概要
This procedure describes how to visualize, track, and quantify Acanthamoeba spp. motility.
Abstract
Acanthamoeba keratitis is a severe ocular infection that poses treatment challenges and can lead to blindness. Despite its ubiquity and potential contamination of contact lenses after water exposure, the natural behavior of this pathogen remains elusive. Understanding Acanthamoeba’s movement patterns can inform us about how it colonizes contact lenses and contaminates the patient’s cornea. It is fortunate that Acanthamoeba spp. are visible via brightfield microscopy starting at 4x magnification. Previous techniques have been developed to quantify Acanthamoeba motility in regard to cytopathic effects or under-exposure to the electric field. Here, we describe a method to track and quantify Acanthamoeba spp. motility long-term (hours to days), which is a protocol applicable to multiple amoeba strains, surfaces, and nutritional status of amoeba. This procedure is germane to determining many core motility quantifications, such as distance, speed, confinement, and directionality, which are necessary to monitor different stages of infection, proliferation, or behavioral change.
Introduction
Acanthamoeba research has increased dramatically in recent years due to the increased prevalence of Acanthamoeba keratitis (AK), which is a parasitic infection following the attachment of Acanthamoeba to the cornea1. While outbreaks of AK can be attributed to inappropriate contact lens care or ineffective contact lens care solutions2,3,4,5, there are currently no requirements to demonstrate Acanthamoeba disinfection efficacy for any product on the market. However, there is an ongoing effort in the scientific community and within standards organizations to examine the protocols necessary for quantifying the disinfection efficacy of contact lens care products6,7. Further, due to its resemblance in cellular and functional aspects to human macrophages, Acanthamoeba has been noted as having a significant role in hosting and disseminating other human pathogens8, in addition to the pathogenicity the amoeba itself brings.
Recent techniques have been described that have been able to quantify Acanthamoeba motility – which is generally not highly subject to Brownian movement9,10 – in regard to cytopathic effects or under-exposure to the electric field9,11, as well as advancements in motility analysis in giant virus research using Acanthamoeba as the trackable viral vector12,13. There have also been constant and significant improvements in cell and particle tracking occurring over the last 20 years using new software programs such as the imaging software used here, and new algorithms and deep learning technologies14. However, this is a relatively new and growing field of science regarding benchtop research, clinical application, and industrial standards, and there has been a paucity of data published regarding methods to visualize and track this amoeba, particularly in order to quantify behavioral changes following adherence to contact lenses or during or after contact lens disinfection. Other fields expanding into long-term visual monitoring have supported this effort15,16,17. Due to Acanthamoeba's inherently challenging nature – including a general resistance to plasmids (which could confer fluorescence), the ability of the amoeba to consume and destroy standard cell dyes, and a unique outer protein makeup – making antibody tagging difficult – methods available to other cells which make them visible in settings other than brightfield imaging have been unusable in this organism. Thus, quantifying the motility of this amoeba has demonstrated a significant addition to the field. Using the method described here, we have been able to ascertain that amoeba remain motile for at least 12 h without nutrients18 and that amoeba that is challenged with a disinfection process and ceases motility during disinfection can recover their motility post-disinfection if they are not fully lysed19.
This protocol details how to visually track and quantify the motility of amoeba microscopically. The primary steps are to record amoeba in brightfield using the appropriate focus and timing between images, transform the images into binary using imaging software, and then use an imaging software's tracking plugin to set the experimental parameters and follow each amoeba to determine the required measurements such as speed, distance, and confinement. Following this, it is possible to quantify the chemotaxis of an amoeba or an amoeba population in order to define directionality. The key contribution of this method is to visualize and quantify amoeba behavior during different states of nutritional support, surface adherence, disinfection challenge, or other environmental alterations such as cohabitation with mammalian cell culture.
Protocol
1. Acanthamoeba preparation
NOTE:This protocol has been verified for ATCC 50655, 30872, 50702, 30010, 30461, 50370, 50703, 30137, PRA-115, and PRA-411. This is a BSL2 pathogen and should be worked within a BSL2 hood and lab.
- Create a master culture from a sample vial or prepared plug20 by filling a T75 flask with 30 mL of AC6 medium and aseptically add one plug or the contents of a sample vial to it. Incubate the flask for 3-5 days at 26-30 °C until the flask is 50% to 80% confluent.
- Passage the cells the day before they are needed to create a homogenous population of trophozoites.
- According to the needs of the strain, shake and/or briskly strike the master culture 2x to dislodge adhered trophozoites.
- Fill a T75 flask with 30 mL of AC6 medium. Add 2 mL of the master culture to the passage. Incubate the flask for 18-24 h at 26-30 °C.
- Prior to harvest, visually inspect the trophozoite population under 4x magnification in the microscope. Ensure trophozoites are adherent and uniform.
- Briskly strike the flask 2x to dislodge adhered trophozoites. Pour the contents into a 50 mL conical tube.
- Spin the tube at 500 x g for 5 min in a balanced centrifuge to pellet the amoeba. Pour or pipette off the supernatant media and discard. Dilute the pellet in 2-10 mL of ¼ Ringer's solution.
- Vortex the sample. Add 10 µL of sample to a disposable hemocytometer and count the CFU/mL of the Acanthamoeba.
- Based on the hemocytometer count, dilute the amoeba pellet to a concentration of 7.5 x 103 cells/mL in ¼ Ringer's solution.
