Isolation and Quantitative Evaluation of Brush Cells from Mouse Tracheas
概要
Brush cells are rare cholinergic chemosensory epithelial cells found in the naïve mouse trachea. Due to their limited numbers, ex vivo evaluation of their functional role in airway immunity and remodeling is challenging. We describe a method for isolation of tracheal brush cells by flow cytometry.
Abstract
Tracheal brush cells are cholinergic chemosensory epithelial cells poised to transmit signals from the airway lumen to the immune and nervous systems. They are part of a family of chemosensory epithelial cells which include tuft cells in the intestinal mucosa, brush cells in the trachea, and solitary chemosensory and microvillous cells in the nasal mucosa. Chemosensory cells in different epithelial compartments share key intracellular markers and a core transcriptional signature, but also display significant transcriptional heterogeneity, likely reflective of the local tissue environment. Isolation of tracheal brush cells from single cell suspensions is required to define the function of these rare epithelial cells in detail, but their isolation is challenging, potentially due to the close interaction between tracheal brush cells and nerve endings or due to airway-specific composition of tight and adherens junctions. Here, we describe a procedure for isolation of brush cells from mouse tracheal epithelium. The method is based on an initial separation of tracheal epithelium from the submucosa, allowing for a subsequent shorter incubation of the epithelial sheet with papain. This procedure offers a rapid and convenient solution for flow cytometric sorting and functional analysis of viable tracheal brush cells.
Introduction
Brush cells belong to a class of chemosensory epithelial cells characterized by the expression of bitter taste receptors and the taste receptor transduction machinery found in taste bud cells. Unlike taste bud cells, chemosensory epithelial cells are scattered in epithelial surfaces and are referred to as solitary chemosensory cells (SCCs) and microvillous cells in the nasal epithelium1,2, brush cells in the trachea3,4, and tuft cells in the intestine5,6. Epithelial cells expressing bitter taste receptors and the bitter taste transduction machinery are also found in the urethra7,8 and the auditory tube9. Airway brush cells have unique functions in neurogenic and immune airway responses. They are acetylcholine-producing chemosensory cells that evoke protective respiratory reflexes upon activation with bitter compounds and bacterial metabolites like quorum-sensing substances10. Airway brush cells are also the dominant airway epithelial source of IL-25, which regulates aeroallergen-elicited type 2 inflammation in the airways3.
Characterization of the full transcriptome of lower airway brush cells and their response to environmental stimuli has been limited by their low numbers in the tracheal epithelium and very limited numbers beyond the large bronchi10. Techniques used for the isolation of chemosensory cells from the intestinal epithelium have not yielded proportionally high numbers from the trachea, possibly because of the intimate contacts of tracheal brush cells with nerve endings10 or other tissue-specific factors in the respiratory mucosa such as the composition of adherens and tight junction proteins. Recent reports of successful isolation of tracheal brush cells in higher numbers for single cell RNA sequencing analysis employed either a 2 h incubation with papain or an 18 h incubation with pronase11,12. Since longer incubations with digestive enzymes can decrease cell viability and alter the transcriptional profile of cells from digested tissues13, this could bias comparative analysis with other chemosensory epithelial populations.
Here, we report a method for the isolation of tracheal brush cells for RNA sequencing3. Treatment of the trachea with high-dose dispase separates the epithelium from the submucosa. Subsequent digestion of the epithelial sheet with papain allows for excellent recovery of this structural cell.
Protocol
Before conducting the following experiments, ensure that all animal care use and protocols are approved by the Institutional Animal Care and Use Committee (IACUC) and performed in accord with the National Research Council's "Guide for the Care and Use of Laboratory Animals" (8th Edition, 2011) and the ARRIVE guidelines. All procedures described below have been reviewed and approved by the Institutional Animal Care and Use Committee at the Brigham and Women's Hospital.
1. Preparation of Reagents
- Prepare Dispase digestion solution, which is a PBS solution containing 16 U/mL dispase and 20 µg/mL DNase I. Ensure that the dispase powder is fully dissolved before warming up the solution in a water bath at 37 °C.
- Add 5% heat-inactivated fetal bovine serum (FBS) to Dulbecco Modified Eagle Medium (DMEM) to make a stopping solution.
- Prepare Tyrode I buffer: add 26 U/mL papain (20 µL/mL of 48 U/mg papain solution) and 10 µL/mL L-cysteine to HEPES-Tyrode's buffer without calcium.
