ACT1-CUP1 Assays Determine the Substrate-Specific Sensitivities of Spliceosomal Mutants in Budding Yeast
概要
The ACT1-CUP1 assay, a copper growth assay, provides a quick readout of precursor messenger RNA (pre-mRNA) splicing and the impact mutant splicing factors have on spliceosomal function. This study provides a protocol and highlights the customization possible to address the splicing question of interest.
Abstract
Mutations introduced in the spliceosome or its substrate have significantly contributed to our understanding of the intricacies of spliceosomal function. Whether disease-related or functionally selected, many of these mutations have been studied using growth assays in the model organism Saccharomyces cerevisiae (yeast). The splicing-specific copper growth assay, or ACT1-CUP1 assay, provides a comprehensive analysis of mutation at the phenotypic level. The ACT1-CUP1 assay utilizes reporters that confer copper tolerance when correctly spliced. Thus, in the presence of copper, changes in yeast viability correlate to changes in mRNA production through splicing. In a typical experiment, the yeast spliceosome is challenged with different non-consensus splicing reporters and the splicing factor mutation of interest to detect any synergetic or antithetical impact on splicing. Here a full description of copper plate preparation, plating of yeast cells, and data evaluation are given. A selection of complimentary experiments is described, highlighting the versatility of the ACT1-CUP1 reporters. The ACT1-CUP1 assay is a handy tool in the splicing toolbox thanks to the direct read-out of mutational effect(s) and the comparative possibilities from the continuing use in the field.
Introduction
The spliceosome is a large, biological machine that catalyzes the removal of introns, non-coding regions in precursor messenger RNA (pre-mRNA)1,2. Characterizing the effect of a single point mutant in 1 of the nearly 100 proteins and 5 non-coding RNAs is often ambiguous when studying the protein or RNA in isolation. The change in the mutated component's function can best be evaluated in vivo in the context of the full, functioning spliceosome.
The copper growth assay described here is a quick gauge of splicing efficiency in Saccharomyces cerevisiae or budding yeast. Developed by C.F. Lesser and C. Guthrie and published in 1993, this assay combines the ease of working with a simple model organism and the straightforward readout of cell viability3. The viability correlates with how well the spliceosomes in these cells can recognize and splice the reporter transcript.
This copper growth assay is more commonly called the ACT1-CUP1 assay. The name ACT1-CUP1 originates from the two genes fused to create a reporter of splicing efficiency. ACT1 is yeast's actin gene, which is highly expressed and has an efficiently spliced intron4,5. Cup1p is a copper chelator that sequesters copper in the cell to prevent interference with regular cellular functions6,7,8. The ACT1-CUP1 reporter contains these genes in sequence such that CUP1 is in the proper reading frame only if pre-mRNA splicing of ACT1's intron occurs (Figure 1). The resulting fusion protein contains the first 21 amino acids of actin and the full length Cup1p protein, which increases yeast viability in a copper-rich environment3. Thus, an increase in the amount of splicing of the reporter results in a higher concentration of Cup1p and a higher copper resistance (Figure 1). In comparison to other reporter genes, CUP1 impacts cell viability even at low levels, has a wide sensitivity range, and can be used to directly select for splicing mutations3,6,7. In addition, CUP1 is non-essential for standard yeast growth, and thus cellular homeostasis is not impacted during the setup for this assay. Complementary to deletion or temperature growth assays, ACT1-CUP1 provides information about the effects on splicing under otherwise optimal yeast growth conditions.
The spliceosome recognizes its substrate through three intronic sequences, namely the 5' splice site (5' SS), branch-site (BS), and 3' splice site (3' SS). Numerous ACT1-CUP1 reporters have been generated containing non-consensus sequences at these sites. A selection of the most common ACT1-CUP1 reporters is shown in Figure 1 and Table 1. As the spliceosome interacts with each splice site uniquely at different points in the splicing cycle, the robustness of the spliceosome can be tested at different steps based on which non-consensus reporter is used. Non-consensus reporters are named for the mutated position within the intron and the base it was mutated to. For example, A3c is a reporter with a mutation at the 5' SS, specifically position 3 from the consensus adenosine to a cytosine. This reporter will interact strongly with spliceosome mutations that impact 5' SS selection and use. In their initial study, Lesser and Guthrie determined which 5' SS mutations inhibited splicing3. Later the same year, non-consensus reporters at all three splice sites were published by Burgess and Guthrie in a suppressor screen of mutations in the ATPase Prp16p9. Comparing consensus to non-consensus reporters, the ACT1-CUP1 assay has been an important key to understanding the robustness and selectivity of the yeast spliceosome and to infer the function of other eukaryotes' spliceosomes.
As non-consensus ACT1-CUP1 reporters sensitize the spliceosome to further perturbation, the impact of a single splicing factor mutation can be characterized through the reporters it positively or negatively impacts. This has been applied to splicing research questions in a variety of ways. First, the ACT1-CUP1 assay can and has been used as a genetic screen for mutations in splicing factors. For example, Prp8p, the largest splicing protein, serves as a platform upon which the RNA core of the spliceosome catalyzes the splicing reaction. This was deduced, in part, through how Prp8p mutants improved or reduced the splicing of different ACT1-CUP1 reporters10,11,12,13,14,15,16,17. Other protein components of the spliceosome have also been investigated using ACT1-CUP1, including Hsh155p, Cwc2p, Cef1p, and Ecm2p18,19,20,21,22,23,24,25. The energetic thresholds for Prp16p and four other ATPases involved in spliceosomal transition have also been studied with this assay9,26,27,28,29,30. The small nuclear RNAs (snRNAs) have also been extensively studied utilizing ACT1-CUP1 to identify the pre-mRNA sequences they coordinate and the changes in secondary structure the snRNAs undergo during splicing3,31,32,33,34,35,36,37.
The ACT1-CUP1 assay requires a yeast strain where all copies of the CUP1 gene have been knocked-out. As CUP1 can have a high copy number6,38, preparation of a full knock-out strain can require multiple rounds or extensive screening. As a result, cup1Δ yeast strains have often been shared between labs, as have the reporters.
