Determining the Likelihood of Variant Pathogenicity Using Amino Acid-level Signal-to-Noise Analysis of Genetic Variation
概要
Amino acid-level signal-to-noise analysis determines the prevalence of genetic variation at a given amino acid position normalized to background genetic variation of a given population. This allows for identification of variant "hotspots" within a protein sequence (signal) that rises above the frequency of rare variants found in a population (noise).
Abstract
Advancements in the cost and speed of next generation genetic sequencing have generated an explosion of clinical whole exome and whole genome testing. While this has led to increased identification of likely pathogenic mutations associated with genetic syndromes, it has also dramatically increased the number of incidentally found genetic variants of unknown significance (VUS). Determining the clinical significance of these variants is a major challenge for both scientists and clinicians. An approach to assist in determining the likelihood of pathogenicity is signal-to-noise analysis at the protein sequence level. This protocol describes a method for amino acid-level signal-to-noise analysis that leverages variant frequency at each amino acid position of the protein with known protein topology to identify areas of the primary sequence with elevated likelihood of pathologic variation (relative to population "background" variation). This method can identify amino acid residue location "hotspots" of high pathologic signal, which can be used to refine the diagnostic weight of VUSs such as those identified by next generation genetic testing.
Introduction
The rapid improvement in genetic sequencing platforms has revolutionized the accessibility and role of genetics in medicine. Once confined to a single gene, or a handful of genes, the reduction in cost and increase in speed of next generation genetic sequencing has led routine sequencing of the entirety of the genome's coding sequence (whole exome sequencing, WES) and the entire genome (whole genome sequencing, WGS) in the clinical setting. WES and WGS have been used frequently in the setting of critically ill neonates and children with concern for genetic syndrome where it is a proven diagnostic tool that can change clinical management1,2. While this has led to increased identification of likely pathogenic mutations associated with genetic syndromes, it has also dramatically increased the number of incidentally found genetic variants, or unexpected positive results, of unknown diagnostic significance (VUS). While some of these variants are disregarded and not reported, variants localizing to genes associated with potentially fatal or highly morbid diseases are often reported. Current guidelines recommend reporting of incidental variants found in specific genes which may be of medical benefit to the patient, including genes associated with the development of sudden cardiac death-predisposing diseases such as cardiomyopathies and channelopathies3. Although this recommendation was designed to capture individuals at risk of a SCD-predisposing disease, the sensitivity of variant detection far exceeds specificity. This is reflected in a growing number of VUSs and incidentally identified variants with unclear diagnostic utility that far exceed the frequency of the respective diseases in a given population4. One such disease, long QT syndrome (LQTS), is a canonical cardiac channelopathy caused by mutations localizing to genes which encode cardiac ion channels, or channel interacting proteins, resulting in delayed cardiac repolarization5. This delayed repolarization, seen by a prolonged QT interval on resting electrocardiogram, results in an electrical predisposition to potentially fatal ventricular arrhythmias such as torsades de pointes. While a number of genes have been linked to the development of this disease, mutations in KCNQ1-encoded IKs potassium channel (KCNQ1, Kv7.1) is the cause of LQTS type 1 and is utilized as an example below6. Illustrating the complexity in variant interpretation, the presence of rare variants in LQTS-associated genes, so called "background genetic variation" has been previously described7,8.
In addition to large compendium-style databases of known pathogenic variants, several strategies exist for predicting the effect different variants will produce. Some are based on algorithms, such as SIFT and Polyphen 2, which can filter large numbers of novel non-synonymous variants to predict deleteriousness9,10. Despite broad use of these tools, low specificity limits their applicability when it comes to "calling" clinical VUSs11. "Signal-to-noise" analysis is a tool which identifies the likelihood of a variant being associated with disease based on the frequency of known pathologic variation at the loci in question normalized against rare genetic variation from a population. Variants localizing to genetic loci where there is a high prevalence of disease-associated mutations compared to population-based variation, a high signal-to-noise, are more likely to be disease-associated themselves. Further, rare variants found incidentally localizing to a gene with a high frequency of rare population variants compared to disease-associated frequency, a low signal-to-noise, may be less likely to be disease-associated. The diagnostic utility of signal-to-noise analysis has been illustrated in the latest guidelines for genetic testing for cardiomyopathies and channelopathies; however, it has only been employed at the whole gene level or domain-specific level12. Recently, given increased availability of both pathologic variants (disease databases, cohort studies in the literature) and population-based control variants (Exome Aggregation Consortium, ExAC and the Genome Aggregation Database, GnomAD13), this has been applied to the individual amino acid positions within the primary sequence of a protein. Amino acid-level signal-to-noise analysis has proven useful in categorizing incidentally identified variants in genes associated with LQTS as likely "background" genetic variation rather than disease-associated. Among the three major genes associated with LQTS, including KCNQ1, these incidentally identified variants lacked a significant signal-to-noise ratios, suggesting that the frequency of these variants at individual amino acid positions reflect rare population variation rather than disease-associated mutations. Furthermore, when protein-specific domain topology was overlaid against areas of high signal-to-noise, pathologic mutation "hotspots" localized to key functional domains of the proteins14. This methodology holds promise in determining 1) the likelihood a variant is disease- or population-associated and 2) identifying novel critical functional domains of a protein associated with human disease.
