Quantitative Real-Time Polymerase Chain Reaction Evaluation of MicroRNA Expression in Kidney and Serum of Mice with Age-Dependent Renal Impairment
概要
We present a method for evaluating microRNA expression in the kidney and serum of mice with age-dependent renal impairment by quantitative reverse-transcription polymerase chain reaction.
Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs consisting of 21-25 bases. They are not translated into proteins but rather work to impede the functioning of their target messenger RNAs (mRNAs) by destabilizing them and disrupting their translation. Although the miRNA expression profiles in various mouse organs and tissues have been investigated, there have been no standard methods for purifying and quantifying mouse kidney and serum miRNAs. We have established an effective and reliable method for extracting and evaluating the miRNA expression in the serum and kidney of mice with age-dependent renal impairment.
The method uses quantitative reverse-transcription-polymerase chain reaction (qRT-PCR), and the protocol requires six steps: (1) preparing senescence-accelerated mouse resistance 1 (SAMR1) mice and senescence-accelerated mouse prone (SAMP1) mice; (2) extracting serum samples from these mice; (3) extracting a kidney sample from each mouse; (4) extracting total RNA (including miRNA) from kidney and serum samples from each mouse; (5) the synthesis of complementary DNA (cDNA) with reverse transcription from the miRNA; (6) conducting a qRT-PCR using the cDNA obtained.
This protocol was used to confirm that, compared to the controls, the expression of miRNA-7218-5p and miRNA-7219-5p was significantly changed in the kidney and serum of a mouse model of age-dependent renal impairment. This protocol also clarified the relationship between the kidney and serum of the mouse model of age-dependent renal impairment. This protocol can be used to determine miRNA expression in the kidney and serum of mice with age-dependent renal impairment.
Introduction
The expression of various mRNAs that play important roles in both physiology and disease (e.g., inflammation, fibrosis, metabolic disorders, and cancer) is known to be regulated by miRNAs, which are short, noncoding RNAs that cause the degradation and inhibit the transcription of mRNA1. It is, therefore, possible that certain miRNAs could serve as new candidatebiomarkers and/or therapeutic targets for various diseases2,3,4,5. Research has been conducted on miRNA expression profiles in a variety of mouse organs and tissues (including brain6, heart7, lung8, liver9, and kidney10). However, there are no standard or established methods for extracting and evaluating miRNAs in the kidneys or serum of mice with age-dependent renal impairment.
Therefore, we established a protocol that can be used to reliably purify and detect miRNA expression in the serum and kidney of mice with age-dependent renal impairment. There are six main steps in the protocol: (1) preparation of both 50-week-old SAMR1 male mice and SAMP1 male mice; (2) extraction of blood samples from the inferior vena cava of both strains of mice, with the subsequent use of a spitz tube with heparin, followed by centrifugation, to obtain a serum sample; (3) kidney sample extraction from the mice-a silicon homogenizer is used to homogenize the kidney sample separately, and the sample is then transferred to a biopolymer-shredding system on a microcentrifuge spin column11; (4) total RNA (containing miRNA) extraction from the serum samples with the use of a silica membrane-based spin column12 and total RNA containing miRNA extraction from the kidney samples with the use of a silica membrane-based spin column11; (5) synthesis of complementary DNA (cDNA) from the total RNA using reverse transcriptase, poly(A) polymerase, and oligo-dT primer13,14; and (6) finally, determination of miRNA expression using qRT-PCR and an intercalating dye13,14.
This new protocol was based on studies that succeeded in extracting and evaluating miRNAs in various types of tissue11,12,13. The protocol's biopolymer-shredding system was demonstrated to be able to purify high-quality total RNA from tissues11. The accuracy and sensitivity of aspects of this protocol used for the evaluation of miRNA expression by qRT-PCR with an intercalating dye have been established13,14, for example, the cDNA synthesis with reverse transcriptase, poly(A) polymerase, and oligo-dT primers from the extracted total RNA. The new protocol has several advantages: simplicity, time savings, and reduced technical errors. It can, thus, be used for investigations that require accurate and sensitive identification of kidney and serum miRNA profiles. Studies of many pathological conditions can also utilize the new protocol.
The miRNA expression profiles in SAMP1 mice, which are a model of age-dependent renal impairment, can be determined as shown below. In humans, age-dependent renal impairment is associated with the progression of renal failure and is characterized by both an increase in the area of renal interstitial fibrosis and the progression of glomerulosclerosis15,16. Age-dependent renal impairment is also an important and frequent feature of chronic kidney disease and end-stage renal disease15,16.