If mutation(s) in a splicing factor are being assessed from a plasmid copy, the wild-type gene for this factor should be knocked-out. In addition, the yeast background should allow for the selection of at least two plasmids, one containing an ACT1-CUP1 reporter, historically on a leucine nutrient-selection plasmid, and one containing a mutation or perturbation in the splicing machinery that will be studied (Figure 2). Usually, in a single assay, multiple yeast strains, each carrying the query splicing perturbation (QSP) and a different reporter, will test the query's impact on splicing.
The independent variables in the ACT1-CUP1 assay allow a researcher to assess the severity of a QSP. These independent variables are the concentration of copper and the selection of multiple non-consensus splicing reporters. First, as the yeast strains are grown on plates containing a range of copper concentrations (Figure 2), setting up the assay includes selecting the gradient of concentrations used. Studies can utilize a course copper concentration gradient to get an initial readout of viability and then repeat the assay with a finer gradient to identify subtle viability differences. The second variable is the wide range of ACT1-CUP1 reporters possible to test (Figure 1 and Table 1). If the QSP impacts yeast viability differently in the presence of a non-consensus reporter versus wild-type, a conclusion can be made that the QSP affects a step in splicing or a region of the spliceosome important during the recognition or processing of that region of the intron.
The yeast toolbox is extensive, and the ACT1-CUP1 assay is an integral part of splicing research. The ACT1-CUP1 assay is often performed alongside a more in-depth genetic, structural, and/or biochemical analysis on the impact of a QSP. As these more detailed studies generally have a lengthier procedure and/or higher price tag, a frequent approach is screening for interesting mutants with ACT1-CUP1 first.
Provided here is an ACT1-CUP1 assay protocol, including copper plate preparation. This assay provides researchers with an initial answer to a QSP's effect on splicing and which intronic regions are most impacted by the perturbation.
Protocol
1. Yeast strain construction
- Generate or obtain an S. cerevisiae strain whose background includes leu2 and cup1Δ. To generate this background, use the well-established yeast method that employs lithium acetate and single-stranded DNA39.
NOTE: Haploid yeast strains may contain one, two, or more copies of CUP16,38. Refer to genomic information for the selected yeast strain when designing knock-out primers to flank the CUP1 gene location(s). - Perform a yeast transformation to incorporate the QSP either via genomic incorporation or on a plasmid. Use a well-established protocol such as those described in previous research40,41,42.
- Perform a yeast transformation with the resulting strain(s) from step 1.2. to add the desired ACT1-CUP1 reporter plasmid.
NOTE: Cells must be maintained on leucine drop-out (-Leu) plates and media to ensure retention of the ACT1-CUP1 reporter plasmids following this transformation. - Perform steps 1.2. and 1.3. for each QSP and each ACT1-CUP1 reporter plasmid to be tested, including control strains.
2. Copper plate preparation
- Select a copper concentration range that suits the reporters to be tested (see Table 1 for frequently used reporters' lethality).
NOTE: An example of a comprehensive copper concentration range is 30 different copper concentrations of 0 mM, 0.025 mM, 0.05 mM, 0.075 mM, 0.1 mM, 0.15 mM, 0.2 mM, 0.25 mM, 0.3 mM, 0.35 mM, 0.4 mM, 0.45 mM, 0.5 mM, 0.6 mM, 0.7 mM, 0.8 mM, 0.9 mM, 1.0 mM, 1.1 mM, 1.2 mM, 1.3 mM, 1.4 mM, 1.5 mM, 1.6 mM, 1.7 mM, 1.8 mM, 1.9 mM, 2.0 mM, 2.25 mM, and 2.5 mM Cu2+. - Make a stock solution of 1 M CuSO4 and sterile filter through a 0.22 µm PES (polyethersulfone) sterile filter.
- Per desired copper plate, prepare a 2 mL dilution of the CuSO4 stock in sterile water.
NOTE: As the plate with 0 mM Cu2+ will always be analyzed and imaged as a reference, it is recommended to make two 0 mM Cu2+ plates, one at the beginning and one at the end of the plating step (step 3.4.).- Calculate the amount of stock for the final desired copper concentration in 40 mL of the plate volume (Supplemental Table 1).
- Add the calculated amount of sterile water and 1 M CuSO4 stock to a sterile, 2 mL tube.
- Pour plates for the ACT1-CUP1 assay.
NOTE: An alternative to the protocol below is to initially combine the media and agar in a large container and aliquot after autoclaving into smaller containers to achieve different copper concentrations for each plate. Whichever method is taken, it is important to ensure that the media concentration is consistent between all of the plates despite each having a different copper concentration.- Label each empty plate to be poured with the final copper concentration it will contain. Prepare at least one square plate per copper concentration to be tested.
- Label a 100 mL bottle per copper concentration to be tested.
- To each bottle, add 790 mg of agar (2% w/v agar) and a stir bar.
- In a large beaker, combine the -Leu growth media for all the copper plates to be poured. Per plate to be made, dissolve 265 mg of yeast nitrogen base (YNB) and 64 mg of drop-out mix minus leucine (and any other nutrients that may be required to maintain the QSP plasmid in the cells) in 34 mL of deionized water.
- Add 34 mL of the -Leu growth media solution to each prepared 100 mL bottle and cap with aluminum foil. Label the foil with the intended copper concentration.
- Autoclave to sterilize and dissolve the agar using the recommended liquid cycle for the autoclave.
- As promptly as possible, add 4 mL of 20% w/v glucose (sterile filtered) to each bottle.
- Match the labels and add the 2 mL dilutions of CuSO4 to its intended bottle.
NOTE: As tens of copper plates can be made at the same time, each with a different concentration, labeling all bottles, tubes, and plates clearly with the intended copper concentration will prevent confusion during plate pouring. - Use a stir plate to mix for ~30 s and pour or pipet 35 mL into the labeled plate, avoiding bubbles. Allow to cool before storing or use.
NOTE: Frequently, the plates are made 1 day or 2 days in advance of the assay and stored at 4 °C until a few hours before use. The plates should be at room temperature (RT) before plating begins (step 3.4.).
3. ACT1-CUP1 assay
- Streak out the desired strains on -Leu plates.