Protocol
1. Identify the Gene and Specific Splice Isoform of Interest
NOTE: Here, we demonstrate the use of Ensembl15 to identify the consensus sequence for the gene of interest which is associated with the pathogenesis of the disease of interest (i.e. KCNQ1 mutations are associated with LQTS). Alternatives to Ensembl include RefSeq via the National Center for Biotechnology Information (NCBI)16 and the University of California, Santa Cruz (UCSC) Human Genome Browser17 (see Table of Materials).
- In the Ensembl homepage, select the species (i.e. human) in the dropdown menu, and enter the gene of interest acronym in the field (i.e. KCNQ1). Click "Go"
- Select the link corresponding to the gene of interest (i.e. "KCNQ1 (Human Gene)"
- Select the link corresponding to the transcript of interest ID of interest from the "Transcript table" (i.e. TranscriptID ENST00000155840.10, NM_000218 [RNA transcript], NP_000209 [protein product of RNA transcript]).
NOTE: Review of the pertinent literature is needed to ensure the correct transcript consensus sequence is selected. - Note the transcript-specific NM and NP identification numbers for future reference found in the "RefSeq" column of the "Transcript Table".
- Select the link associated with the NP ID number to open a new webpage from the NCBI Protein database.
- Scroll down to the "Origin" section to obtain the protein (primary) sequence for the gene transcript of interest.
- Scroll up to the "Features" section to obtain a list of the protein features (functional domains, binding domains, post-translational modification sites).
NOTE: This information can also be obtained via the NCBI Protein database or from primary sources in the literature. This will be further discussed in step 5.
2. Create the Experimental Genetic Variant Database (the "Signal")
NOTE: Here, we demonstrate how to create a database of disease-associated variants in the gene of interest with the frequency of the disease-associated variants among individuals with the disease of interest. This database can take many forms and represents the "signal" (phenotype-positive genetic variation) which will be normalized against the control variant database. This can include 1) disease-associated variants for comparison against VUSs to identify novel functional domains of the protein and/or 2) VUSs, including incidentally identified VUSs, to compare against disease-associated variants to determine likelihood of pathogenicity. Disease-associated variants in KCNQ1 will be presented for illustration; however, the method is the same for analysis of incidentally-identified VUSs or any other set of experimental variants.
- Identify cohort(s) of unrelated index cases/probands with the disease of interest for which the gene of interest was comprehensively genotyped for all probands (i.e. a study identifies 24 unrelated probands hosting variants in KCNQ1 out of 200 individuals with LQTS who were subjected to KCNQ1 genetic interrogation).
NOTE: These cohorts can be identified from the literature, from experimental genetic analysis, or a combination of both.- Exclude studies which are not cohort-based (i.e. a case report describing a single mutation-positive individual), do not provide the total number of individuals genotyped for the gene of interest, or do not comprehensively genetically analyze the gene (i.e. a "targeted" genetic screening of only KCNQ1 exons 2-4) These preclude calculation of the frequency of a variant.
- Include individuals who are unrelated probands and exclude related individuals as this can over-estimate variant frequencies (i.e. a study identifies 4 unrelated individuals with KCNQ1 mutations in a cohort of 20 patients with LQTS. One of these probands is part of a family with 5 other mutation-positive kindred. Exclude all family members and include only the 4 unrelated probands).
- Compile all experimental genetic variants found in identified cohort(s)
- Assign nomenclature that contains the wild-type amino acid, amino acid position, and variant amino acid (i.e. alanine at amino acid number 212 changed to valine, Ala212Val or A212V). One such type of nomenclature is demonstrated in Figure 1.
- Confirm that variant nomenclature of all experimental genetic variants is based on the same reference gene transcript as noted in step 1.4. If experimental genetic variants are not annotated on the same reference gene transcript, then reannotate variant position to a reference transcript using transcript alignment (see step 1.2)
- Exclude variants that are not applicable depending on the question being explored.
- Exclude variants localizing to non-coding regions of the genome or variants which do not alter the protein sequence such as synonymous, intronic variants, 5' or 3' untranslated region [UTR], and intergenic region variants (i.e. a reported pathologic variant in KCNQ1 which localizes to the 5' UTR of the coding region would be excluded as it is not predicted to alter the protein sequence).
- Exclude variants that do not meet inclusion criteria for the study. For disease-associated variants, this includes variants that are no longer deemed pathologic.
- Confirm that each variant is currently considered pathogenic, likely pathogenic, or at least not benign, by cross-referencing variants with the ClinVar database (see Table of Materials).
- Enter the gene and variant of interest into ClinVar search field (i.e. KCNQ1-Y111C), select "Search"
- Identify the variant of interest under the "Variation/Location" column.
- Note the consensus interpretation of pathogenicity under "Clinical Significance" column (i.e. KCNQ1-Y111C is interpreted as "pathogenic").
- Include variants which are "likely pathogenic" or "pathogenic."
- Include variants with designations of "conflicting interpretations of pathogenicity," "uncertain significance," or when no record is available ("not provided") if warranted by the study.
- Exclude variants designated as "likely benign" (i.e. KCNQ1-A62T).