- Seed amoeba on a surface, which can be glass, plastic, or non-nutrient agar, and a variety of well shapes depending on the experimental needs as described below.
- 96-well plate: Seed each well with 200 µL of Acanthamoeba suspension and enclose the plate with the lid.
- 48-well plate: Seed each well with 1 mL of Acanthamoeba suspension and enclose the plate with the lid.
- Flow cell: Slowly add 4 mL of Acanthamoeba suspension through the sterile ports of a sterile aluminum flow cell, avoiding bubbles in the chamber. Clamp the ports closed after the suspension has been added.
- Prior to imaging, allow amoeba at least 30 min to adhere to the surface before initiating microscopic recording.
2. Visualizing and recording Acanthamoeba
- Visualize the amoeba at 4x magnification in brightfield. The background should appear light grey, and the amoeba should be black. Adjust the light and focus so that the amoeba are solid dark circles instead of translucent (so, slightly out of focus, Figure 1.).
NOTE: The optimal magnification is 4x for tracking for several reasons: Amoeba that leave or enter the field of view during tracking should not be included in data analysis, so the lower magnification best supports the maximum number of amoeba included in the analysis in a time period. Amoeba should appear as solid black circles in brightfield in order to convert them to binary for tracking analysis. This is easiest to achieve at a lower magnification, as higher magnifications easily reveal the translucent nature and internal structures of the amoeba. In general, more amoeba can be captured in a single field of view in a lower magnification, supporting more robust data analysis. - Set the recording programs in the imaging software to record an image at regular intervals to track the amoeba. The longest time between images that will allow for accurate tracking is 30 s, optimal time for file size and tracking detail is suggested at either 12 s or 24 s.
- Amoebas can be tracked for multiple days at a time, but be aware of the file size this kind of recording will create and the inherent difficulty of manipulating extremely large files. To avoid this, record amoebas for sections of time at certain intervals. For instance, record for 1 h every 12 h for 5 days.
- If the microscope and program allow, record multiple wells in a single session using the XY coordinates of the plate wells or multiple locations of a flow cell. If needed, stitch together multiple sections of one well or flow cell to create a larger field of view at 4x magnification.
NOTE: The only constraint on how many locations can be recorded is how fast the microscope stage can move between locations within the image interval time (e.g., taking an image every 12 s, 24 s, etc.). However, this is not required, and tracking videos made from a single location can still have a sufficient number of tracks for robust statistical analysis. A Nikon Eclipse Ti-U Microscope was used in this study, and the automated moveable stage was utilized to record images in multiple wells at a time. However, any microscope with a programmable recording capability will work. The microscope must connect to a computer and be able to record images to a program or hard drive.
3. Amoeba size analysis
- Open the microscope file in the imaging software. Bio-formats import options will open. Within the dialog box, make sure the following are true: Stack viewing; View with: Selection-Hyperstack; Memory Management: check Use virtual stack; Color mode: Default.
- Ensure that none of the other drop-down menus or checkboxes are activated. Click Ok.
- Open bio-formats series options. Choose which Series is needed. If only one location is recorded at a time, there will likely only be one option here. Click Ok.
- Choose Image > Duplicate to duplicate the single well that is being worked with at a time by choosing only one C channel and only one Z channel.
- From here on, only work with the duplicated image for manipulation and analysis, not the original. Choose Image > Type > 8-bit. Choose Process > Subtract Background.
- Set the rolling ball to 10.0. Check Light Background. Check Sliding Paraboloid.
- Choose Process > Enhance Contrast. Set saturated pixels to .1% for trophozoites or 0.3% for a group of cells and aggregates.
- Choose Image > Adjust > Threshold (Default > B&W). Make Binary process will open. Select Default, and check Black background (of Binary Masks).
- Select the threshold aggressively so that most background spots are not visible, but some portion of each amoeba is visible.
- If the imaging software inverted the image and the background is white, and the amoeba is black, go to Edit > Invert to fix it to a black background and white amoeba.
- Choose Process> Binary > Close, in case the cells have a slight space in the outer membrane due to being out of focus or motility of the cell.
- Choose Process > Binary > Fill Holes. Remove non-amoeba artifacts by using the shape-drawing tools and Edit > Fill. If, by this point, the background is white and the amoeba is black, choose Edit > Invert. At this point, the image should represent the binary pictures in Figure 2.
- To record the size of each amoeba, choose Analyze > Analyze Particles. Set Size: 10-Infinitiy, Circularity: 0-1, Show: Nothing. Check Only Display Results and Summarize.
- Save the CSV files that appear. Choose File > Save As > Tiff and edit the name to the desired specification.
4. Preparation of microscope files for tracking (motility analysis)
- Open the microscope file in the imaging software. Bio-Formats Import Options will open.
- Within the dialog box, make sure the following are true: Stack viewing; View with: Selection-Hyperstack. For memory management, check Use Virtual Stack. Click Ok.
- Bio-Formats Series Options will open. Choose which Series is needed. If only one location is recorded at a time, there will likely only be one option here. Click Ok.
- Choose Image > Duplicate. From here on, only work on the duplicated section of the video instead of working on the original file.
NOTE: Duplicate the section of the video being tracked at that time. To do this, one needs to know the frames that correspond to the minutes required for the experiment. For instance, if one wants to track the 1st hour and takes images every 24 s, there should be 150 frames in the 1st hour. Therefore, the 1st hour will be frames 1-150, the 2nd hour will be 151-300, and so on. - Choose Image > Type > 8-bit. Choose Process > Subtract Background. Set rolling ball to 10.0.
