbors the T7 RNA polymerase gene driven by the lacUV5 promoter (Blokesch, unpublished). To test for the functionality of the construct, namely the T7 RNA polymerase-dependent expression of tfoX, we performed natural transformation assays in LB medium. The parental strain is not naturally transformable under these conditions due to the lack of tfoX transcription in rich medium and in the absence of its natural inducer chitin (Figure 6). This phenotype was independent of whether the T7 RNA polymerase was artificially induced by IPTG or not (Figure 6, lanes 1 and 2, respectively). However, the TransFLP-generated strain ADVCH-T7POL-T7-tfoX::FRT, which contains the T7 RNA polymerase-dependent promoter upstream of the competence gene tfoX, was indeed naturally transformable in rich medium (Figure 6, lanes 3 and 4). Due to the leakage of the lacUV5 promoter in LB medium15 the produced T7 RNA polymerase already transcribed tfoX from the T7 RNA polymerase-dependent promoter without induction. This initiated natural competence and transformation confirming the functionality of the integrated T7 RNA polymerase-dependent promoter sequence (Figure 6, lane 3). This phenotype was significantly enhanced by full induction of the T7 RNA polymerase due to addition of the inducer of the lacUV5 promoter, IPTG (Figure 6, lane 4).
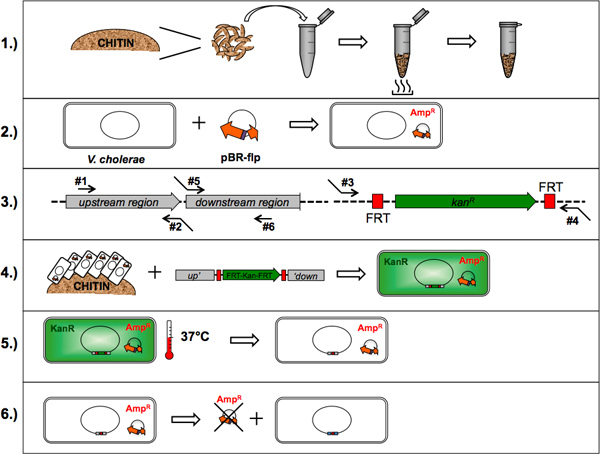
Figure 1. Flow chart of the TransFLP method. The six points described in this protocol are drafted: 1.) Preparation of chitin flakes; 2.) Optional: Artificial transformation of V. cholerae with Flp recombinase-encoding plasmid; 3.) Oligonucleotide design and polymerase chain reaction; 4.) Chitin-induced natural transformation; 5.) Removal of selective cassette(s) by flp-recombination; 6.) Plasmid curing. Click here to view larger figure.
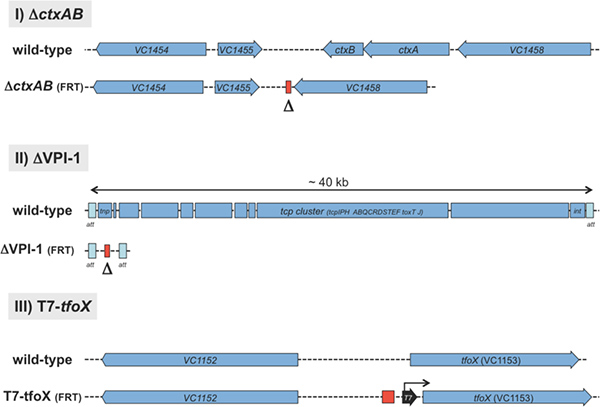
Figure 2. Schematic representation of three application possibilities for the TransFLP method. I) A 1458 bp DNA fragment, containing the cholera toxin encoding genes ctxA and ctxB, was deleted as indicated by Δ. II) The ~ 40 kb Vibrio pathogenicity island 1 (VPI-113) was deleted from the wild-type strain. III) A T7 RNA polymerase-dependent promoter sequence (28 bp) was integrated upstream of the tfoX gene. The red rectangle indicates the left behind short nucleotide sequence of a single FRT site10. Click here to view larger figure.