- Prepare Tyrode II buffer: add 2 μL/mL leupeptin (5 mg/mL) to HEPES-Tyrode's buffer with calcium.
- Prepare FACS buffer: use Hanks' Balanced Salt Solution (HBSS) without calcium, magnesium & phenol red, supplemented with 2 mM ethylenediaminetetraacetic acid (EDTA) and add 2% FBS.
2. Dissection of Mouse Trachea
NOTE: Mice used in this protocol are ChAT(BAC)-eGFP (B6.Cg-Tg(RP23- 268L19-EGFP)2Mik/J), 3-6 months of age of both sexes. Minimize the exposure of the tissue to direct light to reduce photobleaching of eGFP.
- Euthanize the mouse with 100 mg/kg pentobarbital euthanasia solution injected intraperitoneally or using standard protocols approved by the IACUC.
- Fix the mouse on a surgical board in the supine position with 21 G needles with extended upper and lower extremities. Spray the fur with 70% EtOH to sanitize the area.
- Using straight forceps, lift the skin and fur of the abdomen and make an incision in the center with dissecting scissors (straight scissors, 3 cm). Using the scissors, separate the skin from the subcutaneous tissue from the abdomen to the mandibula. While holding the subcutaneous tissue up with the forceps, make a small incision with the scissors in the center of the abdominal wall.
- Open the peritoneum with a V-shaped incision. Using the forceps, gently move the small intestine to the side, locate the abdominal aorta and vena cava and make an incision with the dissecting scissors to allow for rapid exsanguination.
- Locate the diaphragm. Using an 18 G needle, make an opening in the diaphragm just below the sternum to deflate the lungs. Carefully separate the diaphragm from the rib cage using sharp-pointed straight dissecting scissors to cut along the base of the ribs.
- Using the forceps, lift the exposed end of the sternum and cut the sternum longitudinally from the base of the rib cage to the neck. Make a central cervical incision with short straight scissors (2 cm) and separate the two lobes of the submandibular gland.
- Carefully remove the surrounding connective tissue and the thymus overlying the carina with a pair of fine point high precision forceps.
- Dissect the trachea free first by separating the proximal end at the level of the epiglottis and then by dissecting the distal end at the level of the bifurcation of the trachea.
- Locate the epiglottis and cut the trachea longitudinally from the epiglottis to the carina.
3. Tracheal Epithelial Digestion
- Place the trachea into a 1.5 mL tube containing 750 µL of pre-warmed (to 37 °C) dispase digestion solution. Incubate on a shaker at 200 rpm for 40 min at room temperature. Cover the tube with aluminum foil to reduce the exposure to direct light.
- Add 750 µL of cold DMEM with 5% FBS to stop the reaction. Place on ice.
- Transfer the trachea to a Petri dish (100 mm x 15 mm) and place under a dissecting microscope. Orient the trachea with the epithelial side facing up. The longitudinally dissected trachea has a semi-cylindrical shape maintained by the cartilaginous rings. The epithelium is on the concave surface.
- Tether the epiglottis area of the trachea with straight forceps to the Petri dish and using a size 22 disposable scalpel, scrape the epithelium off the trachea. The epithelial layer separates as a translucent sheet.
- Mince the epithelium with the scalpel. Transfer the epithelial layer to a 2 mL tube.
- Rinse the Petri dish with 750 µL of Tyrode I buffer and transfer to the 2 mL tube containing the epithelial layer.
- Incubate the tracheal epithelial layer in Tyrode I buffer for 30 min at 37 °C on a shaker at 200 rpm. Cover the tube with aluminum foil to reduce the exposure to direct light.
- Add 750 µL of cold Tyrode II buffer. Vortex the digested tissue vigorously for 20-30 s. Triturate the homogenate with a syringe attached to an 18 G needle 10 times. Switch to a 21 G gauge needle and triturate 10-20 more times.
- Filter the cells through a 100 µm strainer into a 50 mL conical tube. Add 30:1 vol/vol of cold FACS buffer.
- Spin at 350 x g for 10 min at 4 °C and discard the supernatant. Resuspend the pellet in cold FACS buffer and transfer the suspension to a 12 mm x 75 mm (5 mL) polystyrene tube. Spin again at 350 x g for 10 min at 4 °C and discard the supernatant.
- Re-suspend the pellet in 100 µL of FACS buffer.