- Check Light Background. Check Sliding Paraboloid. Choose Process > Enhance Contrast and set it at 0.1%.
- Check All x# Slices (for example, all 150 slices). Uncheck Normalize. Choose Image > Adjust > Threshold (Default > B&W).
- Make Binary process will open. Select Default. Check the Black Background (of Binary Masks).
- Threshold aggressively, so most background spots are not visible, but some portions of the amoeba are visible. At this step, if the background is white and the amoeba is black, choose Edit > Invert. The background should be black, and the amoeba should be white.
- Choose Process > Binary > Close. Choose Process > Binary > Fill Holes. Choose File > Save As > Tiff and edit the name to the desired specification. The file is now ready for tracking.
5. Analyzing motility through tracking
- From the imaging software with the tiff file that is needed to track open, go to Plugins > Tracking > Trackmate. The version of Trackmate will open.
- Choose Next. Select the LoG detector from the detector drop-down menu. Set estimated blob diameter: 35.0 microns, threshold 1.0. Uncheck Use median filter and Do sub-pixel localization.
- Click プレビュー at the first, middle, and end slices. Ensure that all amoebas are captured by a purple circle and that background defects or artifacts are not included.
- Choose Next. It will begin processing and this step may take a few minutes to do. A detection bar at the top of the screen will indicate how much of the process is complete.
- Choose Next when prompted. In initial thresholding, choose Next again without selecting anything. Select Hyperstack Displayer when selecting a view. Choose Next.
- Set filter on spots and choose Next without selecting anything. Select a tracker; to do so, select LAP Tracker (instead of Simple LAP Tracker). Choose Next.
- On frame-to-frame linking, set the max distance to 40 µm. On track segment gap closing check Allow Gap Closing. Set the maximum distance for this at 100 µm and the maximum frame gap to 4. Do not select anything for Track segment splitting or Track segment merging.
- Choose Next. Tracking may take significant time to execute. When it is completed, the window should say: Tracking done in x s at the bottom. Choose Next.
- Choose Next again when tracking is completed. Set filters on tracks: Select the + sign on the bottom left to add a filter for the number of spots in tracks. Set the filter to Above for at least 93% of the frames (e.g., 150 frames would need a minimum of 140 spots).
NOTE: Some versions of this software will require selecting the opposite equivalent, such as Below at least 7%, which can be done by dragging a line left or right to reach the desired number. - Choose Next. Display Options will appear. Make sure the tracks in each frame are not deformed or odd in the way the amoeba is moving before saving the Trackmate XML file or the Tracks CSV file.
- If a track needs to be deleted, click on TrackScheme. TrackScheme will show all the tracks and points of contact for the amoeba across all the frames.
NOTE: Possible reasons to delete a track are an amoeba has walked on/off the field of view during the recorded, a track is inappropriately shaped compared to the actual path of the amoeba, or for some reason, the track or the amoeba is an outlier in the data according to the experimental parameters, etc. - Click on the amoeba to highlight the spot green on the screen and in Trackscheme the spot will have a green square surrounding it. This will indicate which track or spot needs to be removed.
- Right-click on the track that needs to be removed and select Whole Track. Press the Delete button.
- After reviewing all the tracks, save the tracking file by clicking Save at the bottom left of the tracking software popup. Save the XML file using the video name. Click Resume to get back to the tracking software popup.
- Click Tracks and select Tracks on the left. Click Export to CSV and save the CSV file.
- If the spreadsheet does not recognize the resulting CSV, execute the following steps.
- Open the saved CSV file in Notepad. Click Control + A and Control + C to copy all of the contents.
- Open the spreadsheet and click Control + V to paste it into the spreadsheet. All of the data will be in one cell.
- With the cells with data still selected, go to Data > Text to Columns. Choose Delimited > Next. Choose Comma > Next > Finish. Save this file as the new CSV.
- The tracking CSV will have many parameters available for analysis (Figure 3). Copy and paste the desired parameters to a new CSV file (Figure 4) to analyze. The parameter used here are described below.
- Total distance traveled: total distance traveled by an amoeba in µm.
- Max distance traveled: the maximum distance on an amoeba's path from its start point – this may not necessarily be the endpoint
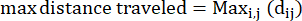
Where dij is the distance to any spot I to any spot j on the track. - Confinement ratio: The confinement ratio indicates how efficient an amoeba was at getting away from its start point: did it travel in a straight line away from its starting point (a value close to 1), or did it stay relatively close to its start point (a value close to 0).
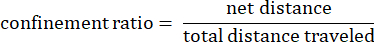
- Track mean speed: Average speed of the amoeba over the total distance traveled in microns per second.
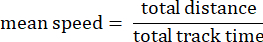
- Track displacement: The distance between the start point of an amoeba and the end point in microns.
NOTE: All of the imaging software analysis parameters and their definitions can be found here: https://imagej.net/plugins/trackmate/analyzers/. Remember that each saved file is a summary of the tracks analyzed at that time. To produce a robust data analysis, one will need to appropriately combine the results from multiple replicates, hours, etc., as is suitable for the data (Figure 5 and Figure 6).
Representative Results
To achieve success using this method, there are several crucial overall pieces to consider. The first is the physical setup of the amoeba and the microscope, the second is proper utilization of the imaging software, and the third is exporting and analyzing the imaging data in a meaningful way.