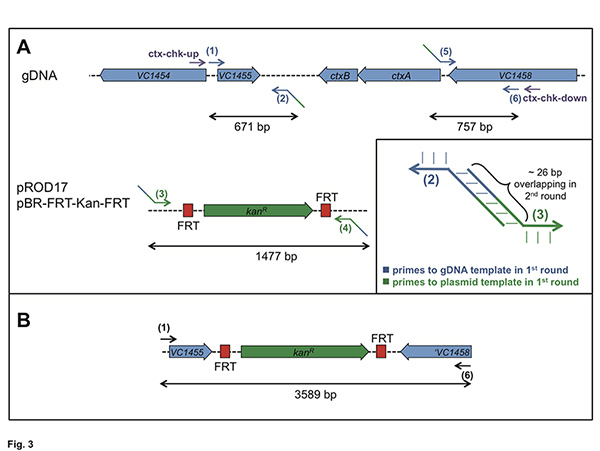
Figure 3. Explanation for primer design and performance of PCR based on the deletion of ctxAB. Panel A: For the 1st round of PCR genomic DNA (gDNA) and plasmids pROD1712 and pBR-FRT-Kan-FRT, respectively, served as templates. The required oligonucleotides are #1 to #6. The primer #2 / #3 and #4 / #5 (example in inset) should also possess significant overlap to each other at their 5’ends. Panel B: After the 2nd round of PCR, which is based on the 1st round PCR fragments as templates, a long fragment should be obtained using primer pair #1 + #6. Click here to view larger figure.
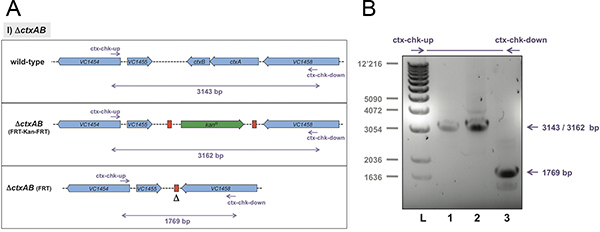
Figure 4. Expected result exemplified for the ctxAB deletion strategy. The original strain (wild-type), the insertional mutant ΔctxAB::FRT-Kan-FRT, and the final strain ΔctxAB::FRT were tested for their genotypes. Panel A: Schematic representation of the differentiation power of the primer pair ctx-chk-up and ctx-chk-down, most notably with respect to the excision of the antibiotic cassette. Panel B: Whole bacterial cells were used as template in PCR reactions. To check for the changed DNA locus primers chk-up and chk-down were utilized. The amplified PCR fragments were separated by agarose gel electrophoresis and visualized using SYBR safe DNA stain. Lane 1: Wild-type V. cholerae strain; lane 2: strain ΔctxAB::FRT-Kan-FRT; lane 3: ΔctxAB::FRT. Expected PCR fragment sizes according to panel A are indicated on the right. L, 1kb ladder (Invitrogen). Click here to view larger figure.
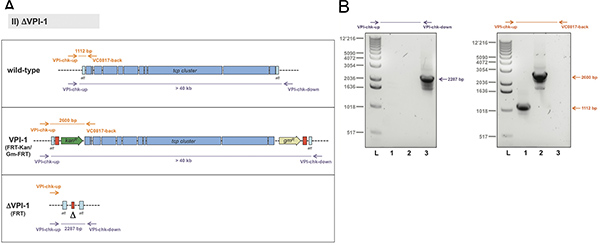
Figure 5. Strategy to verify the deletion of the genomic island. Panel A: Scheme of Vibrio pathogenicity island 1 (VPI-1) (freely adapted from Ref.16; not to scale). For the wild-type strain the region of interest is non-amplifiable by means of standard PCR and the VPI-1-specific primers chk-up and chk-down (~ 40 kb). Therefore an additional oligonucleotide annealing within the region to-be-deleted was applied (VC0817-back) together with VPI-chk-up. Panel B: Representative result using genomic DNA of the respective strains as template and the primer pairs indicated above each image. Strains tested: Wild-type V. cholerae strain (lane 1), strain VPI::FRT-Kan/Gm-FRT (lane 2), and ΔVPI-1::FRT (lane 3). Sizes of the PCR fragments as predicted in panel A are indicated on the right. L, 1kb ladder (Invitrogen). Click here to view larger figure.