NOTE: If working from cryo stocks, care should be taken to ensure the cells are sufficiently revived from storage before plating. A recommended procedure for this is to streak from the cryogenic stock and allow it to grow for 3-5 days at 30 °C. Then, restreak a small swatch and allow it to grow for another 2-3 days at 30 °C. - Grow overnight cultures in 10 mL of media.
- Prepare -Leu growth media using the same ratios as described in step 2.4.2. Per 10 mL of media, add 66 mg of yeast nitrogen base (YNB) and 16 mg of drop-out mix minus leucine to 9 mL of deionized water. Pass through a 0.22 µm PES sterile filter.
- Per yeast strain, add 9 mL of -Leu growth media and 1 mL of 20% w/v glucose (sterile filtered) to a sterile 50 mL conical tube.
- Using a sterile stick or pipet tip, gather a small (~1 mm round) swatch of yeast and inoculate the media.
- Shake all the overnight cultures at 180 rpm and 30 °C.
NOTE: If available, rotators can be used instead of a shaker.
- Dilute the strains to an OD600 0.5 ± 0.05 in 10% glycerol.
- Per strain, add 100 µL of culture to a cuvette containing 900 µL of water.
- Measure the OD600 with a spectrophotometer.
- Calculate the dilution required to be at an OD600 of 0.5 in a final volume of 2 mL.
- Dilute each strain to OD600 0.5 in 10% glycerol (sterile).
- Remeasure the OD600 to confirm the cell density is within the desired range of 0.5 ± 0.05.
- Plate the strains on the copper plates.
NOTE: A variety of methods can be used to plate the strains, including hand pipetting 5-10 µL volumes, using a repeat or multi-channel pipettor, or stamping with a pin replicator. This last method is described below, though most steps will be similar regardless of the method.- Set up a sterile working location and a lit Bunsen burner.
- For a 48-pin replicator, pipet 200 µL of each diluted strain into a separate well of a 96-well plate. Fill the empty spaces in the 6 x 8 grid with 200 µL of 10% glycerol (sterile).
NOTE: An example of a plating scheme for nine yeast strains is in Supplemental Table 2. - Dip the replicator in a shallow dish of 95% (v/v) ethanol and flame to sterilize. Let it cool for at least 2 min after the flame extinguishes to avoid heat shocking the cells.
- Place four plates near the burner and remove the lids.
- Dip the replicator in the 96-well plate and lift it up in one quick motion.
- Place gently onto the plate and rock lightly back and forth to facilitate a good transfer.
- Lift up in one swift motion and place in the exact same orientation in the 96-well plate.
- Repeat for up to three other plates. Repeat the process of dipping the replicator in ethanol, flaming to sterilize, and waiting to cool every four plates.
- After a plate has been plated, move with a smooth motion to the side but still within the sterilization umbrella of the flame.
NOTE: It is recommended to do the yeast dilutions and plating near a flame. The plates can easily become contaminated while drying. - Let the plates dry completely before placing the lids on, usually 3-5 min.
- Incubate the plates for 3 days at 30 °C.
4. Data collection and analysis
- Remove the plates from the incubator and visually inspect them.
- Record the images of the plates with an available camera or other digital imaging system.
- Record (or score) for each strain the last copper concentration visible growth is observed.
NOTE: The cells are able to splice and remain viable up until that concentration. For consistency, always use the same method, either by eye or from the plate images, to score the last viable copper concentration. Very small colonies are sometimes visible by the eye but not on the image. The difference between direct visual inspection or recording from images is small, usually a step in the gradient. As images of the colonies are often used in publications, scoring by the images is recommended. - Combine data from multiple ACT1-CUP1 assays for the same strain to draw conclusions about how the QSP affects splicing.
NOTE: Publication figures customarily show images of the yeast colonies at 0 mM Cu2+ concentration, the last viable copper concentration, and the subsequent concentration where the colony has died. Data can also be displayed as a bar graph with error bars for the standard deviation between replicates. Data do not need to be normalized but can be by setting the viability of the WT splicing factor control to 1 and comparing the effect of the mutation(s) introduced.
Representative Results
Growth assays, like ACT1-CUP1, require the visual, comparative assessment of multiple colonies. Here, each strain was grown to saturation overnight, diluted to an OD600 of 0.5, and plated on 20 plates containing a range of copper concentrations from 0 mM to 1.1 mM CuSO4 (Figure 3). This range is smaller than that listed in the protocol as it allowed for the full assessment of the impact of the QSPs and ACT1-CUP1 reporters used and described below. The plates were imaged and scored (Figure 3 and Supplemental Table 3).
For this representative experiment, the yeast background includes a disrupted ADE2 gene, resulting in the colonies being varying shades of red (Figure 3A,B). This common yeast disruption in the adenosine production pathway causes a buildup of a red-pigmented precursor for adenosine. Thus, the red color is an indicator of yeast colony maturity (i.e., the amount and age of the cells present). For the purposes of an ACT1-CUP1 assay, the red color can serve as an indicator of contaminating fungal species if white or yellow in color. The color can be a secondary confirmation of copper tolerance as more viable colonies will be a deeper red shade.
When plating with a pin replicator, there are several common aberrations. First, it is possible to create colonies that are oval-shaped if the replicator is lifted up while moving left or right (Figure 3B, orange arrow). In addition, microcolonies can also form from small droplets of cell solution if the pin replicator is brought in at an angle or if shaken above the plate (Figure 3B, red arrow). Often innocuous, occasionally, the microcolonies can mix with a stamped colony, and this prevents interpretation of that colony. Additional plating issues include insufficient wait time after sterilization for the replicator pins to cool and insufficient contact between the plate and the pins such that poor transfer of the culture media occurs. In both cases, few, if any, cells will grow on the plate, including for the control strains containing the wild-type reporter. As with any growth assay, regardless of the method of plating, it is important to perform the ACT1-CUP1 in triplicate to confirm the results are consistent and repeatable. Ideally, these different replicates should be performed on copper plates prepared at different times to ensure reproducibility.
For these representative experiments, the A3c reporter was selected to highlight the impact of a non-consensus sequence on copper tolerance in the absence of additional spliceosome perturbations. A3c significantly reduces the amount of CUP1 mRNA produced as it perturbs the ability of the spliceosome to recognize and utilize the 5' SS15. Yeast with the A3c reporter survived to 0.15 mM Cu2+, while the wild-type reporter cells maintained viability to the end of the range of copper concentrations tested (Figure 3C). The wild-type reporter containing cells grow to 2.5 mM Cu2+ without an impact on viability (data not shown).