Before microscopic recording begins, it is critical to ensure that the amoeba is properly focused on the microscope stage (Figure 1). Rather than focusing on the amoeba at the most optimal Z plane for individual details, here it is ideal to view them very slightly out-of-focus so that they become solid black dots as opposed to their translucent state when viewed in detail. Viewing them in this way will allow the imaging software and the tracking software to find each individual amoeba much more easily when the file is converted to binary (Figure 2). Knowing what the goal is in the imaging software – that is, for each amoeba to appear as a well-defined white circle, may help the reader find the proper focus when taking the original images in brightfield. Then, with each circle clearly defined, the tracking software will be able to properly track the motility of and gather the individual motility metrics for each amoeba.
After following the step-by-step directions in the protocol listed above for the imaging software and the tracking software and referring to the associated video to assist in the crucial first steps of imaging software usage, it is time to export the imaging software data for quantification and data analysis. The raw output from the tracking software will be similar to what is seen in Figure 3. Here, it is important to check that the number of spots in the tracks it exported is correct for what is expected (e.g., if one asked for 93% of frames to be present in every track and one had 100 total frames in the video, every exported track should have greater than or equal to 93 spots). If the spots do not match the requirements, then one may need to go back through the tracking software steps to ensure the required parameters were set correctly.
From the raw export, start building the data analysis sheet (Figure 4). It is important to remember that every track represents a singular amoeba within a singular replicate. Therefore, as seen in Figure 4, Figure 5, and Figure 6, all of the tracks for a frameset or time point must be averaged (Figure 4), and this process must be done for every frame set or time point in an entire series (i.e., if a total of 3 h of video are taken and the data points are divided up by the hour, then there will be three frame sets/hours to analyze per replicate, Figure 5). Finally, from this combination of averages in each frame set in each replicate, the averaged data must be combined from each replicate into one space so the data across multiple replicates can be analyzed to determine the true average and standard deviation per time point (Figure 6).
The averages from each replicate will be used for data analysis, and the combined average and standard deviation or standard error from all the replicates will be used for graphical representation (as seen in an example in Figure 7). If analyzing data as we have done here, which is multiple samples being followed over multiple time points, it is most appropriate to analyze them using a 2-way repeat measure ANOVA with a post hoc Tukey's test. This allows us to determine the differences between samples at every time point and also to determine the differences within each sample over time.
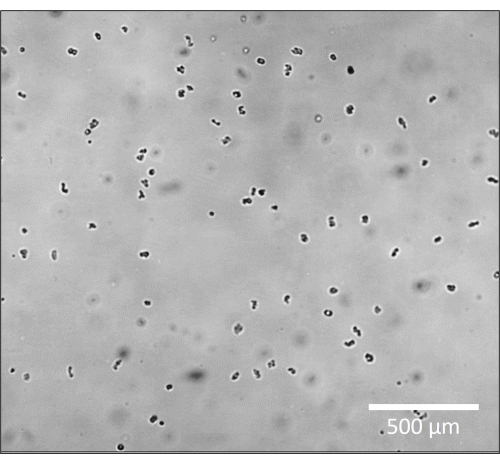
Figure 1: Amoeba appears as opaque dark forms in brightfield microscopy. Zoomed in representative image of amoeba at 4x magnification, slightly out of focus to appear as solid dark circles. Scale bar = 500 µm. Please click here to view a larger version of this figure.
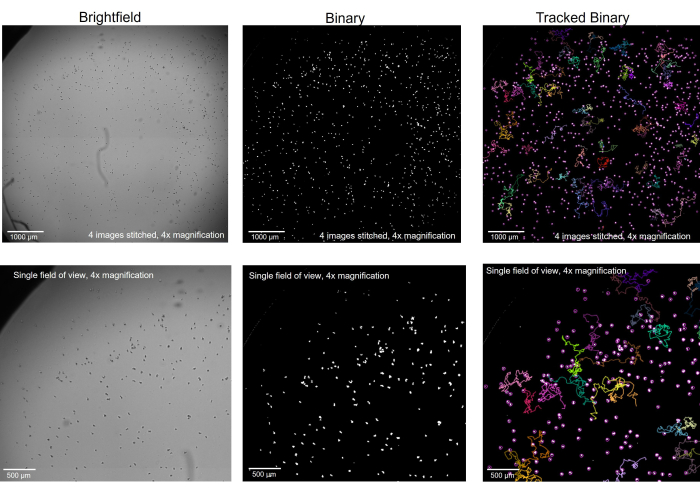
Figure 2: Amoeba imaged in brightfield microscopy converted to binary and tracked. Amoeba at 4x magnification, in brightfield (left), was converted to binary with artifacts removed (middle) and, after tracking, with select tracks showing (right). Top row: a whole well imaged by stitching 4 images together, scale bar = 1000 µm. Bottom row: the size of a single image before stitching, scale bar = 500 µm. Please click here to view a larger version of this figure.