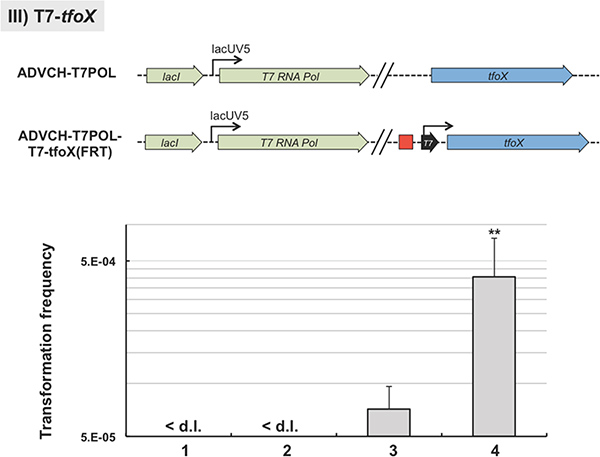
Figure 6. Representative phenotype after insertion of an artificial promoter sequence. The T7 RNA polymerase-dependent promoter sequence was integrated upstream of the competence regulatory gene tfoX using the TransFLP method (sketch above graph). The parental V. cholerae strain (ADVCH-T7POL) contains a T7 RNA polymerase gene driven by the lacUV5 promoter. Graph: The parental strain ADVCH-T7POL (lanes 1 and 2) and the strain manipulated by TransFLP (ADVCH-T7POL-T7-tfoX::FRT; lanes 3 and 4) were tested for natural transformability in the absence (lanes 1 and 3) or presence of 1mM IPTG (lanes 2 and 4). Transforming genomic DNA was derived from strain A1552-LacZ-Kan8. Transformation frequencies are given on the Y-axis. < d.l., below detection limit of ~5 x 10-8. **P < 0.01 (as determined by student’s t test of log-transformed data). Black arrow with text ‘T7’: T7 RNA polymerase-dependent promoter sequence. Click here to view larger figure.
Discussion
The TransFLP method described above and elsewhere5 has been extensively used in our laboratory. The kind of genetic manipulations that are feasible include amongst others: deletion of single genes and gene clusters, deletion of genomic islands (e.g., VPI-1), insertion of sequences upstream of a gene of interest (e.g., promoter sequences) and insertion of sequences at the end of a specific gene (e.g., encoding affinity tags).
An import prerequisite for TransFLP is that the respective V. cholerae strain is naturally transformable. This is sometimes not the case, especially in isolates derived from cholera patients. Such strains are frequently mutated within the quorum sensing circuit e.g., within the gene encoding the major regulator of quorum sensing, HapR17. Providing back a copy of hapR restores natural transformability in these strains6 as exemplified for the HapR-defective strain N1696118.
Possible modifications can be incorporated, such as the enrichment of the bacteria after natural transformation8 in case the frequency is too low. Please note that the transformation frequency is in general lower in case the pBR-flp plasmid is already present while doing the chitin-induced natural transformation assay. This is most likely caused by partial leakage of the temperature sensitive expression of the flp gene from pBR-flp, which would result in premature excision of the antibiotic resistance cassette and the inability to select for these transformants. We therefore recommend for critical cases to insert the pBR-flp plasmid after the natural transformation step.
Another critical step is the 2nd round of PCR, which will combine the three 1st-round PCR fragments. It can occur that partial fragments are present after the PCR reaction (e.g., a partial assembly of only fragments ‘up’ and ‘FRT-Kan-FRT’ according to Figure 1). This is negligible, as double homologous recombination with the targeted chromosomal region cannot occur in this situation. As only transformants harboring the antibiotic resistance cassette are selected for, all other transformants with non-desired DNA exchanges are excluded.