Protocol
The experimental protocol was approved by the Animal Ethics Committee of Jichi Medical University and performed in accordance with the Jichi Medical University Guide for Laboratory Animals and its guidelines concerning the use and care of experimental animals. This protocol uses four 50-week-old SAMR1 male mice and SAMP1 male mice (40-45 g).
1. Serum sample collection
- Prepare the following for each mouse: 30 G needles with a 1.0 mL syringe, a 1.0 mL spitz centrifugation tube with heparin, 1.5 mL microcentrifuge tubes, isoflurane anesthetic, a cork sheet, 70% ethanol, two cotton swabs moistened with phosphate-buffered saline (PBS), a Petri dish with PBS, tweezers, and surgical scissors.
- Anesthetize the mouse with 1.5% isoflurane and maintain at 1.5%. Administer an analgesic (Meloxicam 5 mg/kg) subcutaneously, then inject 1.0 mL of 70% ethanol into its abdomen and place it in the supine position on the cork sheet.
- Confirm the depth of anesthesia by the disappearance of the pedal withdrawal reflex. With the tweezers and surgical scissors, incise the skin of the abdomen. Cut the muscles and the peritoneal membrane from the bladder to the lower-left edge of the ribs.
- Use the tweezers to lift the peritoneal membrane, and make a lateral incision in the upper edge of the peritoneal membrane with the surgical scissors. Continue the incision along the lowest edge of the ribs.
- Identify the inferior vena cava by using the two PBS-moistened cotton swabs. Insert one of the 30 G needles with the 1.0 mL syringe into the inferior vena cava and then pull the syringe. Pull the needle out of the inferior vena cava slowly to avoid hemolysis. Transfer the blood to the 1.0 mL spitz tube with heparin and then mix it by inverting the tube several times.
NOTE: The mouse is euthanized by cervical dislocation. - Spin the spitz tube at room temperature (RT), 3,000 × g for 10 min.
- Aspirate the supernatant slowly, make sure that it does not contain sediment, and then transfer it to an unused 1.5 mL microcentrifuge tube.
- Store the tube at −80 °C before use.
NOTE: If proceeding to Step 3 immediately, the tube does not need to be stored at −80 °C.
2. Kidney sample collection
- Prepare the following for each mouse: 2.0 mL cryotubes, isoflurane anesthetic, a cork sheet, 70% ethanol, a Petri dish with PBS, tweezers, and surgical scissors.
- Anesthetize the mouse with 1.5% isoflurane and maintain at 1.5%. Administer an analgesic (Meloxicam 5 mg/kg) subcutaneously, then inject 1.0 mL of 70% ethanol into its abdomen, and place it in the supine position on the cork sheet.
- Confirm the depth of anesthesia by the disappearance of the pedal withdrawal reflex. Use the tweezers and the surgical scissors to make an incision in the abdominal skin. Cut the muscles and peritoneal membrane from the bladder to the lower-left edge of the ribs.
- Use the tweezers to lift the peritoneal membrane and make a lateral incision in the peritoneal membrane's upper edge with the surgical scissors. Continue the incision along the lowest edge of the ribs.
- Identify the left kidney. Reflux it with PBS to flush the blood from the vessels until the kidney turns a yellowish-white color. Excise the entire kidney first using surgical scissors to sever the left renal artery and vein. Place the kidney in a Petri dish and wash it carefully with PBS.
NOTE: The mouse is euthanized by cervical dislocation. - Use the tweezers and the surgical scissors to cut the kidney into 10 mg samples (10 mg is a suitable sample size for the next step). Place each sample of the kidney in its own 2.0 mL cryotube. Close the tube's cap.
- For long-term storage, transfer each cryotube to liquid nitrogen and store it at −80 °C.
3. Total RNA extraction from a serum sample
- Prepare the following items first: a vortex mixer, phenol/guanidine-based lysis reagent, 80% ethanol, 100% ethanol, 100% chloroform, biopolymer spin columns (in 2.0 mL collection tubes11), membrane-anchored spin columns (in 2.0 mL collection tubes11), wash buffer #1 (i.e., wash buffer containing guanidine and 100% ethanol in a ratio of 1:2), wash buffer #2 (i.e., wash buffer containing guanidine and 100% ethanol in a ratio of 1:4), RNase-free water, 1.5 mL microcentrifuge tubes, and 2.0 mL microcentrifuge tubes.
