Multicolor Flow Cytometry-based Quantification of Mitochondria and Lysosomes in T Cells
概要
This article illustrates a powerful method to quantify mitochondria or lysosomes in living cells. The combination of lysosome- or mitochondria-specific dyes with fluorescently conjugated antibodies against surface markers allows the quantification of these organelles in mixed cell populations, like primary cells harvested from tissue samples, by using multicolor flow cytometry.
Abstract
T cells utilize different metabolic programs to match their functional needs during differentiation and proliferation. Mitochondria are crucial cellular components responsible for supplying cell energy; however, excess mitochondria also produce reactive oxygen species (ROS) that could cause cell death. Therefore, the number of mitochondria must constantly be adjusted to fit the needs of the cells. This dynamic regulation is achieved in part through the function of lysosomes that remove surplus/damaged organelles and macromolecules. Hence, cellular mitochondrial and lysosomal contents are key indicators to evaluate the metabolic adjustment of cells. With the development of probes for organelles, well-characterized lysosome or mitochondria-specific dyes have become available in various formats to label cellular lysosomes and mitochondria. Multicolor flow cytometry is a common tool to profile cell phenotypes, and has the capability to be integrated with other assays. Here, we present a detailed protocol of how to combine organelle-specific dyes with surface markers staining to measure the amount of lysosomes and mitochondria in different T cell populations on a flow cytometer.
Introduction
The activation and proliferation of T cells are critical steps for mounting successful immune responses. Recent advances suggest that the cellular metabolism is tightly associated with both the development and functions of T cells. For example, naive T cells rely largely on oxidative phosphorylation (OXPHOS) to meet the energy demand during the recirculation among secondary lymphoid organs. Upon activation, naive T cells undergo drastic metabolic reprogramming, including the induction of aerobic glycolysis to increase ATP production and to fulfill the tremendous metabolic demands during cell differentiation and proliferation. The cells that fail to follow through the metabolic needs die by apoptosis1,2. During the metabolic reprogramming, mitochondria play important roles since they are the organelles largely responsible for the production of ATP to supply energy for the cell, and the cellular mitochondria content fluctuates during metabolic switches throughout T cell development and activation3. However, the accumulation of superfluous or damaged mitochondria can produce excess ROS that damage lipids, proteins, and DNA, and can eventually lead to cell death4
The excessive or damaged mitochondria resulting from metabolic alterations are removed by a specialized form of autophagy5, known as mitophagy. Mitochondria are wrapped by autophagosomes and then fused with lysosomes for degradation. These close communications between mitochondria and lysosomes have generated great interest6,7. For example, oxidative stress stimulates mitochondria to form mitochondria-derived vesicles (MDVs) that are targeted to lysosomes for degradation in a phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) and parkin (an E3 ubiquitin ligase) dependent manner8. It was also found that mitophagy is essential for beige-to-white adipocyte transition9,10. More importantly, lysosomes are not merely a degradation compartment, but also a regulator of cellular signaling. Excessive substrate accumulation due to enzyme deficiencies results in lysosomal dysfunction by disrupting lysosomal membrane permeability and affects Ca2+ homeostasis11. The T cell functional defects in lysosomal acid lipase (LAL)12 or lysosomal metabolite transporter knockout mouse model13 further showed the importance of lysosomes in maintaining T cell homeostasis. Both mitochondria and lysosomes are inseparable parts of cellular metabolic regulation. Therefore, measurement of cellular mitochondrial content has been a crucial indicator to evaluate the T cell metabolic and functional status.
Assays commonly used to quantify cellular mitochondrial or lysosomal contents include immunoblot, electron microscopy, immunofluorescent (IF) staining, and PCR analysis of mitochondrial DNA copy numbers14,15,16. While immunoblot can quantitatively compare protein levels across different samples and electron or IF microscopy can visualize the morphological hallmarks of these organelles17, these assays bear certain technical drawbacks. For example, it is time consuming to acquire sufficient number of cell images with high magnification and resolution or to compare the expression levels of a protein across dozens of samples, making these assays considered to be low throughput methods. Moreover, these assays can only be applied to homogenous cell population, such as cell lines, but not to tissue samples that are composed of mixed populations.