- Add 1 µL of anti-mouse CD16/32 blocking antibody to block non-specific binding and incubate for 15 min on ice. Do not wash.
- Add the following antibodies and the respective isotype controls: pacific blue anti-mouse CD45 or rat IgG2a, k (0.25 µg/106 cells in 100 µL volume) and allophycocyanin (APC) anti-mouse EpCAM or rat IgG2b, k (0.5 µg/106 cells in 100 µL volume) monoclonal antibodies. Incubate for 45 min on ice protected from direct light. Add 4.5 mL of cold FACS buffer, mix and spin at 350 x g for 10 min at 4 °C. Discard the FACS buffer and re-suspend the pellet in 300 µL of cold FACS buffer.
- Add propidium iodide (PI) (5 µg/mL) immediately before flow cytometric sorting.
NOTE: Brush cells have an irregular shape with several processes that could potentially increase their adherence to filters (Figure 3C). We compared 30 μm to 70 μm and 100 μm filters and found that larger pore filters ensured better yields. In addition, thorough trituration of the cell suspension after papain digestion significantly improves the brush cell yields. We recommend using an 18G needle for initial trituration followed by a smaller bore (21 or 23 G needle) for finer dissociation of cells.
4. Flow Cytometry Gating Strategy
- Identify cells from debris by forward and side scatter angle. Exclude the doublets using forward scatter height and width and side scatter height and width. The doublets would be the cells that have high width values.
- Within the single cells, identify the live cells as the population that is PI negative.
- Within the live single cells, identify the CD45 low to negative cells based on the isotype control.
- Within the CD45 low/negative cells, identify the EpCAM positive cells that are also eGFP positive (in the FITC channel). This is the population of brush cells (Figure 2A).
Representative Results
This procedure has been successfully implemented to isolate tracheal brush cells for RNA sequencing3. After isolation of the trachea and digestion of the tissue with a 2-step protocol (Figure 1), cells were collected and stained with fluorescently-labeled CD45 and EpCAM after exclusion of dead cells with PI. After gating out doublets based on forward and side scatter characteristics, we defined brush cells as low/negative for CD45, positive for EpCAM and positive for eGFP (Figure 2A). Brush cells represented ~0.16-0.42% of CD45low/neg cells by flow cytometry and accounted for 250-600 cells per trachea (Figure 2B). We compared these counts to an estimate of the number of ChAT-eGFP positive cells in whole tracheal mounts from ChAT(BAC)-eGFP mice based on our published extensive immunohistological evaluation of tracheal brush cells3 and illustrated here (Figure 3). Our previous studies of tracheal brush cells in wild type, ChAT(BAC)-eGFP mice and Il25F25/F25 mice suggested that cholinergic brush cells are 90% overlapping with DCLK1+ and IL25+ cells and account for 600-1000 brush cells per trachea3. Notably, this number is lower than the number of cholinergic brush cells reported by other groups10. As chemosensory cell numbers are altered by exposure to microbial metabolites and protozoa5,14, these numbers might reflect a variability in interinstitutional microbiota. Therefore, we would suggest estimating the number of brush cells by fluorescence microscopy to gauge the expected number of isolated brush cells by flow cytometric sorting.
While establishing our protocol, we attempted several previously published methods of isolation of chemosensory cells (Figure 4). When we incubated minced trachea with 650 U/mL collagenase IV and 1.84 U/mL dispase supplemented with DNase I for 45 min, a combination frequently used to obtain single cell suspensions from lung homogenates15, we achieved a single cell suspension of epithelial cells, but we recovered only few brush cells (Figure 4A). Tuft cells in the intestine have been successfully isolated using 2.5 mM EDTA and 0.75 mM Dithiothreitol (DTT) with DNase I for 20 min to dislodge the epithelial cells followed by digestion of the released epithelial cells with 0.01 U/mL dispase with DNaseI for 10 min6. In our hands, using this procedure also led to suboptimal tracheal brush cell recovery (Figure 4B). A protocol for successful isolation of tracheal brush cells for RNA analysis was described by Krasteva et al.10. Following this protocol, we incubated minced trachea with 35 U/mL papain in Tyrode buffer for 45 min and did not achieve good brush cell recovery (Figure 4C). Rock et al. described a method of digestion of tracheal epithelium for basal cell isolation with two steps: separation of the epithelium from the mesenchymal layer with high dose dispase (16 U/mL) at room temperature followed by incubation of the stripped epithelium with 0.1 % trypsin and 1.6 mM EDTA for 30 min at 37 °C16. Following these steps led to a better recovery of brush cells, but the numbers achieved per mouse were insufficient for in depth functional analyses (Figure 4D). To optimize the protocol, we decided to combine the last two approaches. By separating the tracheal epithelium with high dose dispase, we aimed to allow for better access of papain to tracheal brush cells. Thus, using 16 U/mL of dispase at room temperature on whole trachea to separate the epithelium and subsequent incubation of the epithelium with 26 U/mL papain in Tyrode buffer led to the best brush cell recovery (Figure 4E).