- First, take a 200 µL serum sample in a 1.5 mL microcentrifuge tube, and then add 1,000 µL of the phenol/guanidine-based lysis reagent. Vortex the mixture for 5 s.
- Incubate the sample at RT for 5 min.
- Add 200 µL of chloroform to the serum sample in the tube and close the tube cap tightly. Invert the tube 15x to mix the chloroform and the serum sample.
- Each sample is incubated at RT for 3 min. Next, spin the sample at 4 °C for 15 min at 12,000 x g.
- Transfer the 300 µL of supernatant to a new 1.5 mL microcentrifuge tube without disturbing the pellet. Add 450 µL of 100% ethanol, and vortex the tube for 5 s.
NOTE: In all the following steps, place the membrane-anchored spin column for the separation of RNA and DNA in a 2.0 mL collection tube to centrifuge it. - Then, remove 700 µL of the sample, load it onto a membrane-anchored spin column, and close the cap. Spin the column at RT for 15 s at 8,000 × g, and leave the supernatant in the column. Discard the pellet remaining in the collection tube.
- Add 700 µL of wash buffer #1 which is part of the serum/plasma kit (see the Table of Materials) to the membrane-anchored spin column to clean the sample thoroughly. Close the column cap and spin the column at RT for 15 s at 8,000 × g, and leave the supernatant in the column. Discard the pellet remaining in the collection tube.
- For the removal of trace salts, take 500 µL of wash buffer #2 and load it onto a membrane-anchored spin column. After closing the column cap, spin the column at RT for 15 s at 8,000 × g, and leave the supernatant in the column. Discard the pellet in the collection tube.
- For the removal of trace salts, take 500 µL of 80% ethanol and load it onto a membrane-anchored spin column. After closing the column cap, spin the column at RT for 15 s at 8,000 × g, and leave the supernatant in the column. Discard the pellet in the collection tube.
- Spin the membrane-anchored spin column again at RT for 5 min at 15,000 × g.
- After transferring the membrane-anchored spin column to a new 1.5 mL collection tube, add 14 µL of RNase-free water to the column to dissolve the total RNA. After closing the column cap, wait 5 min with the tube left at RT. Spin the column again for 1 min at 15,000 × g at RT.
- Store the tubes with samples at −80 °C before use.
4. Extraction of total RNA from a kidney sample
- Prepare the following items first: a silicon homogenizer, ice, a vortex mixer, 100% ethanol, 100% chloroform, biopolymer spin columns (in 2.0 mL collection tubes11), membrane-anchored spin columns (in 2.0 mL collection tubes11), phenol/guanidine-based lysis reagent, wash buffer #1 (the same buffer as Step 3.1.), wash buffer #2 (the same buffer as Step 3.1.), RNase-free water, 1.5 mL microcentrifuge tubes, and 2.0 mL microcentrifuge tubes.
- Place a 10 mg kidney sample in the silicon homogenizer, and add 700 µL of the phenol/guanidine-based lysis reagent.
- Set up the homogenizer. Gently press and twist the homogenizer's pestle against the kidney sample to homogenize the sample. Continue to press and twist the pestle until the kidney sample is completely homogenized in the phenol/guanidine-based lysis reagent.
- For further homogenization, take the homogenized lysate (in a 2.0 mL collection tube) and transfer it to a biopolymer spin column.
- Centrifuge the homogenized lysate at RT for 3 min at 14,000 × g and then transfer the entire precipitated pellet to an unused 1.5 mL microcentrifuge tube to invert the precipitated pellet
- Mix the pellet with 140 µL of chloroform in the tube; then, close the tube cap tightly. Invert the tube 15x to mix the lysate and chloroform.
NOTE: The chloroform can be used safely without a hood. - Incubate the sample at RT for 2-3 min. Centrifuge the sample at 4 °C for 15 min at 12,000 × g.
- Transfer the supernatant (which is usually ~300 µL) to a new 1.5 mL microcentrifuge tube, without disturbing the precipitate. Add 1.5 times its volume (which is usually ~450 µL) of 100% ethanol. Vortex the mixture for 5 s.