- Calculate the minor allele frequency (MAF) of each experimental variant position.
- Calculate how any alleles were positive for each respective variant (i.e. if a KCNQ1-Y111C heterozygous mutation is found in 2 unrelated individuals, the number of variant-positive alleles is 2).
- Calculate the total number of alleles sequenced within the cohort
- Note the total number of individuals sequenced in each cohort study (step 2.1)
- Multiply the total number of individuals by 2 to determine the total number of alleles.
NOTE: This presumes diploid genomes whereby each individual hosts 2 of each allele.
- Calculate the total number of variant-positive individuals for each amino acid position (alleles in step 2.4.1/alleles in step 2.4.2). For example, if 2 unrelated individuals each host heterozygous KCNQ1-Y111C mutations in cohorts of 100 and 200 LQTS-afflicted individuals, respectively, then the frequency of experimental variants at amino acid position 111 is 2 variants/((100+200 individuals)*2 alleles/individual) (i.e. combined MAF 0.0033).
- Calculate this value for each variant as the respective MAF of each experimental variant. For additional details see step 4.2.
3. Create the Control Genetic Variant Database (the "Noise")
NOTE: Here, we demonstrate how to create a database of control variants in the gene of interest with an associated frequency in a control population. This database represents the "noise" (phenotype-negative, population-based genetic variation) which is the background against which the experimental variant database will be normalized. This is referred to as "control" variation.
- Identify a cohort(s) of healthy, unrelated probands or utilize large population-based studies to identify rare variants among a given population.
NOTE: Sources for this database are diverse and include: 1) healthy individuals and/or otherwise phenotype-negative individuals subjected to Sanger sequencing, or publicly held databases of population-based individuals for which the disease in question is rare in frequency such as 2) 1000 Genome Project (N = 1,094 subjects)18, 3) National Heart, Lung, and Blood Institute GO Exome Sequencing Project (ESP, N = 5,379 subjects)19, 4) Exome Aggregation Consortium (ExAC, N = 60,706 subjects)13, and/or 5) Genome Aggregation Database (GnomAD, N = 138,632 individuals)13 (see Table of Materials). The GnomAD database will be utilized as an illustrative example.- Enter the gene of interest in the search box on the GnomAD homepage (i.e. KCNQ1).
- Confirm that the browser selected the correct gene and transcript of interest (step 1.4).
- Confirm that there is appropriate coverage of sequencing of the locus by reviewing "mean coverage" and "coverage plot."
- Select for coding sequence genetic variation by selecting "Missense + LoF."
- Select "Export table to CSV," which will generate a TextEdit file named "Unknown."
- Relabel the file and include a new extension "*.csv" (i.e. "KCNQ1 Control Variation.csv").
- Open the file using an appropriate software program for analysis of *.csv files (see Table of Materials).
- Identify the protein changing genetic variation in the column labeled "Protein Consequence."
- Apply same exclusion criteria to these control genetic variants as the experimental genetic variants (step 2.3.1).
- Identify the MAF of each control variant.
- Locate the "Allele Count" column, which denotes the number of alleles found to harbor the variant.
- Locate the "Allele Number" column, which denotes the total number of alleles sequenced at this given amino acid position.
NOTE: The total number of alleles sequenced will vary depending on coverage at that location. Areas of high coverage will approach 2*total number of individuals within GnomAD (i.e. for 138,632 individuals, complete coverage encompasses 277,264 total alleles genotyped). Conversely, areas of lower coverage will have a reduced total allele number - Locate the variant MAF which is pre-calculated in the "Allele Frequency" column and represents "Allele Count" divided by "Allele Number."
NOTE: Human genomes have two of each allele (i.e. 1 subject found to have a heterozygous variant in 10 people has a MAF of 1/20) - Note the MAF for each variant as the respective MAF of each control variant.
NOTE: Variant specific MAF for each racial/ethnic group comprising GnomAD can be seen in the columns to the right of "Allele Frequency."
- Apply a MAF threshold for rare variants above which control variants are excluded as "common."
- Set the MAF threshold to the maximal value at which all truly disease-associated variants (see step 2) also observed in the control database are included below the threshold (i.e., among all disease-associated KCNQ1 variants also found in GnomAD the highest common variant MAF is 0.009, then all GnomAD variants above a threshold of 0.01 should be excluded).
- Ensure that the experimental variant nomenclature is identical to control (see step 2.2).
- Save the file. In some cases, this may require changing the file type/extension.
4. Amino Acid Level Signal-to-noise Calculation and Mapping
- Calculate a MAF for each amino acid position with a control variant (see Figure 1 containing example KCNQ1 GnomAD variants).
- In a graphing-capable spreadsheet, create a column of the positions of all experimental variants.
- Remove variant text to leave only the variant position.
NOTE: Various functions/formulas can be utilized to automatically delete these text elements within cells (Figure 1, column C; see Table of Materials). - Sort the variants in ascending value to identify which positions have more than 1 variant associated with it (Figure 1, column E; i.e. amino acid position 10 is listed twice in column E which denotes 2 unique variants at the position).
- Combine the MAF for each variant associated with a given position by taking the sum of all MAFs for a given position (Figure 1, column G and H).