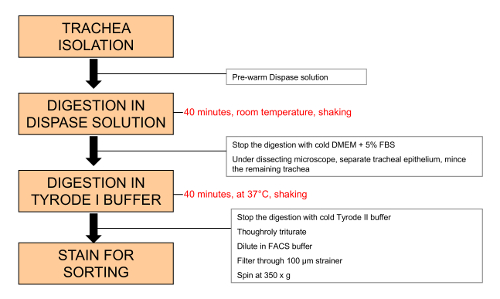
Figure 1: Schema of proposed steps for isolation of tracheal brush cells. The protocol includes 2 major steps: after dissection, the whole trachea is incubated in a high-dose dispase solution to separate the epithelium; this is followed by digestion of the epithelial sheet with papain in calcium containing Tyrode buffer (Tyrode I) and antibody staining. Please click here to view a larger version of this figure.
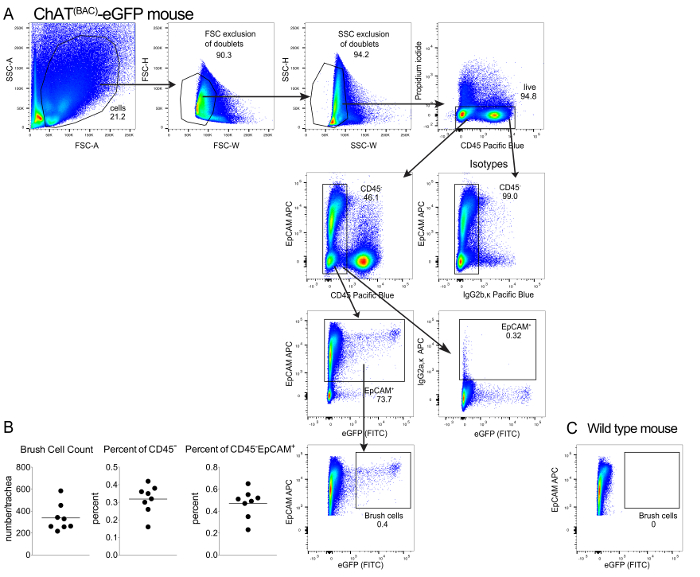
Figure 2: Representative flow cytometry analysis of tracheal brush cells. (A). Schematic representation of the flow cytometry gating strategy. Single cells were gated based on their forward and side scatter characteristics and live cells were chosen based on the exclusion of Propidium Iodide. CD45 low/negative cells were chosen based on the isotype control and evaluated for their expression of EpCAM. EpCAM positive cells were considered brush cells if they expressed eGFP fluorescent in the FITC channel. (B). Number of cholinergic tracheal brush cells per mouse trachea and frequency presented as percent of all CD45low/- cells and as a percent of CD45low/-EpCAM+ cells. Each dot represents a separate mouse. Data are from 3 separate experiments with 2-3 mice each. (C). Absence of eGFP positive brush cells in a tracheal epithelial digest from a wild type mouse stained for CD45 and EpCAM. Please click here to view a larger version of this figure.