NOTE: In all the following steps, place the membrane-anchored spin column for separating RNA and DNA in a 2.0 mL collection tube to centrifuge it. - Load 700 µL of sample onto one of the membrane-anchored spin columns. Close the cap and centrifuge the column at 15,000 × g for 15 s. Discard the precipitated lysate remaining in the collection tube.
- Add 700 µL of wash buffer #1 to the spin column to thoroughly wash it. Close the cap and centrifuge the column at 15,000 × g for 15 s. Discard the precipitated lysate remaining in the collection tube.
- For the removal of trace amounts of salt, load 500 µL of wash buffer #2 onto the membrane-anchored spin column. After closing the column cap, centrifuge the column at 15,000 × g for 15 s. Discard the precipitated lysate in the collection tube.
- Repeat Step 4.11.
- Centrifuge the membrane-anchored spin column again for 1 min at 15,000 × g. Discard the precipitated lysate in the collection tube.
- Take the membrane-anchored spin column and transfer it to a new 1.5 mL collection tube. Add 30 µL of RNase-free water to the column to dissolve the total RNA. After closing the column's cap, wait for 5 min with the tube at RT, and then centrifuge the column for 1 min at 15,000 × g.
- Transfer the total volume of the sample containing total RNA to a new microcentrifuge tube. Place the tube on ice, and measure the total RNA concentration by spectrophotometry. Make sure that the total RNA concentration is ~300-1,500 ng/µL.
- Store tubes containing samples at −80 °C before use.
5. Synthesis of cDNA with the reverse transcription of total RNA in serum
NOTE: The minimum information for the publication of quantitative real-time PCR experiments (MIQE) guidelines recommend using better experimental practices to obtain reliable, unequivocal results17. In this protocol, cDNA is synthesized from the total RNA purified in a two-step procedure using reverse transcriptase, poly(A) polymerase, and oligo-dT primers.
- Prepare the following first: a vortex mixer, a thermal cycler, eight-well strip tubes, the cap of each eight-strip tube, distilled water, ice, the reverse transcriptase kit (see the Table of Materials)13,14 in the melted state, and 1.5 mL microcentrifuge tubes.
- Start the thermal cycler.
- Prepare the master mix solution; to obtain a total of 8.0 µL of master mix per eight-well strip tube, add 2.0 µL of reverse transcriptase mix (included in the kit) and 2.0 µL of 10x nucleic acid mix to 4.0 µL of reverse-transcription buffer in a 1.5 mL microcentrifuge tube.
- Place 8.0 µL of the master mix solution in each tube of an eight-well strip tube.
- Place a 12 µL aliquot of total RNA in each tube of the eight-well strip tube, and close the tube's cap. Centrifuge the tube for 15 s at RT and 2,000 × g.
- Place the tube in the thermal cycler and incubate for 60 min at 37 °C. Incubate the sample for an additional 5 min at 95 °C min to synthesize the cDNA.
- Following the incubation, transfer the cDNA to a new 1.5 mL microcentrifuge tube. Dilute the cDNA tenfold (1:10) with distilled water. Vortex and centrifuge the tube for 5 s at RT and 2,000 × g.
- Store the diluted cDNA temporarily on ice. Store the diluted samples at −80 °C before use.
6. Synthesis of cDNA with the reverse transcription of total RNA in kidney
NOTE: The MIQE guidelines encourage better experimental practices to ensure reliable and unequivocal results17. This protocol uses reverse transcriptase, poly(A) polymerase, and oligo dT primers to synthesize cDNA from 1.0 µg of purified total RNA in a two-step procedure.
- First, prepare the following: a vortex mixer, a thermal cycler, 1.5 mL microcentrifuge tubes, eight-well strip tubes, the cap of each eight-strip tube, distilled water, ice, and a reverse transcriptase kit (see the Table of Materials)13,14 in the melted state.
- Start the thermal cycler.
- Prepare the master mix solution; to obtain a total of 8.0 µL of master mix solution per tube, add 2.0 µL of the reverse transcriptase mix that is included in the kit and 2.0 µL of 10x nucleic acid mix to 4.0 µL of the reverse-transcription buffer.
- Add 8.0 µL of the master mix solution to each tube of an eight-well strip tube.
- Adjust the total RNA density as follows. For the separation of 1.0 µg of total RNA from a kidney sample with 12 µL of RNase-free water, take a suitable amount of total RNA and transfer it to distilled water by using the concentration measured as described above (in Step 4.15.).