- Calculate a MAF for each amino acid position with an experimental variant (see Figure 2 containing mock KCNQ1 pathologic variants).
- In a similar fashion to 4.1.1, create a column of amino acid positions which have experimental variants (Figure 2, column B).
- For each variant position, calculate the MAF of all variants associated with that position from step 2.4 (Figure 2, column C-G).
- Create a rolling average of MAF for both experimental and control variants.
- Expand the columns created in 4.1 and 4.2 to include cells for amino acid positions that have no variant as a MAF = 0. (Figure 3).
- Create a column containing all amino acid positions in the gene of interest (i.e. 1 to 676 for KCNQ1, Figure 3, column C and I).
- Add a MAF of 0 for all positions that do not have variants for both control and experimental data sets.
NOTE: This can be done automatically by utilizing the "VLOOKUP" function in a commonly utilized software program (Figure 3, column D and J, see Table of Materials).
- Create a rolling average for each experimental and control prevalence column.
NOTE: This allows for inference of adjacent position pathogenicity and can be modified, or even excluded, to fit the needs of the study.- Create a column representing a rolling average of the MAF for both the for both control and experimental data sets (Figure 3, column E and K).
- In the rolling average column, place the average of the respective MAF for the 5 variant positions N-terminal and 5 variant positions C-terminal to the given position.
NOTE: This creates a rolling average of +/- 5. For positions with less than 5 amino acid residues preceding, or following, a rolling average location (i.e. the N- or C-terminus), the rolling average will only take into account those residues that are present (i.e. the rolling average at amino acid position 3 will be an average of the MAF at amino acid positions 1 though 8, calculated as the sum of these MAFs divided by 8).
- Expand the columns created in 4.1 and 4.2 to include cells for amino acid positions that have no variant as a MAF = 0. (Figure 3).
- Calculate the minimum control frequency by dividing the lowest rolling MAF by 2.
- Change any cell with a control MAF of 0 to the minimum frequency to avoid dividing by 0 when calculating a signal-to-noise ratio.
- Calculate the amino acid level signal-to-noise ratio (Figure 4).
- Divide each amino acid position experimental rolling average by the respective control rolling average.
- Graph this ratio (Y-axis) vs. amino acid position (X-axis).
5. Protein Domain Topology Overlay
- Identify the consensus amino acid locations of functional domains/features, or areas of post-translational modification, of the protein of interest (step 1.7).
NOTE: A number of resources can be utilized to identify these domains. These resources, as well as resources for identifying putative domains in novel proteins, have been well reviewed in the literature20. This protocol will describe the protein database available through NCBI, which is widely utilized and robust (see Table of Materials). - Identify amino acid positions associated with protein domains/features.
- Open the NCBI webpage.
- Enter the NP of the protein of interest into the search field.
- Identify known protein domains and features are catalogues under "Features."
- Identify and note the domain name/type and amino acid positions.
- Select the link corresponding to the feature to visualize the region on the protein of interest primary sequence.
- Create a column containing the boundaries of the domains/features.
- Create a column next to the signal:noise column so that the amino acid position column can be referenced (Figure 5A, column C).
- Identify the cells corresponding at the N-terminal or C-terminal aspect of each domain/feature and place a 1 in each cell (i.e. if the N-terminal domain of the S1 transmembrane domain of KCNQ1 is amino acid position 122, and the C-terminal domain is position 142, then a 1 is placed in the row for amino acid position 122 and 142).
- For overlapping domains/features, display multiple domains by changing the 1 to other values (i.e. 1.5, 2, 2.5); this can assist in distinguishing domains.
- Create a graph with these boundaries as a Y-axis and amino acid position on the X-axis (Figure 5B).
- Overlay this graph with the signal-to-noise graph created in step 4.4.
- Identify correlations between known protein domains/features and the signal-to-noise analysis.
6. Variant Position Overlay
- Map individual variant positions for overlay of graphs produced in steps 4.4 and 5.4.
- Create a column next to the domain/feature column such that rows in the column will correspond to amino acid positions (Figure 5A, column D).
- Place a 1 in each cell in the added row corresponding to a position containing a respective variant.
- Create a graph with this column as a Y-axis and amino acid position on the X-axis (Figure 5C).
- Overlay this graph with the signal-to-noise graph created in step 4.4 and domain graph created in step 5.4.
Representative Results
A representative result for amino acid-level signal to noise analysis for KCNQ1 is depicted in Figure 6. In this example, rare variants identified in the GnomAD cohort (control cohort), incidentally-identified WES variants (experimental cohort #1), and LQTS case-associated variants deemed likely disease-associated (experimental cohort #2) are depicted. Further, the signal-to-noise analysis comparing the WES and LQTS cohort variant frequency normalized against GnomAD variant frequency is depicted. LQTS-associated variants demonstrated high signal-to-noise ratios in domains corresponding with the channel pore, selectivity filter, and the KCNE1-binding domain. In comparison, incidentally identified variants in the WES cohort did not clearly demonstrate specific regions of high signal-to-noise elevation, suggesting that these variants reflect background genetic variation. This example did not utilize variant MAFs as noted above; however, it demonstrates all of the same principles as described.