U6 snRNA is an essential catalytic component of the spliceosome. Multiple studies have used ACT1-CUP1 to study the effect mutations can have on this RNA16,32,34. Duplicated for this study, three U6 snRNA sequences were studied, namely the wild-type sequence (WT), position 57 substituted from a uridine to a cytosine (U57c), and position 57 substituted from a uridine to an adenosine (U57a) (Figure 3 and Figure 4). Combined with ACT1-CUP1 reporters that impact the catalytic steps in splicing, it was determined that U57c favors the first catalytic step, and U57a favors the progression to the second step16,32.
To set up for the ACT1-CUP1 assay, a strain was created with the genomic copy of U6 snRNA deleted and the U6 snRNA wild-type or mutated sequences included on plasmids. As U6 snRNA is an essential component of the cell, three separate transformations were used to first knock-out the genomic U6 snRNA while maintaining cell viability, subsequently introduce mutated U6 snRNAs on plasmids, and, finally, add the ACT1-CUP1 reporters. For the first transformation, a cup1Δ yeast strain was used in the knock-out of the genomic copy of U6 snRNA while simultaneously adding WT U6 snRNA on a URA selection marker plasmid to maintain viability. A subsequent transformation added either wild-type or mutated U6 snRNA on a TRP selection marker plasmid, selecting against the URA marker via 5-FOA selection. Thus, three cup1Δ yeast strains were generated, each with one of the U6 snRNA sequences, namely WT, U57a, and U57c. A final yeast transformation for each strain added one of the three different ACT1-CUP1 reporter plasmids to be used in this experiment. The reporters selected were the wild-type reporter, A3c, and a mutation of the branch-site adenosine to guanine (BS-G). A total of nine strains were generated for this experiment, each containing one U6 snRNA sequence and one ACT1-CUP1 reporter (Supplemental Table 4 and Supplemental Table 5).
The results showed that the different bases at U6 snRNA position 57 have unique impacts on the spliceosome in combination with a 5' SS or BS mutation (Figure 4). Both the A3c and BS-G reporters primarily inhibit splicing by stabilizing the first catalytic step conformation14,15. Thus, U57c is an additive mutation decreasing copper tolerance in combination with either of these reporters (Figure 4B). In contrast, U57a increases copper tolerance because it promotes progression to the second step (Figure 4B)16,32. The BS-G reporter strain's decreased copper tolerance with U57a compared to U6 snRNA WT highlights a likely secondary impact of BS-G on the second step of splicing16.
These results also highlight the qualitative nature of this assay and why wild-type query and reporter sequences should be tested. While the general pattern of increased or decreased copper tolerance holds true for these U6 snRNA mutations compared to wild-type U6 snRNA, the exact concentration of copper to which the cells survive can differ between studies (Table 1) and did differ between these representative data and other published results. This is likely due to the background of the strain containing the CUP1 deletion but can also be due to the general health of the strains before plating (i.e., how soon after generation of the strain or restreak from cryo the assay was performed), differences in how the plates are prepared, and variability between different incubators. A similar variance was noted in Mayerle et al. for Prp8 query mutations in the literature43. Thus, comparing copper tolerance trends can be performed for different ACT1-CUP1 assays, but the numeric comparison of the copper concentrations should be done only within the same lab and, at times, with the same set of copper plates.
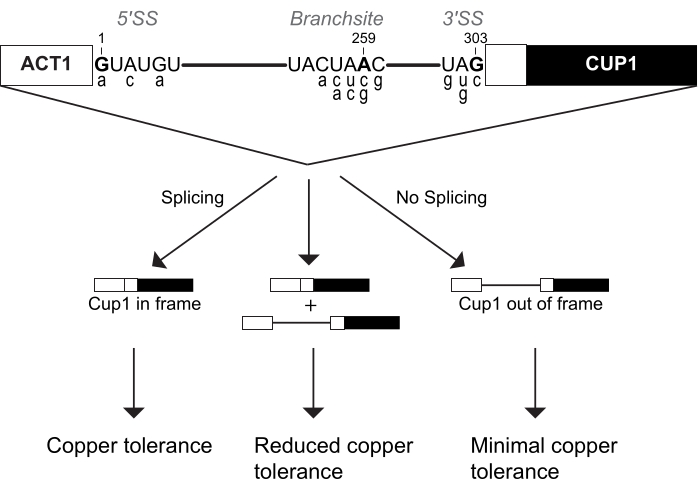
Figure 1: The ACT1-CUP1 reporter design. The concentration of spliced reporter directly correlates with yeast copper tolerance. The diagram of the reporter includes the three splice sites of the 5' splice site (5' SS), branch-site (BS), and 3' splice site (3' SS). The yeast consensus sequences for these sites are shown, with those targeted for cleavage indicated in bold and numbered based on their location in the intron. Commonly used non-consensus ACT1-CUP1 reporter sequences are shown in the lower case below their corresponding consensus sequence locations and are listed in Table 1. Please click here to view a larger version of this figure.
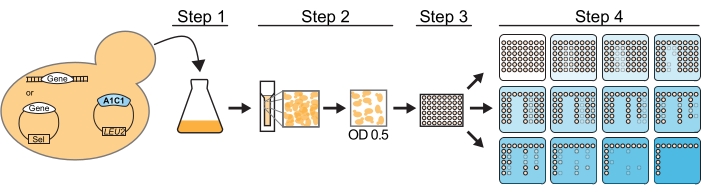
Figure 2: ACT1-CUP1 assay workflow. For this assay, a yeast strain must be cup1Δ leu2 and contain the desired ACT1-CUP1 reporter plasmid and the QSP gene, labeled Gene in the figure. The QSP gene must either be genomically inserted or on a plasmid. The protocol using a pin replicator involves four steps to prepare the yeast cells. Step 1 is to grow the cells to saturation. Step 2 is to measure the OD600 of the culture and dilute to an OD600 of 0.5. Step 3 is to distribute over a 96-well plate. Step 4 is to plate on plates containing increasing concentrations of copper (indicated with increasing blue color). Step 1, step 2, and step 4 would be identical for hand pipetting. Once plated, the plates are incubated for 3 days at 30 °C and then scored for viability. Please click here to view a larger version of this figure.