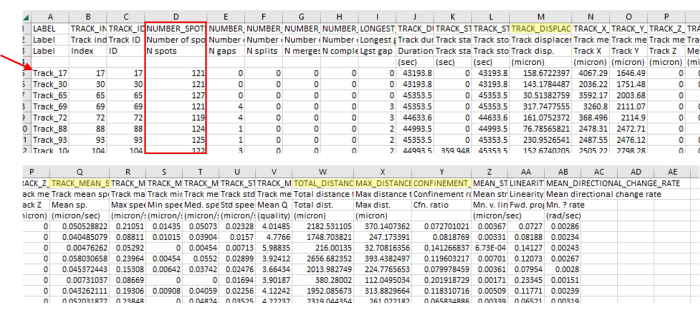
Figure 3: Immediate data analysis output from the imaging and tracking software contains a wide variety of data types. The representative output from tracking is shown in Figure 2, which is a singular video. Note the track numbers (red arrow) skip numbers, indicating the tracking software correctly removed tracks that did not align with the parameters. In this example, only 71 out of 5385 identified tracks aligned with the parameters. This ratio is normal, given such strict guidelines. This number of tracks is also in line with the number of amoebae used in this protocol (for instance, if a 96-well plate is seeded with 200 µL of 7.5 x 103 CFU/mL per well, then one should expect around 1,500 amoebae in the well and roughly that many visible in a 4-stitch field of view. There will likely be many more tracks than there is amoeba as amoeba walks on and off the field of view, generating new tracks). This track number is again apparent in Figure 2, where only a few tracks are visible compared to a large number of spots. Always check the number of spots in the track (red box) to ensure only the correct track lengths are included. For instance, here, we include more than 93% of frames, and there were 127 frames in this section, so all numbers of spots should be above 118. Highlighted in yellow are the measurements that were mentioned in the protocol. Please click here to view a larger version of this figure.
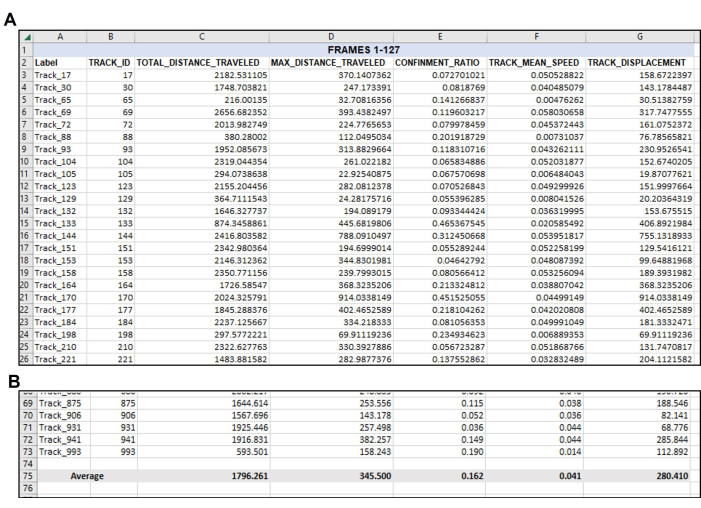
Figure 4: Data re-organized as needed following the initial data output. Organization of wanted data after export. (A) The pertinent measurements chosen to quantify were copied from the tracking software export (Figure 3) and pasted into a new spreadsheet.(B) After the desired measurements have been organized in the new spreadsheet, the average of each track should be calculated. Thus, in this representative example, the Total Distance, Max Distance, Confinement, Mean Speed, and Displacement of every useable track in this singular video have been averaged to a singular number for each measurement. Note again here that a total of 71 tracks met the parameters for Frames 1-127. There will likely be a different number of suitable tracks from each video or frame set analyzed, as evident in Figure 5. Please click here to view a larger version of this figure.
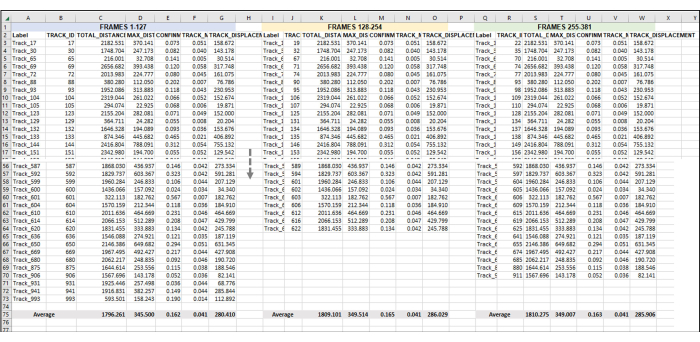
Figure 5: Sequential frames set for analysis of an entire image series or video. This is a representative image of the addition of sequential videos to data analysis. Assuming that frames 1-127 constitute Hour 1 of the video (here, 1 h is made up of taking an image once every 28 s). Frames 128-254 will be Hour 2 of video, and Frames 255-381 will be Hour 3 of video. Combine the exported data (as was exported in Figure 3) from all the hours into one sheet to work with all at once. As was done in Figure 4, the data from all the usable tracks for each frame set/h will be averaged. Everything in Figure 5 is the data from one replicate or one sample. If there are 3 replicates, then there will be 3 identically laid-out separate spreadsheet tabs, and the data from these will be fed into a new tab, as demonstrated in Figure 6. Please click here to view a larger version of this figure.