NOTE: In the presence of DNA contamination, the contaminated DNA is co-amplified by qRT-PCR. - Perform the same process as that described above in Steps 5.5.-5.8.
7. The qRT-PCR of miRNA
NOTE: An intercalator method is used for the qRT-PCR of the miRNAs. Primers are used for RNA: U6 small nuclear 2 (RNU6-2), miRNA-223-3p, miRNA-423-5p, miRNA-7218-5p, and miRNA-7219-5p.
- Prepare the following: a vortex mixer, a real-time PCR system, a 96-well reaction plate for qRT-PCR, adhesive film for the 96-well reaction plate, adhesive film applicator, a 96-well centrifuge rotor, miRNA-specific primers, a green dye-based PCR kit containing 2x PCR master mix and 10x universal primer (see the Table of Materials)13,14, and a 1.5 mL microcentrifuge tube.
- Vortex the following after mixing them in a 1.5 mL microcentrifuge tube: 6.25 µL of distilled water, 1.25 µL each of 5 µM miRNA primer dissolved in nuclease-free water, 12.5 µL of 2x PCR master mix, and 2.5 µL of 10x universal primer.
- Prepare and melt the cDNA synthesized as described in Step 5 (serum) or Step 6 (kidney). Vortex and centrifuge the cDNA for 5 s.
- Take 22.5 µL aliquots of the reagent (as described in Step 7.2. above) and place them separately in each well of the 96-well plate.
- Add a 2.5 µL aliquot of cDNA to each well of the plate.
- To secure the adhesive film to the plate, use the adhesive film applicator. Centrifuge the plate for 30 s at 1,000 x g in the 96-well centrifuge rotor. Stabilize the reaction at the bottom of each well.
8. Using the real-time PCR system and software to run the PCR cycling program
- Begin the real-time PCR system. Place the plate created as described in Step 7.6. in the real-time PCR system. Modify the settings; provide a name for the experiment, and then select 96-well (0.2 mL) as the system's experiment type, Comparative CT (ΔΔCT) as the quantitation method, standard as the system run mode, and SYBR Green Reagents as the reagents for detecting the target sequence.
- Provide names for the sample and the target miRNAs, and name the sample and target miRNA in each well. Assign duplicate samples to obtain data suitable for confirming the results, and choose a reference sample and the endogenous control. For the dye to use as the passive reference, select none. To eliminate reagent cross-contamination, set up negative reverse transcriptase and a non-template control for miRNA expression.
- Next, make sure that the reaction volume is set to 20 µL and the PCR cycling conditions are set as follows: 95 °C for 15 min, then 40 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s, and lastly extension at 70 °C for 30 s.
- Click on analyze in the system's software program to analyze the qRT-PCR data after the process is complete. Confirm that the threshold line automatically selected by the program is appropriate for each well.
- Check the threshold cycle (CT) value of the endogenous control and target miRNAs analyzed in each sample. Determine the CT values by the intersection of the amplification curve and threshold line.
NOTE: In the present study, RNU6-2 and miRNA-423-5p were used as the endogenous controls for target miRNA expression levels, and the ΔΔCT method was used to determine the relative expression levels of each target miRNA18.
Representative Results
For the age-dependent renal impairment mouse model, we used 50-week-old SAMP1 male mice weighing 40-45 g. Approximately 0.8 mL of blood was collected per mouse and transferred into a 1.0 mL spitz tube with heparin, inverted, and centrifuged. Each kidney was rinsed with PBS, dissected, and stored in liquid nitrogen for further analysis. Fifty-week-old SAMR1 mice served as controls. Based on the miRNA qRT-PCR data obtained using this age-dependent renal impairment model, we observed that the kidney level of miRNA-7219-5p was significantly increased and the kidney level of miRNA-7218-5p was considerably decreased in the SAMP1 mice compared to the controls (Figure 1). The serum levels of both miRNA-7219-5p and miRNA-7218-5p were considerably increased in the SAMP1 mice compared to the controls (Figure 2). The expression levels of miRNA-223-3p did not change in either strain and between kidney and serum (Figure 1 and Figure 2).