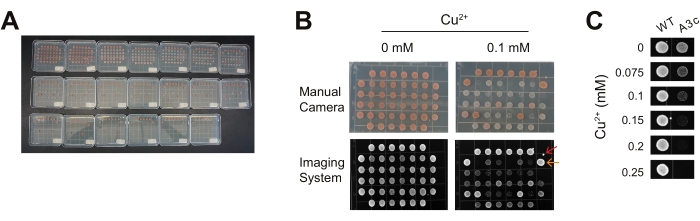
Figure 3: Representative views of ACT1-CUP1 data. For this experiment, yeast spliceosomes are challenged with two different U6 snRNA mutations and tested in the presence of three non-consensus reporters. Three replicates of this assay are presented on the same plates for demonstration purposes. It is recommended for this assay to do replicates on separate plates and on separate days. (A) A completed assay with a copper gradient of 0 mM to 1.1 mM over 20 plates is shown. As the copper concentration increases, strains with lower splicing show decreased confluency to the point where the copper concentration becomes lethal. The yeast background was ade2, and, thus, mature colonies are red in color. (B) Comparison of the same plates at 0 mM and 0.1 mM CuSO4 imaged with a handheld camera versus a digital imaging system. This is an example of how a number of non-consensus reporters can be compared side by side and with the wild-type reporter. Common observations on plates include a spurious drop of culture media that fell from the pin replicator (red arrow) and slight ovaling of the colonies due to sliding of the pin along the surface of the plate or movement of the plate before the drop of culture has dried sufficiently (orange arrow). (C) Example of the A3c reporter's effect on cell viability compared to the wild-type reporter. Images such as those shown in (B) are cropped and aligned to highlight the growth differences at different copper concentrations. The copper tolerance of the A3c reporter strain decreases to 0.15 mM Cu2+ compared to the wild-type reporter strain's viability to the end of the range tested. Please click here to view a larger version of this figure.
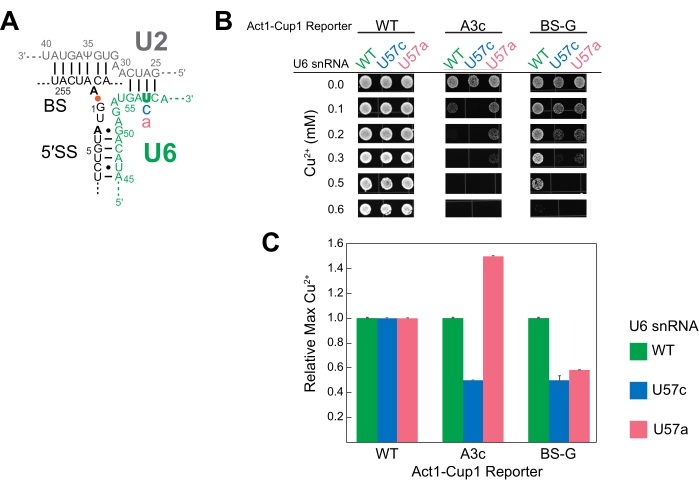
Figure 4: Representative ACT1-CUP1 results monitoring splicing in yeast with splicing component mutations. (A) Schematic of the RNA components of the active site immediately after the first catalytic step. The U2 and U6 snRNAs are duplexed, bringing the 5' SS and BS of the intron in close proximity. The intronic bond between A259 (BS) and G1 (5' SS) is indicated by the orange dot. The locations of the non-consensus sequence substitutions in the ACT1-CUP1 reporter tested in this experiment are indicated in bold black. The mutated position in U6 snRNA (U57) is in bold green, and the substituted bases are in either blue or pink. (B) One possibility for the presentation of ACT1-CUP1 data in a publication includes several images of colonies from relevant Cu2+ concentrations. The WT reporter survived past the copper concentration tested for all three U6 snRNA strains queried. (C) A bar graph comparing the effects per reporter and per U6 snRNA mutant. Normalization is performed for each ACT1-CUP1 reporter by setting the copper tolerance of the U6 snRNA WT to 1 and calculating the ratio for the U57c and U57a mutations. Error bars represent the standard deviation from three replicates. Please click here to view a larger version of this figure.
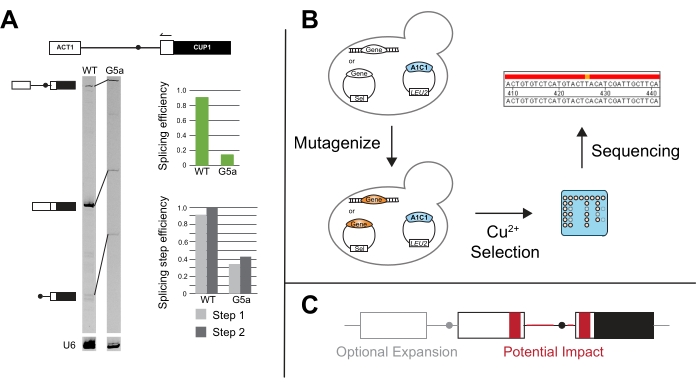
Figure 5: Additional methods to compliment ACT1-CUP1 results. (A) Primer extension is performed via a primer annealing in the 3' ACT1 exon and then elongating into the intron. In this example, the primer is end-labeled with IR700 dye, and the primer extension was performed as described in 20,21. Primer extension of U6 snRNA is performed in the same reaction to serve as a loading control. The 7% 19:1 bis/acrylamide, denaturing gel resolves the pre-mRNA, mRNA, and lariat products after primer extension using a near infra-red gel imaging device as described in van der Feltz et al.22. The intensity of the different bands can be used to measure overall the splicing efficiency as well as distinguish the splicing differences that occur at the first or second step, as quantified with ImageJ44 or other gel band quantifying software. Both the splicing efficiency and the splicing step efficiency are calculated as described in Query and Konarska14. (B) Mutational screens with ACT1-CUP1 reporters can utilize copper selection to identify mutants that impact splicing. The gene(s) of interest can then be sequenced in the resulting strains to determine if the mutation occurred in that gene. (C) Alterations can be made to the reporter outside of the splice sites to monitor splicing changes in relation to intron structure, exonic sequences, number of exons, or nuclear export of unspliced RNA. Regions in grey are optional expansion regions to include on a reporter, and areas in red are regions in which the sequence and structural changes could be tested for their potential impact on splicing using the ACT1-CUP1 assay. Please click here to view a larger version of this figure.