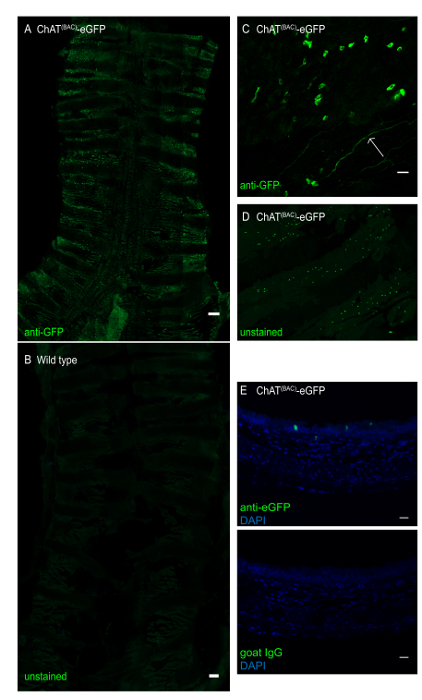
Figure 3: Whole mount of mouse trachea of a ChAT(BAC)-eGFP mouse. The trachea was opened longitudinally and stained with anti-GFP antibody to enhance the eGFP green fluorescence signal. (A) Whole tracheal mount of ChAT(BAC)-eGFP mouse, intensely green fluorescent cells represent brush cells; scale bar 1 mm; (B) whole tracheal mount of a wild type mouse demonstrating absence of brush cells; Scale bar = 1 mm; (C) magnification of the epithelial layer demonstrating irregularly shaped brush cells (green cells) as wells as a cholinergic nerve ending (arrow); (D) whole tracheal mount of a trachea of a ChAT(BAC)-eGFP imaged for GFP without antibody-enhancement of the fluorescence signal; (E) cross-section of a paraffin-embedded mouse trachea of a ChAT(BAC)-eGFP mouse stained with anti-GFP (green) or goat IgG control and DAPI (blue); Scale bars = 20 μm (C-E). Please click here to view a larger version of this figure.
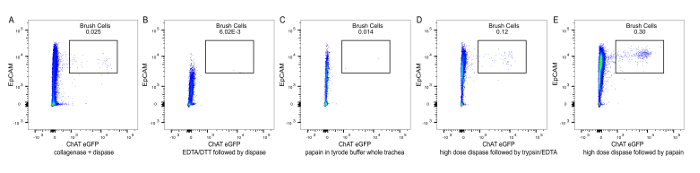
Figure 4: Comparison of different tracheal digestion protocols. (A) The trachea was minced and incubated with 650 U/mL collagenase IV and 1.84 U/mL dispase supplemented with DNase I for 45 min. This experiment was repeated 2 times. (B) The whole trachea was incubated with 2.5 mM EDTA and 0.75 mM DTT supplemented with DNase I for 20 min; the epithelial cells were dislodged and further digested with 0.01 U/mL dispase and DNase I for 10 min. This experiment was repeated 2 times. (C) The trachea was minced and incubated with 35 U/mL of papain in Tyrode I buffer for 45 min; residual papain activity was inhibited with leupeptin. This experiment was performed once. (D) The whole trachea was incubated with high dose dispase (16 U/mL) for 30 min to separate the epithelial sheet, followed by incubation of the epithelial sheet with 0.1% trypsin and 1.6 mM EDTA for 30 min. This experiment was repeated 2 times. (E). The whole trachea was incubated with high dose dispase (16 U/mL) for 30 min followed by digestion of the epithelial sheet with 26 U/mL papain in Tyrode buffer for 30 min. The residual papain activity was inhibited with leupeptin and high volume dilution. Please click here to view a larger version of this figure.
Discussion
We found that a combination of high-dose dispase treatment for 40 min followed by a short papain treatment (30 min) provides an optimal protocol for tracheal digestion and brush cell isolation. This combination both avoids extensive digestion and produces the highest yield of brush cells, compared to alternate protocols.
While lung digestion to extract hematopoietic cells has classically relied on mild digestive enzymes like collagenase IV15, isolation of epithelial cells requires more complex protocols. Single cell suspensions of epithelial cells are more difficult to achieve because of the tight and adherens junctions binding them to each other and the basement membrane and because of the frailty of the cells themselves. Most protocols for epithelial cell digestion involve two steps - a first step is the separation of the epithelium from the basement membrane followed by subsequent steps to disturb the tight junctions and desmosomes which adhere the epithelial cells to each other16,17.
Several groups have successfully isolated chemosensory tuft cells from the intestine using different modifications of a 2-step protocol for isolation of epithelial cells: the first step uses EDTA and DTT to release epithelial cells from the submucosa followed by digestion of the epithelial sheets with dispase. Howitt et al. used a mixture of 5 mM EDTA + 1 mM DTT followed by digestion with 0.5 units/mL Dispase II and DNase14. Gerbe et al. used high concentration 30 mM EDTA alone to separate the epithelial fraction and subsequently incubated with dispase supplemented with DNaseI18. Von Moltke et al. used 2.5 mM EDTA, 0.75 mM DTT and DNaseI followed by dispase digestion6. We found that these techniques allow for isolation of viable epithelial cells in the trachea but the percent of tracheal brush cells was extremely low (Figure 4B) and not consistent with our expectation of isolation of 600-800 brush cells per trachea based on our histology evaluation.