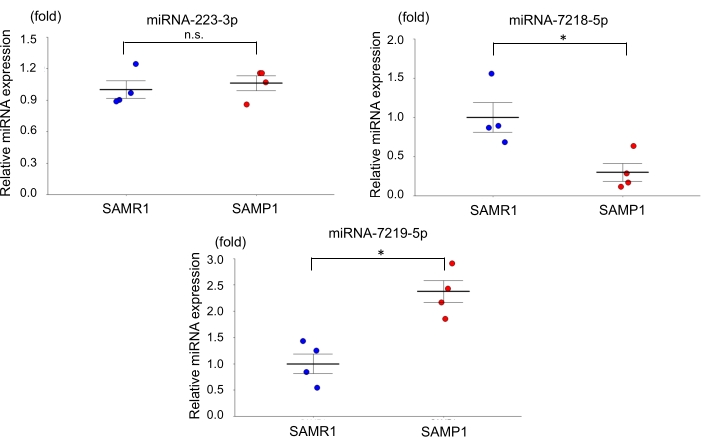
Figure 1: Differentially expressed microRNAs in the kidneys of SAMP1 mice. qRT-PCR analysis of the expression of miRNA-223-3p, miRNA-7218-5p, and miRNA-7219-5p in SAMR1 mice (control, n = 4) and SAMP1 mice (n = 4). The data are mean ± standard error (error bars); t-tests were used to analyze between-group differences; p < 0.05 was considered significant (*p < 0.05), n.s.: not significant. Abbreviations: miRNA = microRNA; SAMP1 = senescence-accelerated mouse prone; SAMR1 = senescence-accelerated mouse resistance 1; qRT-PCR = quantitative reverse-transcription-polymerase chain reaction. Please click here to view a larger version of this figure.
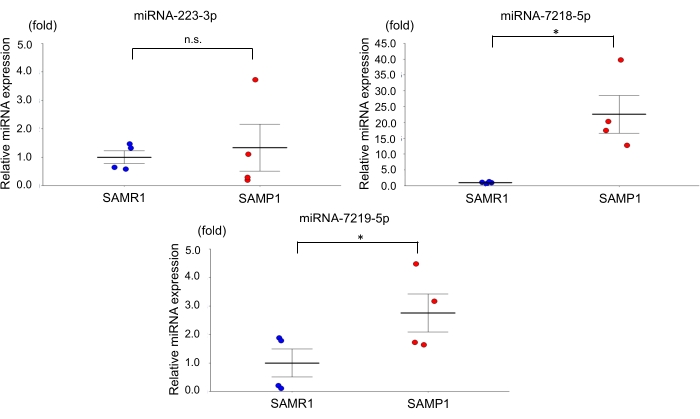
Figure 2: Differentially expressed miRNAs in the serum of SAMP1 mice. qRT-PCR analysis of the expression of miRNA-223-3p, miRNA-7218-5p, and miRNA-7219-5p in SAMR1 mice (control, n = 4) and SAMP1 mice (n = 4). The data are mean ± SE (error bars); t-tests were used to investigate significant differences between groups. *p < 0.05 by t-test. Abbreviations: miRNA = microRNA; SAMP1 = senescence-accelerated mouse prone; SAMR1 = senescence-accelerated mouse resistance 1; qRT-PCR = quantitative reverse-transcription-polymerase chain reaction Please click here to view a larger version of this figure.
Discussion
The expression levels of the target miRNAs were successfully determined by the above-described protocol using qRT-PCR. The evaluation of the extracted miRNAs is an important step in obtaining meaningful qRT-PCR data. To confirm the adequate quality of miRNAs before performing the qRT-PCR, spectrophotometry should be used to determine the ratio of absorbance at 260 nm to that at 280 nm. DNA contamination may occur, and/or primer dimers in each well of the reaction plate may be present if the qRT-PCR does not provide a single PCR amplification of the expected length and melting temperature, or if it provides a monomodal melting curve.
MiRNA expression levels can be evaluated by several methods other than qRT-PCR, including northern blotting, a microarray, and ribonuclease protection assays. However, the qRT-PCR method is a sensitive, accurate, simple, and reproducible procedure that requires a smaller sample volume than those required for northern blotting and ribonuclease protection assays19. Since microarrays can measure the expression of tens of thousands of miRNAs simultaneously, they can be used to identify candidate miRNA markers. Microarray data also show a high overall correlation with data obtained by qRT-PCR20. However, no consensus has been reached regarding the optimal methodology for comparing microarray data obtained in different studies21.