| Reporter name | Intronic region | Sequence | Last viable Cu2+ concentration (mM) | |
| WT | 5’ | GUAUGU | > 2.5 | |
| G1a | 5ʹ | aUAUGU | 0.0133 or 0.0532 | |
| A3c | 5ʹ | GUcUGU | 0.15^,18 or 0.216 | |
| G5a | 5ʹ | GUAUaU | 0.303 or 0.259 | |
| WT | Branch-site | UACUAAC | > 2.5 | |
| C256a | Branch-site | UAaUAAC | 0.1526 or 0.1832 | |
| U257c/a | Branch-site | UACc/aAAC | 0.226 or 0.332 or 0.545 | 0.059,32 |
| A258c/u | Branch-site | UACUc/uAC | 0.826 | 1.045 or 1.646 |
| BS-C | Branch-site | UACUAcC | 0.153 or 0.1832,56 or 0.226 | |
| BS-G | Branch-site | UACUAgC | 0.0516,32 or 0.625 or 0.846 | |
| C260g | Branch-site | UACUAAg | 0.826 | |
| WT | 3’ | UAG | > 2.5 | |
| U301g | 3ʹ | gAG | 0.1518 | |
| A302g/u | 3’ | Ug/uG | 0.0139 | 0.07516 or 0.1824 |
| G303c | 3ʹ | UAc | 0.0532 | |
Table 1: List of common reporters and the Cu2+ concentration lethality reported. There are over 100 citations of Lesser and Guthrie3. While a small selection of these citations was used to create this table, they highlight the general trend of slight to moderate differences between studies in reported copper viability concentrations. In ACT1-CUP1, it is important to compare wild-type protein or RNA to the mutants all with the same yeast strain background using the published concentrations as a guide but anticipating that the observed concentrations may differ. Data gathered from Figure 3 (^) and multiple publications annotated by the following citations3,9,16,18,24,25,26,32,45,46.
Supplemental Table 1: Contents of a single copper plate and an example of calculations done to achieve copper dilutions from 0 mM to 2.5 mM from a 1 M CuSO4 stock. Please click here to download this Table.
Supplemental Table 2: Example of a plating scheme for the 48-pin replicator. Please click here to download this Table.
Supplemental Table 3: Example of viability scored from copper plates incubated for 3 days at 30°C. Please click here to download this Table.
Supplemental Table 4: List of yeast strains used to generate the representative data. Please click here to download this Table.
Supplemental Table 5: List of plasmids used to generate the representative data. U6 snRNA plasmids generated and published in previous research22,47. Please click here to download this Table.
Discussion
ACT1-CUP1 is a growth assay, and care must be taken to ensure that observed growth differences can only be attributed to splicing defects. All strains should be handled in a similar fashion prior to plating, including having a similar length and type of growth and storage conditions. If using temperature-sensitive strains, ACT1-CUP1 assays should only be performed under conditions where those strains grow comparably to wild type. Relatedly, for the QSP component, it is advised to have identical yeast backgrounds and expression levels of QSP gene(s) so as to not cloud the interpretation of the results. When considering the number of QSPs and reporters to use, it is not advised to assay more than 30 strains at a time with the pin replicator method. With the additional time to perform each step, the cells will settle, and the assay results will be inconsistent due to decreased cell viability.
In addition to the limitations ACT1-CUP1 inherently has as a growth assay, a QSP may have a more complex impact on splicing than can be resolved with this method. As splicing is a multi-step process and factors are recycled, the observed phenotype can result in the perturbation affecting more than one splicing step. This holds true even for the subsequent analyses that can follow up ACT1-CUP1, some of which are described below. The data will highlight the most perturbed step of the process, even though other steps may be affected by the altered function of the splicing factor.
Primer extension of ACT1-CUP1 reporters was first utilized in Lesser and Guthrie's original paper and is often performed if the ACT1-CUP1 results show a growth defect3. This assay targets the reporter and uses the length of the PCR products to determine the relative amounts of unspliced, partially spliced, and fully spliced reporter present (Figure 5A). Overall splicing efficiency and whether the defect more strongly impacts the first or second catalytic step are calculated by taking ratios of the PCR product amounts14. For example, 5' SS reporter G5a has reduced splicing compared to the wild-type reporter, but its first and second step efficiencies follow a similar pattern to wild-type (Figure 5A). This points toward a defect prior to the first catalytic step, possibly in spliceosome assembly, because both steps are affected similarly31.
Novel assays have been developed from the canonical ACT1-CUP1 assay, such as screening for mutants that improve splicing in the presence of other splicing factor mutants and/or non-consensus splicing sequences (Figure 5B). For example, exposing yeast containing the ACT1-CUP1 reporter to UV and then selecting in the presence of copper yielded Prp8 and Hsh155 mutants that improved the splicing of non-consensus sequences14,19.
Expanding past the study of the effect of splice sites and constitutive splicing factors, CUP1 growth dependence has been used to study multiple intron splicing, nuclear export control, and the impact of UTR and other peripheral sequences on splicing (Figure 5C). Some of these studies have created reporters with CUP1 and other intron-containing yeast genes that can have more complex splicing patterns to study the effect on intronic secondary structure and multi-intron transcript splicing48,49,50,51,52. The link between splicing and nuclear export was studied by having either spliced or unspliced transcripts encoding CUP1 in frame53,54. The length of the pyrimidine track and splice site selection's distance dependence due to specific factors has also been tested20,55,56,57. These examples and many others highlight the versatility of ACT1-CUP1 and other CUP1 growth assays.
The ACT1-CUP1 assay links perturbations to a complex reaction cycle with a straightforward growth phenotype with a wide sensitivity range. This legacy assay has been used by multiple labs to lay the foundation of the understanding of the splicing cycle. More recently, ACT1-CUP1 has been used to answer questions that arise from the wealth of structural data now available21,22,25. Structural studies of spliceosomes bound to non-consensus sequences could be paired with ACT1-CUP1 results to interpret how altered structure and altered function correlate. ACT1-CUP1 is an ideal first screen for splicing mutations that can complement more complex analysis.