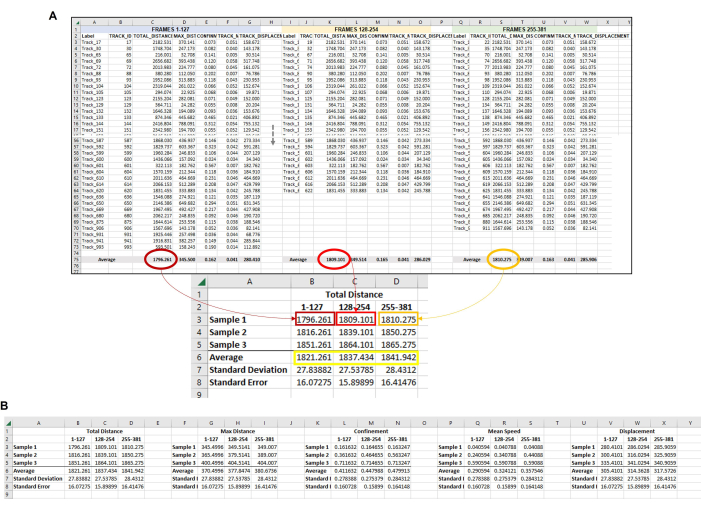
Figure 6: Composite data from all the amoeba in each frameset used to represent a single replicate. (A) All of the averages from each frame set in Figure 5 need to be gathered in a central location so that the final data analysis can be completed. In this representative image, the Total Distance averages from the first three frame sets (or hours) are shown. This must be repeated for each sample/condition/replicate being run. Then, the averages from each sample from each frame set must be averaged (yellow box). This is the final data point from which standard deviation and standard error can be calculated. The individual averages from each Sample (here, the numbers in B3, B4, and B5 for Total Distance Sample 1; C3, C4, and C5 for Total Distance Sample 2, etc.) will be used to run the statistical analysis. This process will be repeated for all required measurements (Max Distance, Confinement, Mean Speed, Displacement, etc.) (B) To make a graphical representation of the quantification, use the numbers in the yellow box as data points to use for the final graph. Please click here to view a larger version of this figure.
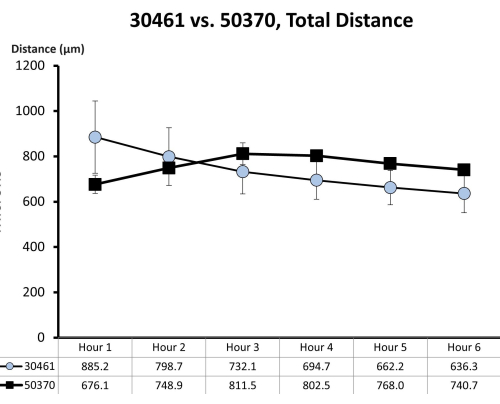
Figure 7: Raw motility data for ATCC 30461 and ATCC 50370. Raw data (represented here as mean ± SE for each hour) was previously collected and not published for prior studies18,19. This motility quantification demonstrates that these two amoebas are similar in their first 6 h of motility (as measured by total distance in microns traveled in that hour) in ¼ Ringer's solution on a glass surface. Each time point represents three separate replicates, each composed of 20-200 amoeba tracks. Please click here to view a larger version of this figure.
Discussion
Being able to track and quantify the motility of amoeba such as Acanthamoeba, which are adherent microscopic organisms unaffected by low-velocity Brownian movement9,10, reveals a substantial amount of information about amoeba behavior and can greatly enhance the enlightenment regarding AK prevention methods. This protocol has been detailed in recent publications, and the data has been confirmed by or combined with other similar data analyses18,19. We note here that the speed of amoeba detailed using this method is similar to what has been published by other groups. This protocol is notable for its ability to be used on basically any surface or amoeba treatment, which is transparent to light, but it is important to note while this protocol could potentially be modified to work with other organisms, it has only been optimized so far for Acanthamoeba. The above protocol has consistently worked well in the lab, but the following alterations for changing the method to suit other needs or troubleshooting difficult situations are provided.
Determining tracking parameters: In the above protocol, the frame-to-frame linking, gap closing, max frame gap, and number of spots on track have been listed, which were the most applicable for recent publications. However, it is possible that other parameters will be more well-suited for other needs. Further, there may be parameters that others would like to use but were not used here (such as the Maximum Distance of a track under track filters). More information about each parameter and what they describe can be accessed here on the website (see reference21). When deciding which parameters and filters are most relevant to other experiments and how they should be set, think about what is statistically rational for a specific project; for instance, would one want data from an amoeba that walks off of and then back into the field of view? Or would one want to keep the data from an amoeba that did not move very much at all but still got tagged as a track? and what makes sense for the number of frames and length of time between frames when the images were recorded.
Changing blob size: We found that it is extremely difficult with any software or any microscope setup – based on current technologies and software available to standard labs – to accurately track the number of amoebas in an aggregate as it changes over time. If tracking the size of a blob and trying to correlate that size with the number of organisms in it, use a standard curve generated after repeated experiments to mathematically predict the size of an aggregate. For instance, aggregates were created by seeding wells with a range of numbers of amoeba, such as 8, 16, 32, 125, 250, 500, 1,000, and 2,000 cells per well. Timelapse images at various time points of each well for 24 h were generated. Each spheroid (which was of a known number of amoeba) was analyzed for a two-dimensional area, and a standard curve was generated as a function of amoeba count versus spheroid size over time. This experiment was repeated in at least triplicate to give us an appropriate standard deviation of any curve generated.
Determining the directionality of the amoeba: While this may not be necessary for particular studies, it can be helpful to understand the directionality of the amoeba. This would provide data about the chemotactic effects of the treatment or experiment. This data can also be used to create both visual (graphic) and quantitative data. This is available via the Chemotaxis and Migration Tool, which is a free plug-in for imaging software. It is available on the website along with the application guide and sample images and movies (see Table of Materials).
Detailed movement dynamics: other groups have examined the spreading dynamics and diffusive trajectories with highly advanced and developed statistical analyses beyond what is discussed here9,10,22. These could be considered based on the user's needs.