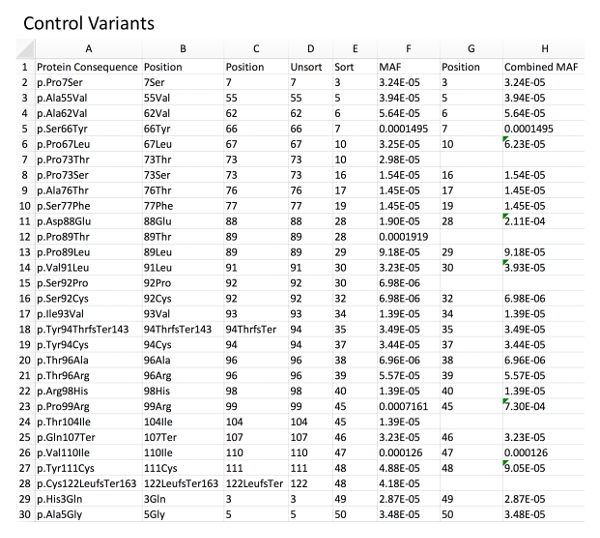
Figure 1: Example of control variant database with MAF calculation. Column A, directly imported GnomAD control rare variants. Column B, deletion of left-sided, non-position-related text from the variant nomenclature using an example formula for character removal (i.e.: for B2 "=RIGHT(A2,LEN(A2)-5", see Table of Materials). Column C, deletion of right-sided, non-position-related text from the variant nomenclature using a related formula (i.e.: for C2 "=LEFT(B2,LEN(B2)-3"). Column D, resultant unsorted amino acid positions. Column E, amino acid positions sorted in an ascending fashion to allow for identification of duplicate positions. Column F, associated MAF for each variant as imported from GnomAD. Column G and H, combined MAF for a given amino acid position (sum of each variant MAF at a specific position). Please click here to view a larger version of this figure.
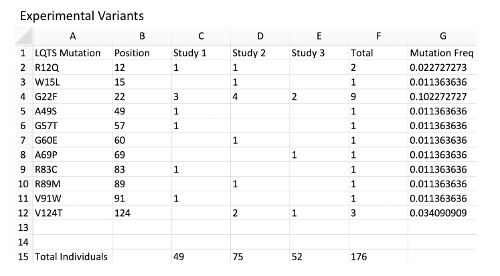
Figure 2: Example of experimental variant database with MAF calculation. Column A, a list of mock LQTS-associated mutations in KCNQ1 representing a disease-associated mutation experimental database. Column B, mutation position corresponding to each variant. Column C, a count of mutation-positive individuals within mock Study 1. Each are presumed to be heterozygous mutation carriers. The total number of individuals genotyped in the study is located at the bottom of the sheet. Column D, count of mutation-positive individual in mock Study 2. Column E, count of mutation-positive individual in mock Study 3. Column F, total mutation-positive individuals hosting the observed mutation across all studies. Note that distinct mutations associated with the same amino acid position should be combined. Column G, MAF of each mutation and amino acid position using an example formula (i.e.: for G2 "=2/(176*2) ", see Table of Materials). Note that since all individuals are presumed to be heterozygous and each individual presumed to carry 2 alleles of the KCNQ1 locus, the total individuals should be multiplied by 2 for the allele frequency. Please click here to view a larger version of this figure.
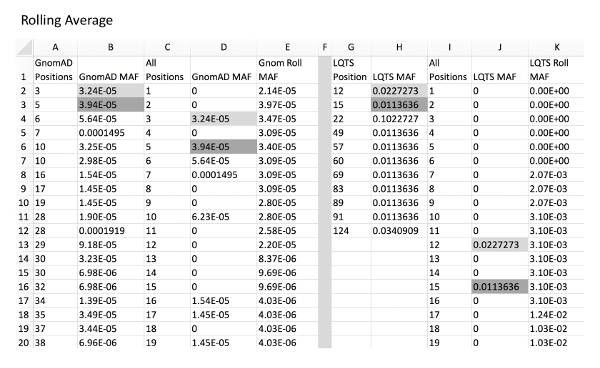
Figure 3: Example of rolling average calculation for control and experimental variants. Column A and B, GnomAD control variant positions and respective MAFs. Column C, all amino acid positions of KCNQ1 from amino acid position to final. Column D, GnomAD variant MAF for all positions with a MAF of 0 in place of positions without a variant. This can be automatically calculated using a VLOOKUP function (i.e. for D2, "=IFERROR(VLOOKUP(C2,A:B,2,),0), see Table of Materials). Column E, rolling average of position MAF using an example formula (i.e. for E2, "=SUM(D2:D7)/6" and for E7, "=SUM(D2:D12)/11"). Column G and H, LQTS experimental variant positions with respective MAFs. Column I, all amino acid positions of KCNQ1. Column J, LQTS variant MAF for all positions. Column K, rolling LQTS MAF. Gray fill cells are examples of where MAF values from columns B and H are expanded into column D and J, respectively, which correlate with respective positions in column C/I. Note that it is critical that all cells are formatted as "Numbers" for proper formula functioning. Please click here to view a larger version of this figure.