開示
The authors have nothing to disclose.
Acknowledgements
Thank you to Aaron Hoskins and the Hoskins lab members at the University of Wisconsin-Madison for use of yeast strains and equipment in the generation of figures 3-5. Thank you to Harpreet Kaur and Xingyang Fu for their insightful comments on the manuscript. Thank you to the supportive students, staff, and faculty at Northwest University during the writing, editing, and filming of this paper. Thank you to Isabelle Marasigan for help in filming this method.
Materials
| 1.5 mL sterile microcentrifuge tubes | Fisher Scientific | 05-408-129 | Or comparable item from a different manufacturer. |
| 2 mL sterile microcentrifuge tubes | Fisher Scientific | 05-408-138 | Or comparable item from a different manufacturer. |
| 50 mL sterile centrifuge tubes | Fisher Scientific | 07-201-332 | Or comparable item from a different manufacturer. |
| 96-well round bottom microplate | Fisher Scientific | 07-200-760 | Or comparable item from a different manufacturer. |
| 190 proof ethanol | Fisher Scientific | 22-032-600 | Or comparable item from a different manufacturer. |
| 500 mL Filter System (0.22 µm) | CellTreat Scientific Products | 229707 | Or comparable item from a different manufacturer. |
| Agar | Fisher Scientific | BP1423-500 | Any molecular grade agar will work. |
| Autoclave | Tuttnauer | 3870EA | Or comparable item from a different manufacturer. |
| Bunsen burner | Humboldt | PN6200.1 | Or comparable item from a different manufacturer. |
| Cell Density Meter | VWR | 490005-906 | Or other spectral device that can measure absorbance at 595 nm. |
| Copper sulfate Pentahydrate | Fisher Scientific | LC134051 | Or comparable item from a different manufacturer. |
| Digital imaging system | Cytiva | 29399481 | ImageQuant 4000 (used for Figure 3), Amersham ImageQuant 800, or comparable item from a different manufacturer. |
| Dropout mix (-Leu) | USBiological Life Sciences | D9525 | Use the appropriate drop out mix for your experiment. It is possible you will be using a yeast nutrient marker for your query perturbation also. In that case, the drop out mix should be for that marker and Leu |
| D-Glucose | Fisher Scientific | AAA1682836 | Or comparable item from a different manufacturer. |
| Gel band quantifying software | Cytiva | 29-0006-05 | ImageQuant TL v8.1 (used for figure 5A) or comparable item from a different manufacturer. |
| Hand held camera | Nikon | D3500 | Or comparable item from a different manufacturer. |
| Near infra-red gel imaging device | Cytiva | 29238583 | Amersham Typhoon NIR (used for Figure 5a) or comparable item from a different manufacturer. |
| Laboratory grade clamp | Fisher Scientific | 05-769-7Q | Or comparable item from a different manufacturer. |
| Laboratory grade stand and clamp | Fisher Scientific | 12-000-101 | Or comparable item from a different manufacturer. |
| Magnetic stir bars | Fisher Scientific | 14-513-51 | Or comparable item from a different manufacturer. |
| Pin replicator | VP Scientific | VP 407AH | |
| Semi-micro disposable cuvettes | VWR | 97000-590 | Or comparable item from a different manufacturer. |
| Shaker | JEIO Tech | IST-3075 | Or comparable item from a different manufacturer. |
| Spectrophotometer | Biowave | 80-3000-45 | Or any spectophotometer that can measure the absorbance at 600 nm. |
| Square plates | VWR | 102091-156 | Circular plates may also be used though are more challenging if using a pin replicator. |
| Stir plate | Fisher Scientific | 11-520-16S | Or comparable item from a different manufacturer. |
| Yeast nitrogen base | USBiological Life Sciences | Y2025 | Or comparable item from a different manufacturer. |
参考文献
- Wahl, M. C., Will, C. L., Luhrmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 136 (4), 701-718 (2009).
- Wilkinson, M. E., Charenton, C., Nagai, K. RNA splicing by the spliceosome. Annual Review of Biochemistry. 89, 359-388 (2020).
- Lesser, C. F., Guthrie, C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. 遺伝学. 133 (4), 851-863 (1993).
- Ng, R., Abelson, J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 77 (7), 3912-3916 (1980).
- Gallwitz, D., Sures, I. Structure of a split yeast gene: Complete nucleotide sequence of the actin gene in I. Proceedings of the National Academy of Sciences of the United States of America. 77 (5), 2546-2550 (1980).
- Fogel, S., Welch, J. W., Cathala, G., Karin, M. Gene amplification in yeast: CUP1 copy number regulates copper resistance. Current Genetics. 7 (5), 347-355 (1983).
- Hamer, D. H., Thiele, D. J., Lemontt, J. E. Function and autoregulation of yeast copperthionein. Science. 228 (4700), 685-690 (1985).
- Winge, D. R., Nielson, K. B., Gray, W. R., Hamer, D. H. Yeast metallothionein. Sequence and metal-binding properties. Journal of Biological Chemistry. 260 (27), 14464-14470 (1985).
- Burgess, S. M., Guthrie, C. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 73 (7), 1377-1391 (1993).
- Collins, C. A., Guthrie, C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes & Development. 13 (15), 1970-1982 (1999).
- Siatecka, M., Reyes, J. L., Konarska, M. M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Development. 13 (15), 1983-1993 (1999).
- Umen, J. G., Guthrie, C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. 遺伝学. 143 (2), 723-739 (1996).
- Grainger, R. J., Beggs, J. D. Prp8 protein: At the heart of the spliceosome. RNA. 11 (5), 533-557 (2005).
- Query, C. C., Konarska, M. M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Molecular Cell. 14 (3), 343-354 (2004).
- Konarska, M. M., Vilardell, J., Query, C. C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Molecular Cell. 21 (4), 543-553 (2006).
- Liu, L., Query, C. C., Konarska, M. M. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nature Structural and Molecular Biology. 14 (6), 519-526 (2007).