As with any successful method, the protocol detailed in this manuscript has been through many rounds of troubleshooting to achieve consistency. While excellent high-quality trackable videos can be made by taking an image every second (or as fast as the microscope is able, it could be less than a second), this creates extremely large files, which can be difficult to work with. This is only realistically suitable for very short-term videos recorded for a maximum of a few minutes. Conversely, by taking an image every few seconds, we have found that amoeba behavior is still very trackable based on the speed of this species, and it is possible to create workable videos over hours or days. We have found that the maximum amount of time between images is 30 s, which is the time that is required for amoeba to be accurately tracked by imaging software. The time interval the user chooses should be considered using the known constraints of how fast the microscope can record images, how many images per well are needed, and how many wells are being recorded at each interval. Similarly, the parameters mentioned in this protocol regarding maximum displacement, allowed frame gaps, minimum frames needed, and so on, have been determined through trial and error by this lab to create track information which includes the most complete amoeba tracks and ignore noise created by incomplete tracks, amoeba which joins or leave the stage mid-video, or errors created by imaging software confusion such as when two amoeba meet mid-track and then separate. These kinds of errors and incomplete tracks are understandably high when creating extremely long (hours to days) videos of microscopic biological organisms that are constantly interacting with each other and are the reason a very large portion of erroneous tracks must be rejected. It should be noted that the fill holes step in the protocol is, in this lab's experience, important to reducing error in how the imaging software tracks amoeba. By making sure each amoeba is a solid circle instead of, at times, a donut or a c shape, the software is much more likely to be able to track each amoeba successfully.
Further, as discussed, there are many parameters available to a user for analyzing an image or video. Based on the experimental needs, we have consistently benefitted the most from analyzing total distance, max distance, speed, and displacement. These are discussed in depth (including using different strains and surfaces) with graphical interpretations in previous publications18,19. These four parameters allow a user to extrapolate an amoeba's ability to travel linear distances and how long it takes them to do so, which aids in understanding their behavior as it relates to contact lens contamination. Retrieving and analyzing these parameters is a large job, as detailed in our figures. While working with large and numerous spreadsheets from imaging software output, we limited unintentional errors by creating locked spreadsheet templates that auto calculated all of the required analyses. However, a possible improvement to this method would be to write a script that can handle this data and sort and analyze it.
In conclusion, an accessible and accurate method to measure Acanthamoeba size and motility in many different conditions is described here. We have demonstrated that this method can be applied to many different strains of amoeba and outlined that while there may be straightforward parameters for attaining motility information, this experimental set up can be highly tailored to any specific needs.
開示
The authors have nothing to disclose.
Acknowledgements
This work was funded by Alcon Research, LLC.
Materials
| ¼ Ringer’s solution | ThermoFisher Scientific | BR0052G | Oxoid Ringers Solution Tablets. Follow directions to make one-quarter strength instead of full strength Ringers. |
| 10 µL pipette | Eppendorf Research | 3123000039 | 2 µL-20 µL single channel |
| 10 µL pipette tips | Neptune Scientific, San Diego, CA, USA | BT10.N | 10 µL Universal Barrier Tip |
| 48 well plate | Millipore Sigma, | CLS3548 | Corning Costar TC-Treated Multiple Well Plate: polystyrene plate, flat bottom wells, sterile, with lid |
| 50 mL conical tubes (1 for each sample, 1 for each pass 2 sample) | Fisher Scientific | Falcon 352098 | Falcon 50 mL High Clarity Conical Centrifuge Tubes, polypropylene |
| 96-well plate | Millipore Sigma, Burlington, MA, USA | CLS3596 | Corning Costar TC-Treated Multiple Well Plate: polystyrene plate, flat bottom wells, sterile, with lid |
| Acanthamoeba | American Type Culture Collection, Manassas, VA, USA | ATCC 30461 | This protocol has been verified for ATCC 50655, 30872, 50702, 30010, 30461, 50370, 50703, 30137, PRA-115, and PRA-411 |
| Axenic culture media (AC6) | Made in house | n/a | Containing 20 g biosate peptone, 5 g glucose, 0.3 g KH2PO4, 10 µg vitamin B12, and 1 glass5 mg l-methionine per liter of distilled deionized water. Adjust pH to 6.6-6.95 with 1 M NaOH and autoclave at 121 °C for 20 min, store at room temperature for up to 3 months. |
| Centrifuge and appropriate rotor | Thermo Scientific, Waltham, MA, USA | Sorvall ST 40R | Any equivalent centrifuge and rotor are acceptable so long as it can spin 50 mL conical tubes at 500 x g for 5 min |
| Chemotaxis and Migration Tool | Free software based on ImageJ, available at https://ibidi.com/chemotaxis-analysis/171-chemotaxis-and-migration-tool.html | ||
| Disposable hemocytometer | Bulldog Bio, Portsmouth, NH, USA | DHC-N01 | Neubauer Improved 2-Chip Disposable Hemocytometer, Individually packaged, Nonpyrogenic |
| ImageJ software with Trackmate plugin (this protocol written with Trackmate version 6.0.2.) | Free software developed by the National Institutes of Health, available at imagej.net. Trackmate plugin available at https://imagej.net/plugins/trackmate/ | ||
| Microscope, preferably with automated moveable stage | Nikon, Tokyo, Japan | A Nikon Eclipse Ti-U Microscope was used in this study and the automated moveable stage was utilized to be able to record images in multiple wells at a time. | |
| NIS-Elements software | Nikon | ||
| Serological pipette | Fisher Scientific, Hampton, NH, USA | BrandTech 26331 | BrandTech accu-jet pro Pipet Controller |
| Serological pipette tips | VWR | 5 mL: 76201-710 10 mL: 170356 25 mL: 89130-900 50 mL: 75816-088 |
VWR Serological Pipette, Non-Pyrogenic |
| Sterile aluminum transmission flow cell | Biosurface Technologies Corporation, Bozeman, MT, USA | FC 81-AL | Anodized aluminum single channel transmission flow cell with 96-well plate footprint for use with an inverted microscope |
| T75 Flasks | VWR, Radnor, PA, USA | 734-2316 | VWR Tissue Culture Flask, 182.5 cm, Surface treated, Plug seal cap, Sterile |
参考文献
- Randag, A. C., et al. The rising incidence of Acanthamoeba keratitis: A 7-year nationwide survey and clinical assessment of risk factors and functional outcomes. PLoS One. 14 (9), e0222092 (2019).