Tracheal epithelial cell isolation protocols have been developed by several groups to study basal epithelial cells. The major differences between these protocols and the ones employed in the gut are in the sequence of digestive enzymes. The first step is the separation of the tracheal epithelium through incubation with a high concentration of dispase (16 U/mL) at room temperature16. This allows for the epithelium to separate as a single sheet from the mesenchymal layer19. Subsequent processing of the tracheal epithelium uses a combination of trypsin and EDTA, one of the most frequently used enzymatic methods of cell detachment, to achieve a single cell suspension. Trypsin cleaves peptides on the C-terminal side of lysine and arginine amino acid residues, whereas EDTA is used as a calcium chelator for cell-cell and cell-matrix interactions20. Using this technique, Rock et al. have been successful in recovering basal epithelial cells for transcriptional profiling16. A similar protocol was recently used to isolate bitter taste receptor expressing cells from a Tas2r143-CreERT2 driven eGFP reporter and from B6.Il25Flare25/Flare25 for RNA sequencing studies5,8. In our experience, digestion of the epithelial sheet with trypsin and EDTA led to excellent epithelial cell recovery but insufficient numbers of cholinergic tracheal brush cells for functional or transcriptional analysis unless multiple mice were pooled for each sample (Figure 4C).
Papain is a potent endolytic plant cysteine protease derived from papaya latex, and it is known to cleave peptide bonds involving basic amino acids, particularly arginine, lysine and residues following phenylalanine21. Tracheal brush cell isolation using papain from ChAT(BAC)-eGFP mice was first reported by Krasteva et al10. Papain digestion was also recently used to dissociate airway epithelial cells for single cell RNA sequencing where brush/tuft cells comprised a small fraction of epithelial cells11. Papain is also the most commonly used enzyme for digestion of brain tissue for isolation of viable neurons22. Dissociation of brain tissue with papain for 30 min results in markedly higher yields, neuronal survival and integrity compared to trypsin digestion23. Since some brush cells are tightly intertwined with nerve endings3,10, the use of papain might be critical for the successful cleavage of brush cell junctions to neurons. However, we found that a 45 min digestion with papain at a concentration of 35-40 U/mL dramatically reduced epithelial cell viability, leading to low brush cell recovery (Figure 4D).
We used 3 methods to reduce papain toxicity and increase our cellular yields. First, by separating the epithelial sheet with dispase, we reduced the subsequent incubation time of papain from 45 to 30 min. Second, we immediately inhibited the papain activity with leupeptin, an inhibitor of cysteine proteases24. Notably, leupeptin is highly unstable at 4 °C and should be aliquoted and frozen at -20 °C. Third, we found that dilution of the enzymatically digested tissue in a large volume of cold PBS also helps to preserve the viability of the digested cells. Another step that is critical to increasing the cell yields is rigorous trituration10. Since prolonged vortexing could also decrease viability, we partially substituted it with trituration. We recommend trituration with an 18G needle to dissociate larger cellular clumps followed by additional trituration with a 21 G needle. Finally, we found that eGFP is highly light sensitive and protection from direct light exposure at any step is helpful. These modifications allowed for robust recovery of viable tracheal brush cells (Figure 4E).
The major limitations of the protocol proposed here are the 2-step procedure and the use of papain, a potent cysteine protease. Some groups have recently substituted liberase for dispase5 when isolating chemosensory cells. Furthermore, liberase digestion was recently used to isolate chemosensory (brush) epithelial cells from nasal polyps25,26. We have not directly compared liberase digestion to our two-step dispase/papain digestion protocol. Papain is a potent cysteine protease that can activate the protease activated receptor PAR2 on epithelial cells27 leading to activation of brush cells themselves or to the generation of mediators by other respiratory cells that could in turn activate the brush cells. Finally, papain can modulate receptor expression through the cleavage of surface receptors making it difficult to validate brush cell receptor expression by flow cytometry28,29.
In summary, we provide a protocol of isolation of tracheal cholinergic brush cells from ChAT-eGFP reporter mice that allows for robust recovery of viable tracheal brush cells. This protocol has been successfully used for isolation and transcriptional analysis of brush cells by RNA sequencing3.
開示
The authors have nothing to disclose.