- MacRae, A. J., et al. Prp8 positioning of U5 snRNA is linked to 5′ splice site recognition. RNA. 24 (6), 769-777 (2018).
- Query, C. C., Konarska, M. M. CEF1/CDC5 alleles modulate transitions between catalytic conformations of the spliceosome. RNA. 18 (5), 1001-1013 (2012).
- Tang, Q., et al. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes & Development. 30 (24), 2710-2723 (2016).
- Carrocci, T. J., Zoerner, D. M., Paulson, J. C., Hoskins, A. A. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Research. 45 (8), 4837-4852 (2017).
- Kaur, H., Groubert, B., Paulson, J. C., McMillan, S., Hoskins, A. A. Impact of cancer-associated mutations in Hsh155/SF3b1 HEAT repeats 9-12 on pre-mRNA splicing in Saccharomyces cerevisiae. PLoS One. 15 (4), 0229315 (2020).
- vander Feltz, C., et al. Saccharomyces cerevisiae Ecm2 modulates the catalytic steps of pre-mRNA splicing. RNA. 27 (5), 591-603 (2021).
- Carrocci, T. J., Paulson, J. C., Hoskins, A. A. Functional analysis of Hsh155/SF3b1 interactions with the U2 snRNA/branch site duplex. RNA. 24 (8), 1028-1040 (2018).
- Hogg, R., de Almeida, R. A., Ruckshanthi, J. P., O’Keefe, R. T. Remodeling of U2-U6 snRNA helix I during pre-mRNA splicing by Prp16 and the NineTeen Complex protein Cwc2. Nucleic Acids Research. 42 (12), 8008-8023 (2014).
- Hansen, S. R., Nikolai, B. J., Spreacker, P. J., Carrocci, T. J., Hoskins, A. A. Chemical inhibition of pre-mRNA splicing in living Saccharomyces cerevisiae. Cell Chemical Biology. 26 (3), 443-448 (2019).
- Xu, Y. Z., Query, C. C. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Molecular Cell. 28 (5), 838-849 (2007).
- Staley, J. P., Guthrie, C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Molecular Cell. 3 (1), 55-64 (1999).
- Bousquet-Antonelli, C., Presutti, C., Tollervey, D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 102 (6), 765-775 (2000).
- Villa, T., Guthrie, C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes & Development. 19 (16), 1894-1904 (2005).
- Mayas, R. M., Maita, H., Staley, J. P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nature Structure and Molecular Biology. 13 (6), 482-490 (2006).
- Lesser, C. F., Guthrie, C. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 262 (5142), 1982-1988 (1993).
- McPheeters, D. S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 2 (11), 1110-1123 (1996).
- Perriman, R. J., Ares, M. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Development. 21 (7), 811-820 (2007).
- Mefford, M. A., Staley, J. P. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 15 (7), 1386-1397 (2009).
- Hilliker, A. K., Mefford, M. A., Staley, J. P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Development. 21 (7), 821-834 (2007).
- Wu, G., et al. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO Journal. 35 (6), 654-667 (2016).
- Crotti, L. B., Bacikova, D., Horowitz, D. S. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes & Development. 21 (10), 1204-1216 (2007).
- Fogel, S., Welch, J. W. Tandem gene amplification mediates copper resistance in yeast. Proceedings of the National Academy of Sciences of the United States of America. 79 (17), 5342-5346 (1982).
- Gardner, J. M., Jaspersen, S. L. Manipulating the yeast genome: Deletion, mutation, and tagging by PCR. Methods Molecular Biology. 1205, 45-78 (2014).
- JoVE. Yeast Transformation and Cloning. In Biology I: yeast, Drosophila and C. Elegant. JoVE Science Education Database. , (2021).
- Gietz, R. D., Woods, R. A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Molecular Biology. 313, 107-120 (2006).
- Gietz, R. D., Woods, R. A. Genetic transformation of yeast. Biotechniques. 30 (4), 816 (2001).
- Mayerle, M., et al. Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proceedings of the National Academy of Sciences of the United States of America. 114 (18), 4739-4744 (2017).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Beier, D. H., et al. Dynamics of the DEAD-box ATPase Prp5 RecA-like domains provide a conformational switch during spliceosome assembly. Nucleic Acids Research. 47 (20), 10842-10851 (2019).
- vander Feltz, C., DeHaven, A. C., Hoskins, A. A. Stress-induced pseudouridylation alters the structural equilibrium of yeast U2 snRNA Stem II. Journal of Molecular Biology. 430 (4), 524-536 (2018).
- Rodgers, M. L., Didychuk, A. L., Butcher, S. E., Brow, D. A., Hoskins, A. A. A multi-step model for facilitated unwinding of the yeast U4/U6 RNA duplex. Nucleic Acids Research. 44 (22), 10912-10928 (2016).
- Stutz, F., Rosbash, M. A functional interaction between Rev and yeast pre-mRNA is related to splicing complex formation. EMBO Journal. 13 (17), 4096-4104 (1994).
- Libri, D., Lescure, A., Rosbash, M. Splicing enhancement in the yeast rp51b intron. RNA. 6 (3), 352-368 (2000).
- Libri, D., Stutz, F., McCarthy, T., Rosbash, M. RNA structural patterns and splicing: Molecular basis for an RNA-based enhancer. RNA. 1 (4), 425-436 (1995).
- Howe, K. J., Kane, C. M., Ares, M. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 9 (8), 993-1006 (2003).
- Cuenca-Bono, B., et al. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Research. 39 (19), 8599-8611 (2011).
- Scherrer, F. W., Spingola, M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA. 12 (7), 1361-1372 (2006).
- Hálová, M., et al. Nineteen complex-related factor Prp45 is required for the early stages of cotranscriptional spliceosome assembly. RNA. 23 (10), 1512-1524 (2017).
- Umen, J. G., Guthrie, C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Development. 9 (7), 855-868 (1995).
- Crotti, L. B., Horowitz, D. S. Exon sequences at the splice junctions affect splicing fidelity and alternative splicing. Proceedings of the National Academy of Sciences of the United States of America. 106 (45), 18954-18959 (2009).
- Perriman, R., Ares, M. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Molecular Cell. 38 (3), 416-427 (2010).