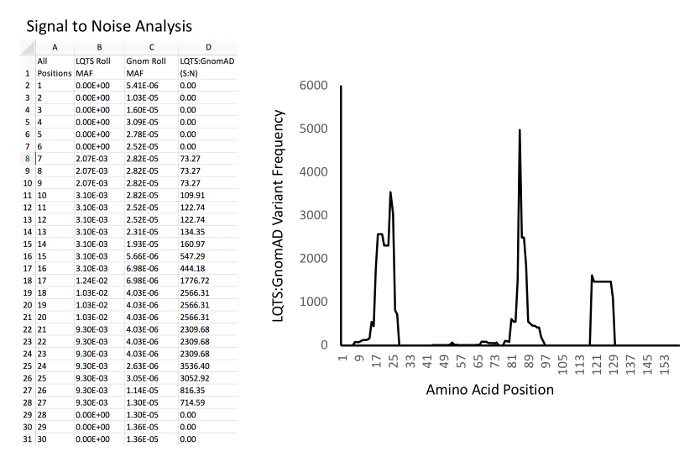
Figure 4: Example of signal-to-noise analysis and graphing. Left, example database and calculations. Column A, all amino acid positions of KCNQ1. Column B, LQTS experimental MAF rolling average for each position. Column C, GnomAD control MAF rolling average for each position. D: Signal-to-noise ratio (i.e. for D2, "=B2/C2"). Right, example of graph of signal-to-noise ratio (Y-axis) versus amino acid position (X-axis). Please click here to view a larger version of this figure.
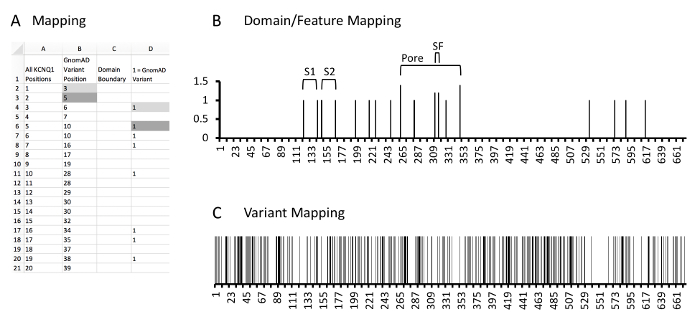
Figure 5: Example of protein and variant position mapping. A, example database and calculations. Column A, all amino acid positions of KCNQ1. Column B, KCNQ1 positions which have a rare control variant identified in GnomAD. Column C, the domain mapping column where cells containing values correspond to the N or C-terminal aspect of identified KCNQ1 protein domains or features. As the most N-terminal domain is the S1 domain has the N-terminal boundary at amino acid 122, no values are noted here. Column D, the variant mapping column where cells containing a 1 correspond to KCNQ1 positions which localize rare variants. Gray fill cells are two examples of where variant positions in column B are expanded into column D which correlate with respective positions in column A. Please click here to view a larger version of this figure.
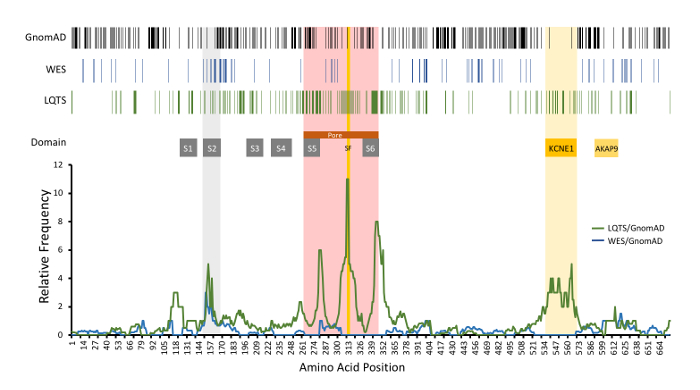
Figure 6: Example of amino acid-level signal-to-noise analysis of KCNQ1-encoded KCNQ1 (Kv7.1). Top, variant positions are demonstrated with vertical lines, including rare GnomAD cohort variants (black), incidentally-identified variants in WES referrals (blue), and variants identified in LQTS cases(green). Functional domains are noted. Relative frequency of LQTS case variants normalized to GnomAD variants (green line) is depicted compared to WES (blue line). S1-S6, transmembrane domains; SF, ion selectivity filter; KCNE1 and AKAP9, respective protein binding domains. Modified and reprinted with permission from previous work14. Please click here to view a larger version of this figure.
Discussion
High-throughput genetic testing has advanced dramatically in its application and availability over the past decade. However, in many diseases with well-established genetic underpinnings, such as cardiomyopathies, expanded testing has failed to improve diagnostic yield21. Further, there is significant uncertainty regarding the diagnostic utility of many identified variants. This is partially due to a growing number of incidentally identified rare variants discovered on WES and WGS, which can lead to misdiagnosis22. Amino acid level signal-to-noise analysis is based on well-established strategies for predicting variant pathogenicity and provides the advantage of leveraging large-scale population-based genome studies to refine variant interpretation.