Acknowledgements
We thank Adam Chicoine at the Brigham and Women’s Human Immunology Center Flow Core for his help with flow cytometric sorting. This work was supported by National Institutes of Health Grants R01 HL120952 (N.A.B.), R01 AI134989 (N.A.B), U19 AI095219 (N.A.B., L.G.B), and K08 AI132723 (L.G.B), and by the American Academy of Allergy, Asthma, and Immunology (AAAAI)/ American Lung Allergic Respiratory Disease Award (N.A.B.), by the AAAAI Foundation Faculty Development Award (L.G.B.), by the Steven and Judy Kaye Young Innovators Award (N.A.B.), by the Joycelyn C. Austen Fund for Career Development of Women Physician Scientists (L.G.B.), and by a generous donation by the Vinik family (L.G.B.).
Materials
| Antibodies | |||
| Anti-GFP (Polyclonal goat Ig) | Abcam | cat# ab5450 | |
| APC anti-mouse CD326 (EpCAM) (G8.8) | Biolegend | cat#118214 | |
| APC Rat IgG2a, k isotype control | Biolegend | cat#400511 | |
| DAPI | Biolegend | cat#422801 | |
| Donkey anti-goat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Life Technologies/Molecular Probes | cat#A-11055 | |
| Normal Goat IgG | R&D Systems | cat#AB-108-C | |
| Pacific Blue anti-mouse CD45 (30F-11) | Biolegend | cat#103126 | |
| Pacific Blue Rat IgG2b, k isotype control | Biolegend | cat#400627 | |
| TruStain FcX (anti-mouse CD16/32) Antibody | Biolegend | cat#101320 | |
| Chemicals, Peptides, and Recombinant Proteins | |||
| Dispase | Gibco | cat# 17105041 | |
| DNase I | Sigma | cat# 10104159001 | |
| HEPES-Tyrode’s Buffer Without Calcium (10 mM HEPES, 135 mM NaCl, 2.8 mM KCl, 1 mM MgCl2, 12 mM NaHCO3, 0.4 mM NaH2PO4, 0.25% BSA, 5.5 mM Glucose. Prepared in 18.2 megohms water and filtered through 0.22 µm filter | Boston BioProducts | cat# PY-912 | |
| Tyrode’s Solution (HEPES-Buffered) 140 mM NaCl, 5 mM KCl, 25 mM HEPES, 2 mM CaCl2, 2 mM MgCl2 and 10 mM glucose. Prepared in 18.2 megohms water and filtered through 0.22 µm filter. ) | Boston BioProducts | cat# BSS-355 | |
| L-Cysteine | Sigma | cat# C7352 | |
| Leupeptin trifluoroacetate salt | Sigma | cat# L2023 | |
| Papain from papaya latex | Sigma | cat# P3125 | |
| Propidium iodide | Sigma | cat# P4170 | |
| Experimental Models: Organisms/Strains | |||
| ChATBAC-eGFP (B6.Cg-Tg(RP23-268L19-EGFP)2Mik/J) | The Jackson Laboratory | 7902 | |
| Equipment | |||
| LSM 800 with Airyscan confocal system on a Zeiss Axio Observer Z1 Inverted Microscope | Zeiss | ||
| LSRFortessa | BD | 647465 | |
| Disposable equipment | |||
| 1.5 mL sterile tubes | Thomas Scientific | 1157C86 | |
| 5 mL Poysterene Round-bottom Tube, 12 x 75 mm style | Falcon | 14-959-1A | |
| 50 mL Polypropylene conical tube, 30 x 115 mm style | Falcon | 352098 | |
| Feather Disposable Scalpel no.12 | Fisher Scientific | NC9999403 | |
| Petri dish, 100 x 15 mm Style | Falcon | 351029 | |
| Sterile cell strainer, 100 μm | Fisherbrand | cat#22363549 |
参考文献
- Tizzano, M., et al. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proceedings of the National Academy of Sciences of the United States of America. 107 (7), 3210-3215 (2010).
- Genovese, F., Tizzano, M. Microvillous cells in the olfactory epithelium express elements of the solitary chemosensory cell transduction signaling cascade. PLoS One. 13 (9), 0202754 (2018).
- Bankova, L. G., et al. The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Science Immunology. 3 (28), (2018).
- Krasteva, G., Canning, B. J., Papadakis, T., Kummer, W. Cholinergic brush cells in the trachea mediate respiratory responses to quorum sensing molecules. Life Sciences. 91 (21-22), 992-996 (2012).