Another point to keep in mind is with respect to the repetition of the procedure for several rounds. We have already successfully combined the deletion of the ctxAB gene cluster with the deletion of the pathogenicity island (VPI-1) and another virulence gene cluster (Blokesch, unpublished). Furthermore, we have effectively integrated the T7 RNA polymerase-dependent promoter within these strains (upstream tfoX and elsewhere). However, the envisioned genotype should be constantly tested for all strain intermediates (e.g., by colony PCR as demonstrated in Figure 4). This will help to exclude the undesired Flp-mediated recombination of the left behind FRT scars (shown as red rectangular in all figures) with each other while the pBR-flp plasmid is still present in the bacteria. This can probably be disregarded for distantly located genes, as the probability that the excision of the in-between region would be lethal due to the deletion of essential genes is very high.
Finally, we would like to discuss the deletion of genes within operons. We cannot exclude that the deletion of an upstream gene, which leaves an FRT scar behind, might change the expression of the downstream gene(s). To ensure that this effect is not causing the observable phenotype, but that this is indeed caused by the deletion, a complementation assay is recommended. Another control that we have already successfully used in the laboratory is the integration of the FRT scar downstream of the gene-of-interest without deleting the latter. We did not observe such problems in our laboratory yet but cannot exclude their existence.
Disclosures
The authors have nothing to disclose.
Acknowledgements
I would like to acknowledge Olga de Souza Silva for technical assistance. This work was supported by Swiss National Science Foundation (SNSF) Grant 31003A_127029.
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Chitin flakes | Sigma | C9213 | Autoclaving required |
| OPTIONAL: Micropulser (in case antibiotic cassette should be excised by Flp recombinase encoded on pBR-flp) | Biorad | 165-2100 | Or comparable electroporators |
References
- Miller, V. L., Mekalanos, J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575-2583 (1988).
- Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P., Kushner, S. R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171, 4617-4622 (1989).
- Kato, C., Ohmiya, R., Mizuno, T. A rapid method for disrupting genes in the Escherichia coli genome. Biosci. Biotechnol. Biochem. 62, 1826-1829 (1998).
- Donnenberg, M. S., Kaper, J. B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310-4317 (1991).
- De Souza Silva, O., Blokesch, M. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid. 64, 186-195 (2010).
- Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C. -. Y., Schoolnik, G. K. Chitin induces natural competence in Vibrio cholerae. Science. 310, 1824-1827 (2005).
- Pollack-Berti, A., Wollenberg, M. S., Ruby, E. G. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. 12, 2302-2311 (2010).
- Marvig, R. L., Blokesch, M. Natural transformation of Vibrio cholerae as a tool-optimizing the procedure. BMC Microbiol. 10, 155 (2010).
- Blokesch, M., Schoolnik, G. K. Serogroup Conversion of Vibrio cholerae in Aquatic Reservoirs. PLoS Pathog. 3, e81 (2007).
- Cherepanov, P. P., Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 158, 9-14 (1995).
- Sambrook, J., Fritsch, E. F., Maniatis, T., Ford, N., Nolan, C., Ferguson, M. . Molecular Cloning: A laboratory manual. 1, (1989).
- Reyes-Lamothe, R., Possoz, C., Danilova, O., Sherratt, D. J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 133, 90-102 (2008).
- Karaolis, D. K. R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA. 95, 3134-3139 (1998).
- Ikeda, R. A., Ligman, C. M., Warshamana, S. T7 promoter contacts essential for promoter activity in vivo. Nucleic Acids Res. 20, 2517-2524 (1992).
- Pan, S. H., Malcolm, B. A. Reduced background expression and improved plasmid stability with pET vectors in BL21 (DE3). Biotechniques. 29, 1234-1238 (2000).
- Faruque, S. M., Zhu, J., Asadulghani, M., Kamruzzaman, J. J., Mekalanos, Examination of diverse toxin-coregulated pilus-positive Vibrio cholerae strains fails to demonstrate evidence for Vibrio pathogenicity island phage. Infect. Immun. 71, 2993-2999 (2003).
- Joelsson, A., Liu, Z., Zhu, J. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 74, 1141-1147 (2006).
- Heidelberg, J. F. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 406, 477-483 (2000).