It follows that one of the most crucial steps to this protocol is the selection of control and experimental cohorts. Many of the publicly-available large genome studies are accessible through aggregate databases, such as GnomAD, that can allow for representative control cohorts in this protocol to be as large as 138,632 individuals at present date. Though not all subjects in these aggregate cohorts are ostensibly healthy, the large sample size in the setting of rare disease makes this resource invaluable and allows for a stringent MAF exclusion threshold. Exclusion of common variants is necessary as they are unlikely to be a cause of highly penetrant Mendelian disease. Based on previous work, a MAF threshold of 0.01 for channelopathy-associated genes and 0.0001 for cardiomyopathy genes may be appropriate and has been validated by independent groups23,24. Importantly, given the importance of the MAF threshold, this should be set and validated for each study independently. A MAF threshold need not be applied to an experimental cohort, given the well-established presence of founder mutations in channelopathies and cardiomyopathies. The size of the experimental cohort needs to be sufficient to identify areas where variants may cluster; however, there is no strict size. Additionally, the experimental cohort should not include variants known to be benign within the literature, as this would diminish the veracity of the pathogenic signal.
Properly selecting exclusion criteria is also crucial for interpretation and applicability of the result. Though this protocol recommends excluding certain mutation classes such as synonymous variants, these could feasibly be included for disease processes in which deleterious synonymous variants have been identified25,26. Additionally, when various exclusion criteria are applied to both experimental and control groups, it can allow for stratification of signal-to-noise mapping by mutation subclass (i.e. comparing missense to truncating variants).
Setting a rolling average for MAFs allow for inference of involvement to neighboring amino acids. For example, if amino acid position 35 contains a pathologic variant and resides in a critical protein domain, then position 36 may have a degree of pathogenicity when mutated. Likewise, should a stretch of primary sequence have a large amount of rare control variants, then amino acids within this region that do not host rare variants may yet have a higher likelihood of containing rare variants found in a population. While the rolling average in this protocol is +/- 5, this range can be vary based on the user's desired level of resolution of signal-to-noise ratio and the specific protein being studied. In the example of LQTS, the interrogated KCNQ1-encoded KCNQ1 channel has several transmembrane domains spanning ~10 amino acids, prompting the authors to adjust their desired resolution to reflect significant findings on that scale14. For proteins with a longer primary sequence and protein length, the span of the rolling average may need to be increased due to larger spans of protein sequence without control variation.
There are several limitations to this method. As previously stated, a sufficient phenotype-positive population hosting putative pathologic variants must be identified in order to drive a clear pathologic signal. Additionally, these pathologic variants may have variable penetrance, thus truly pathologic mutations may not manifest a disease phenotype or may otherwise not be fully penetrant and disease causing. While many publicly held databases, such as GnomAD, are often considered "healthy cohorts," the prevalence of genetic diseases is likely similar in this database as population studies. As detailed, this protocol focuses specifically on amino-acid level changes resulting from exonic gene variants that code for amino acids, which excludes the role that pathogenic intronic splicing variants may play in monogenic disease. Given their recently demonstrated role in cardiomyopathies, expansion of the resolution this approach may be warranted to identify intergenic "hotspots" as well. Furthermore, the application of a MAF threshold may miss certain "risk alleles" that, though existing in the population with a MAF higher than that of disease prevalence, may contribute to disease pathogenesis27,28. Despite these limitations, this analysis is adaptable and can play a key role in providing clinicians a relative probability of disease pathogenicity when appropriate applied.
Finally, given the predilection of this analysis to identify critical regions within a protein, amino acid-level signal-to-noise calculations utilizing pathologic mutations offers the possibility of identifying novel functional domains of the proteins being studied. Given the observation of high pathogenicity signal-to-noise at key locations of ion channels, such as the pore domain, selectivity filter, S2 transmembrane domain, and the KCNE1-binding domain of KCNQ1, identification of a "peak of pathogenicity" within an area of the protein without a known function may suggest a novel critical domain. For example, a marked peak of pathogenicity of LQTS-associated mutations has been identified localizing to amino acid residues 912-930 of KCNH2-encoded KCNH2 (Kv11.1). This region of the protein has no identifiable functional domain yet demonstrates a marked propensity for LQTS-associated mutations14. As the knowledge of protein topology expands, more sophisticated proteomics could feasibly improve the resolution of this method in the future from analyzing signal-to-noise ratio along a protein's primary structure to include its secondary, tertiary, or quaternary structure. Addition of advanced computational sciences to this analysis, such as machine learning and artificial intelligence, affords the opportunity to identify novel patterns among pathologic versus population-based genetic variation, if robust databases of these variants can be generated29,30. In turn, this method could aid in better characterizing and predicting the genotype-phenotype relationship of specific diseases and be used in conjunction with an individual's pre-test probability of disease to improve the diagnostic yield of genetic testing. Further, this analysis may discover novel protein biology and identify novel loci within the human genome which manifest with disease when altered.
開示
The authors have nothing to disclose.
Acknowledgements
APL is supported by the National Institutes of Health K08-HL136839.