The evaluation of serum miRNAs has the following features. First, it is easy to collect serum, and as serum miRNAs are stable against freezing and thawing, temperature, and acid, the miRNAs can be good biomarkers. Second, there is high homology of miRNAs among species, and the results of animal experiments are easily extrapolated to humans. Third, serum miRNAs have shown potential for use as therapeutic drugs3. Several studies have also demonstrated that the level of expression of miRNA in organs is correlated with miRNA in serum22,23,24. In the present study, miRNA-223-3p, the kidney levels of which did not show a significant difference between the SAMR1 and SAMP1 mice, also showed no significant between-strain difference in the serum. In contrast, miRNA-7218-5p and miRNA-7219-5p, the kidney levels of which showed a significant difference between the SAMR1 and SAMP1 mice, showed considerable between-strain differences in the serum.
This protocol has the following limitations. First, its usefulness has not been verified in other organs such as the liver and lung, and second, it has not been tested on other laboratory animals such as rats, dogs, and pigs. Several research groups have used this protocol for the purification and detection of miRNAs by qRT-PCR and reported that this protocol enabled the purification of high-quality RNA from tissues and serum13,14,22,23,24. This method has been demonstrated to have high accuracy and sensitivity for detecting the expression of miRNAs13,14,22,23,24. The present study's results demonstrate that this protocol can successfully detect miRNA expression in the serum and kidney of mice. Therefore, the protocol can be used to determine the serum and kidney miRNA expression profiles in mice with a variety of pathologies. Due to the protocol's simplicity, a large number of samples can be processed simultaneously. Analyses of the expression of many miRNAs in various pathological conditions of the kidney can, thus, use the protocol described herein.
There are certain aspects of the protocol to keep in mind. To avoid the degradation of the purified miRNAs that would occur at room temperature, the miRNAs must be kept on ice. The kidney samples must be homogenized until they are completely dissolved in the lysis reagent. Mouse kidney contains a substantial amount of connective tissue that is insoluble in lysis reagent, and thus a column shredder is required for further homogenization. In addition, the appropriate endogenous control miRNA (with stable expression among the samples) should be validated throughout the setup of a qRT-PCR experiment. This is because the interference of various substances during the performance of this protocol can alter the expression levels of endogenous control miRNAs, possibly compromising the results. In conclusion, this paper describes a qRT-PCR protocol for the detection, purification, and evaluation of miRNA expression in the serum and kidney of mice with age-dependent renal impairment.
開示
The authors have nothing to disclose.
Acknowledgements
None.
Materials
| 1.0 mL spitz with heparin | Greiner-bio-one | 450534 | |
| Buffer RPE (wash buffer #2 containing guanidine and ethanol in ratio of 1:4) | Qiagen | 79216 | Wash buffer 2 |
| Buffer RWT (wash buffer #1 containing guanidine and ethanol in ratio of 1:2) | Qiagen | 1067933 | Wash buffer 1 |
| MicroAmp Optical 96-well reaction plate for qRT-PCR | Thermo Fisher Scientific | 4316813 | 96-well reaction plate |
| MicroAmp Optical Adhesive Film | Thermo Fisher Scientific | 4311971 | Adhesive film for 96-well reaction plate |
| miRNA-223-3p primer | Qiagen | MS00003871 | 5'-CGUGUAUUUGACAAGCUGAGUU G-3' |
| miRNA-423-5p primer | Qiagen | MS00012005 | 5'-UGAGGGGCAGAGAGCGAGACU UU-3' |
| miRNA-7218-5p primer | Qiagen | MS00068067 | 5'-UGCAGGGUUUAGUGUAGAGGG -3' |
| miRNA-7219-5p primer | Qiagen | MS00068081 | 5'-UGUGUUAGAGCUCAGGGUUGA GA-3' |
| miRNeasy Mini kit | Qiagen | 217004 | Membrane anchored spin column in a 2.0 mL collection tube |
| miRNeasy Serum/Plasma kit | Qiagen | 217184 | Membrane anchored spin column in a 2.0 mL collection tube |
| miScript II RT kit (reverse transcription buffer) | Qiagen | 218161 | Reverse transcriptase kit |
| miScript SYBR Green PCR kit | Qiagen | 218073 | Green dye-based PCR kit |
| QIA shredder | Qiagen | 79654 | Biopolymer spin columns in a 2.0 mL collection tube |
| QIAzol Lysis Reagent (phenol/guanidine-based lysis reagent) | Qiagen | 79306 | Phenol/guanidine-based lysis reagent |
| QuantStudio 12K Flex Flex Real-Time PCR system | Thermo Fisher Scientific | 4472380 | Real-time PCR instrument |
| QuantStudio 12K Flex Software version 1.2.1. | Thermo Fisher Scientific | 4472380 | Real-time PCR instrument software |
| RNase-free water | Qiagen | 129112 | |
| RNU6-2 primer | Qiagen | MS00033740 | Not disclosed due to confidentiality |
| SAMP1 male mice | Nippon SLC Corporation | Not assigned | |
| SAMR1 male mice | Nippon SLC Corporation | Not assigned | |
| Takara biomasher standard | Takara Bio | 9790B | Silicon homogenizer |
参考文献
- Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. Journal of Cellular and Molecular Medicine. 22 (12), 5768-5775 (2018).