- Nadjsombati, M. S., et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity. 49 (1), 33-41 (2018).
- von Moltke, J., Ji, M., Liang, H. E., Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 529 (7585), 221-225 (2016).
- Deckmann, K., et al. Bitter triggers acetylcholine release from polymodal urethral chemosensory cells and bladder reflexes. Proceedings of the National Academy of Sciences of the United States of America. 111 (22), 8287-8292 (2014).
- Liu, S., et al. Members of Bitter Taste Receptor Cluster Tas2r143/Tas2r135/Tas2r126 Are Expressed in the Epithelium of Murine Airways and Other Non-gustatory Tissues. Frontiers in Physiology. 8, 849 (2017).
- Krasteva, G., et al. Cholinergic chemosensory cells in the auditory tube. Histochemistry and Cell Biology. 137 (4), 483-497 (2012).
- Krasteva, G., et al. Cholinergic chemosensory cells in the trachea regulate breathing. Proceedings of the National Academy of Sciences of the United States of America. 108 (23), 9478-9483 (2011).
- Montoro, D. T., et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 560 (7718), 319-324 (2018).
- Plasschaert, L. W., et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 560 (7718), 377-381 (2018).
- Dwyer, D. F., Barrett, N. A., Austen, K. F. Immunological Genome Project, C. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nature Immunology. 17 (7), 878-887 (2016).
- Howitt, M. R., et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 351 (6279), 1329-1333 (2016).
- Bankova, L. G., Dwyer, D. F., Liu, A. Y., Austen, K. F., Gurish, M. F. Maturation of mast cell progenitors to mucosal mast cells during allergic pulmonary inflammation in mice. Mucosal Immunology. 8 (3), 596-606 (2015).
- Rock, J. R., et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 106 (31), 12771-12775 (2009).
- Rock, J. R., et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proceedings of the National Academy of Sciences of the United States of America. 108 (52), 1475-1483 (2011).
- Gerbe, F., et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 529 (7585), 226-230 (2016).
- Rock, J. R., et al. Transmembrane protein 16A (TMEM16A) is a Ca2+-regulated Cl- secretory channel in mouse airways. Journal of Biological Chemistry. 284 (22), 14875-14880 (2009).
- Olsen, J. V., Ong, S. E., Mann, M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Molecular and Cellular Proteomics. 3 (6), 608-614 (2004).
- Verma, S., Dixit, R., Pandey, K. C. Cysteine Proteases: Modes of Activation and Future Prospects as Pharmacological Targets. Frontiers in Pharmacology. 7, 107 (2016).
- Huettner, J. E., Baughman, R. W. Primary culture of identified neurons from the visual cortex of postnatal rats. Journal of Neuroscience. 6 (10), 3044-3060 (1986).
- Kaiser, O., et al. Dissociated neurons and glial cells derived from rat inferior colliculi after digestion with papain. PLoS One. 8 (12), 80490 (2013).
- Aoyagi, T., Takeuchi, T., Matsuzaki, A., Kawamura, K., Kondo, S. Leupeptins, new protease inhibitors from Actinomycetes. Journal of Antibiotics (Tokyo). 22 (6), 283-286 (1969).
- Kohanski, M. A., et al. Solitary chemosensory cells are a primary epithelial source of IL-25 in patients with chronic rhinosinusitis with nasal polyps. The Journal of Allergy and Clinical Immunology. 142 (2), 460-469 (2018).
- Patel, N. N., et al. Solitary chemosensory cells producing interleukin-25 and group-2 innate lymphoid cells are enriched in chronic rhinosinusitis with nasal polyps. International Forum of Allergy & Rhinology. , (2018).
- Kouzaki, H., O’Grady, S. M., Lawrence, C. B., Kita, H. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. The Journal of Immunology. 183 (2), 1427-1434 (2009).
- Cambier, J. C., Vitetta, E. S., Kettman, J. R., Wetzel, G. M., Uhr, J. W. B-cell tolerance. III. Effect of papain-mediated cleavage of cell surface IgD on tolerance susceptibility of murine B cells. The Journal of Experimental Medicine. 146 (1), 107-117 (1977).
- Nishikado, H., et al. Cysteine protease antigens cleave CD123, the alpha subunit of murine IL-3 receptor, on basophils and suppress IL-3-mediated basophil expansion. Biochemical and Biophysical Research Communications. 460 (2), 261-266 (2015).