Materials
| 1000 Genome Project | N/A | www.internationalgenome.org | |
| ClinVar | N/A | www.ncbi.nlm.nih.gov/clinvar | |
| Ensembl Genome Browser | N/A | uswest.ensembl.org/index.html | |
| Excel | Microsoft | office.microsoft.com/excel/ | Used for all example formulas and functions |
| Exome Aggregation Consortium | N/A | www.exac.broadinstitute.org | |
| Genome Aggregation Database | N/A | www.gnomad.broadinstitute.org | |
| National Center for Biotechnology Information Domain and Structure Database | N/A | www.ncbi.nlm.nih.gov/guide/domains-structures/ | |
| National Center for Biotechnology Information Gene Database | N/A | www.ncbi.nlm.nih.gov/gene/ | |
| National Center for Biotechnology Information Protein Database | N/A | www.ncbi.nlm.nih.gov/protein/ | |
| National Heart, Lung, and Blood Institute GO Exome Sequencing Project | N/A | www.evs.gs.washington.edu/EVS/ | |
| SnapGene | GSL Biotech LCC | www.snapgene.com | |
| University of California, Santa Cruz Human Genome Browser | N/A | www.genome.ucsc.edu |
参考文献
- Yang, Y., et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. New England Journal of Medicine. 369 (16), 1502-1511 (2013).
- Meng, L., et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. Journal of the American Medical Association Pediatrics. 171 (12), 173438 (2017).
- Kalia, S. S., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genetics in Medicine. 19 (2), 249-255 (2017).
- Landstrom, A. P., Ackerman, M. J. The Achilles’ heel of cardiovascular genetic testing: distinguishing pathogenic mutations from background genetic noise. Clinical Pharmacology and Therapeutics. 90 (4), 496-499 (2011).
- Landstrom, A. P., Tester, D. J., Ackerman, M. J., Lawless, C. Role of genetic testing for sudden death predisposing heart conditions in athletes. Sports Cardiology Essentials. , (2011).
- Wang, Q., et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nature Genetics. 12 (1), 17-23 (1996).
- Kapa, S., et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 120 (18), 1752-1760 (2009).
- Ackerman, M. J., et al. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clinic Proceedings. 78 (12), 1479-1487 (2003).
- Kumar, P., Henikoff, S., Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 4 (7), 1073-1081 (2009).
- Adzhubei, I., Jordan, D. M., Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics. , (2013).
- Flanagan, S. E., Patch, A. M., Ellard, S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genetic Testing and Molecular Biomarkers. 14 (4), 533-537 (2010).
- Ackerman, M. J., et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 8 (8), 1308-1339 (2011).
- Lek, M., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536 (7616), 285-291 (2016).
- Landstrom, A. P., et al. Amino acid-level signal-to-noise analysis of incidentally identified variants in genes associated with long QT syndrome during pediatric whole exome sequencing reflects background genetic noise. Heart Rhythm. 15 (7), 1042-1050 (2018).
- Hubbard, T., et al. Ensembl 2005. Nucleic Acids Research. 33, 447-453 (2005).
- O’Leary, N. A., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Research. 44, 733-745 (2016).
- Kent, W. J., et al. The human genome browser at UCSC. Genome Research. 12 (6), 996-1006 (2002).
- The 100 Genome Projects Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 491 (7422), 56-65 (2012).
- Fu, W., et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 493 (7331), 216-220 (2013).
- Mulder, N. J., Apweiler, R. Tools and resources for identifying protein families, domains and motifs. Genome Biology. 3 (1), (2002).
- Cirino, A. L., et al. A Comparison of Whole Genome Sequencing to Multigene Panel Testing in Hypertrophic Cardiomyopathy Patients. Circulation Cardiovascular Genetics. 10 (5), (2017).
- Landstrom, A. P., et al. Interpreting Incidentally Identified Variants in Genes Associated With Catecholaminergic Polymorphic Ventricular Tachycardia in a Large Cohort of Clinical Whole-Exome Genetic Test Referrals. Circulation Arrhythmia and Electrophysiology. 10 (4), (2017).
- Whiffin, N., et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genetics in Medicine. 19 (10), 1151-1158 (2017).
- Walsh, R., et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in Medicine. 19 (2), 192-203 (2017).
- Buske, O. J., Manickaraj, A., Mital, S., Ray, P. N., Brudno, M. Identification of deleterious synonymous variants in human genomes. バイオインフォマティクス. 31 (5), 799 (2015).
- Wen, P., Xiao, P., Xia, J. dbDSM: a manually curated database for deleterious synonymous mutations. バイオインフォマティクス. 32 (12), 1914-1916 (2016).
- Bagnall, R. D., et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients With Hypertrophic Cardiomyopathy. Journal of the American College of Cardiology. 72 (4), 419-429 (2018).
- Giudicessi, J. R., Roden, D. M., Wilde, A. A. M., Ackerman, M. J. Classification and Reporting of Potentially Proarrhythmic Common Genetic Variation in Long QT Syndrome Genetic Testing. Circulation. 137 (6), 619-630 (2018).
- Sundaram, L., et al. Predicting the clinical impact of human mutation with deep neural networks. Nature Genetics. 50, 1161-1170 (2018).
- Krittanawong, C., Zhang, H., Wang, Z., Aydar, M., Kitai, T. Artificial Intelligence in Precision Cardiovascular Medicine. Journal of the American College of Cardiology. 69 (21), 2657-2664 (2017).