- Yang, C., Dou, R., Yin, T., Ding, J. MiRNA-106b-5p in human cancers: diverse functions and promising biomarker. Biomedicine and Pharmacotherapy. 127, 110211 (2020).
- Lu, T. X., Rothenberg, M. E. MicroRNA. The Journal of Allergy and Clinical Immunology. 141 (4), 1202-1207 (2018).
- McGuire, A., Brown, J. A., Kerin, M. J. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer and Metastasis Reviews. 34 (1), 145-155 (2015).
- Bjorkman, S., Taylor, H. S. MicroRNAs in endometriosis: biological function and emerging biomarker candidates. Biology of Reproduction. 100 (5), 1135-1146 (2019).
- Zhou, C. X., et al. miRNA and circRNA expression patterns in mouse brain during toxoplasmosis development. BMC Genomics. 21 (1), 46 (2020).
- Jing, R., Zhong, Q. Q., Long, T. Y., Pan, W., Qian, Z. X. Downregulated miRNA-26a-5p induces the apoptosis of endothelial cells in coronary heart disease by inhibiting PI3K/AKT pathway. European Review for Medical and Pharmacological Sciences. 23 (11), 4940-4947 (2019).
- Xie, W., et al. miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. Journal of Cellular Physiology. 233 (9), 6615-6631 (2018).
- Bala, S., et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 56 (5), 1946-1957 (2012).
- Ishii, H., et al. MicroRNA expression profiling in diabetic kidney disease. Translational Research. 237, 31-52 (2021).
- Mastropasqua, R., et al. Serum microRNA levels in diabetes mellitus. Diagnostics (Basel). 11 (2), 284 (2021).
- Gordanpour, A., Nam, R. K., Sugar, L., Bacopulos, S., Seth, A. MicroRNA detection in prostate tumors by quantitative real-time PCR (qPCR). Journal of Visualized Experiments: JoVE. (63), e3874 (2012).
- Ahn, J. H., Kwak, J., Lee, J. H., Lee, S. S. Efficient and accurate analysis of microRNA using a specific extension sequence. Molecular Biology Reports. 45 (4), 611-619 (2018).
- Denic, A., Glassock, R. J., Rule, A. D. Structural and functional changes With the aging kidney. Advances in Chronic Kidney Disease. 23 (1), 19-28 (2016).
- Weinstein, J. R., Anderson, S. The aging kidney: physiological changes. Advances in Chronic Kidney Disease. 17 (4), 302-307 (2010).
- Bustin, S. A., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 55 (4), 611-622 (2009).
- Livak, K. J., Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25 (4), 402-408 (2001).
- Rajeevan, M. S., Vernon, S. D., Taysavang, N., Unger, E. R. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. The Journal of Molecular Diagnostics. 3 (1), 26-31 (2001).
- Chen, Y., Gelfond, J. A., McManus, L. M., Shireman, P. K. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 10, 407 (2009).
- Dallas, P. B., et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR — how well do they correlate. BMC Genomics. 6, 59 (2005).
- Petriella, D., et al. miRNA profiling in serum and tissue samples to assess noninvasive biomarkers for NSCLC clinical outcome. Tumour Biology. 37 (4), 5503-5513 (2016).
- Skrzypa, M., et al. miRNA-146a-5p is upregulated in serum and cartilage samples of patients with osteoarthritis. Polski Przeglad Chirurgiczny. 91 (3), 1-5 (2019).
- Farzanehpour, M., et al. Serum and tissue miRNAs: potential biomarkers for the diagnosis of cervical cancer. Journal of Virology Journal. 16 (1), 116 (2019).