- Szentmary, N., et al. Acanthamoeba keratitis – Clinical signs, differential diagnosis and treatment. J Curr Ophthalmol. 31 (1), 16-23 (2019).
- Carnt, N., et al. Acanthamoeba keratitis: confirmation of the UK outbreak and a prospective case-control study identifying contributing risk factors. Br J Ophthalmol. 102 (12), 1621-1628 (2018).
- Verani, J. R., et al. National outbreak of Acanthamoeba keratitis associated with use of a contact lens solution, United States. Emerg Infect Dis. 15 (8), 1236-1242 (2009).
- Tu, E. Y., Joslin, C. E. Recent outbreaks of atypical contact lens-related keratitis: what have we learned. Am J Ophthalmol. 150 (5), 602-608.e2 (2010).
- . ISO 14729:2001/A1. Ophthalmic optics-Contact lens care products-Microbiological requirements and test methods for products and regimens for hygienic management of contact lenses. , (2010).
- . ASC Z80, Parent Committee Meeting, February 27 Available from: https://www.thevisioncouncil.org/sites/default/files/ASCZ80_ParentCommitteeMinutes_February_27_2018_FINALMar19-2018.pdf (2023)
- Rayamajhee, B., et al. Acanthamoeba, an environmental phagocyte enhancing survival and transmission of human pathogens. Trends Parasitol. 38 (11), 975-990 (2022).
- Reverey, J. F., et al. Superdiffusion dominates intracellular particle motion in the supercrowded cytoplasm of pathogenic Acanthamoeba castellanii. Sci Rep. 5, 11690 (2015).
- Thapa, S., Lukat, N., Selhuber-Unkel, C., Cherstvy, A. G., Metzler, R. Transient superdiffusion of polydisperse vacuoles in highly motile amoeboid cells. J Chem Phys. 150 (14), 144901 (2019).
- Rudell, J. C., et al. Acanthamoeba migration in an electric field. Invest Ophthalmol Vis Sci. 54 (6), 4225-4233 (2013).
- Fukaya, S., Aoki, K., Kobayashi, M., Takemura, M. Kinetic analysis of the motility of giant virus-infected amoebae using phase-contrast microscopic images. Front Microbiol. 10, 3014 (2019).
- Fukaya, S., Takemura, M. Kinetic analysis of Acanthamoeba castellanii infected with giant viruses quantitatively revealed process of morphological and behavioral changes in host cells. Microbiol Spectr. 9 (1), e0036821 (2021).
- Cheng, H. J., Hsu, C. H., Hung, C. L., Lin, C. Y. A review for cell and particle tracking on microscopy images using algorithms and deep learning technologies. Biomed J. 45 (3), 465-471 (2022).
- Mathieu, E., et al. Time-lapse lens-free imaging of cell migration in diverse physical microenvironments. Lab Chip. 16 (17), 3304-3316 (2016).
- Svensson, C. M., Medyukhina, A., Belyaev, I., Al-Zaben, N., Figge, M. T. Untangling cell tracks: Quantifying cell migration by time lapse image data analysis. Cytometry A. 93 (3), 357-370 (2018).
- Piltti, K. M., et al. Live-cell time-lapse imaging and single-cell tracking of in vitro cultured neural stem cells – Tools for analyzing dynamics of cell cycle, migration, and lineage selection. Methods. 133, 81-90 (2018).
- Campolo, A., et al. Continuous real-time motility analysis of Acanthamoeba reveals sustained movement in absence of nutrients. Pathogens. 10 (8), 995 (2021).
- Campolo, A., Patterson, B., Lara, E., Shannon, P., Crary, M. Complete recovery of Acanthamoeba motility among surviving organisms after contact lens care disinfection. Microorganisms. 11 (2), 299 (2023).
- Crary, M. J., Walters, R., Shannon, P., Gabriel, M. M. Variables affecting the recovery of Acanthamoeba trophozoites. Pathogens. 10 (2), 221 (2021).
- . Trackmate Available from: https://imagej.net/plugins/trackmate/tutorials/getting-started (2024)
- Cherstvy, A. G., Nagel, O., Beta, C., Metzler, R. Non-Gaussianity, population heterogeneity, and transient superdiffusion in the spreading dynamics of amoeboid cells. Phys Chem Chem Phys. 20 (35), 23034-23054 (2018).