It is also difficult to apply these assays to rare populations, for which the requirement of minimal cell numbers from 106 to 108 is impossible to meet. Finally, cells are usually lysed or fixed during the process, making them incompatible with other methods to extract further information. Compared with the traditional methods, fluorescent-based flow cytometry has a relatively high throughput – the information of all the cells within a sample composed of mixed cell populations can be analyzed and collected simultaneously. In addition, one can detect more than 10 parameters on the same cell and sort the cells based on desired phenotypes for further assays. Reactive fluorescent probes have been used to label lysosomes and mitochondria in live cells and can be detected by flow cytometry18,19. These organelle-specific probes are cell permeable and have physico-chemical characteristics that allow them to be concentrated in specific subcellular locations or organelles. Conveniently, these probes are available in various fluorescent formats, thus enabling their application for multicolor analysis.
This protocol describes in detail how to combine surface marker staining with lysosome- or mitochondria-specific dyes to label lysosomes or mitochondria in live cells. This is especially useful for samples generated from primary tissues and organs, which are often composed of heterogeneous cell populations. Researchers can identify cell populations of interest by their surface marker expression and specifically measure the lysosomal or mitochondrial contents through organelle-specific dyes in these cells. Here, we demonstrate the detailed procedure of flow cytometric analysis that evaluates the lysosomal or mitochondrial mass in the major splenic T cell subpopulations.
Protocol
The mouse tissue harvest procedure described here has been approved by the Institutional Animal Care and Use Committee (IACUC) of National Yang-Ming University.
1. Preparation of Lymphocyte Suspension from Lymphoid Organs
- Euthanize the mouse by an approved method such as CO2 inhalation in a transparent acrylic chamber followed by cervical dislocation to ensure death.
NOTE: Keep the flow rate of CO2 for a 20% displacement per minute of the chamber, and no higher than 5 psi (pound per square inch). It takes 2-3 min for a mouse to lose consciousness. Maintain the CO2 flow at least for 1 min after the animal has stopped breathing (5 – 7 min overall). - Place the mouse on its back on a clean board (e.g. a polystyrene plastic board) and fix the limbs with tape. Rinse with 70% ethanol.
- To harvest spleen, cut and open the abdominal cavity with 11-cm dissecting scissors (the spleen is below the stomach and above the left kidney). Remove the spleen with forceps and clean from connective tissues (use scissors if necessary).
- To harvest the thymus, cut the diaphragm and the rib cage from both sides with dissecting scissors. Use forceps to lift up the sternum to reveal the entire thoracic cavity. Separate the thymus carefully from surrounding tissues with scissors, and remove any residual connective tissue on a paper towel wetted with PBS.
- Mechanically dissociate the tissues with a syringe plunger in a 6-cm dish containing 5 mL ice-cold FACS buffer (PBS supplemented with 2% fetal bovine serum (FBS) and 1 mM EDTA). Transfer the single-cell suspension to a 15-mL centrifuge tube; wash the 6-cm dish with additional 2 mL FACS buffer and combine the wash with cell suspension as well.
- Centrifuge the cell suspension (300 x g, for 5 min at 4 °C) and discard the supernatant. Re-suspend the pellet with 2 mL of Ammonium-Chloride-Potassium (ACK) lysing buffer (155 mM NH4Cl, 10 mM KHCO3 and 0.1 mM Na2EDTA) for 1 min at room temperature (RT) to lyse the red blood cells. Neutralize the ACK buffer by adding 10 mL FACS buffer to the cell suspension.
- Centrifuge the cell suspension (300 x g, for 5 min at 4 °C) and discard the supernatant. Re-suspend cells in 5 mL of complete RPMI medium (RPMI 1640 medium, supplemented with 44 mM NaHCO3, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100 U/mL penicillin, 100 mg/mL streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 1% MEM non-essential amino acids and 10% FBS).
- Filter cell suspension through a cell strainer (pore size 70 µm) to remove cell clumps. Count cells with a hemocytometer or automated cell counter.
2. Quantification of Mitochondria and Lysosomes
NOTE: Perform the organelle-specific dye staining first because the optimal staining condition for these dyes is incubation at 37 °C. The surface marker staining can be significantly reduced because of antibody capping and internalization at that temperature.
- Add 1 x 106 cells to round-bottom FACS tubes, centrifuge cells (300 x g, for 5 min at 4 °C) and discard supernatant.
- Dilute the probe stock solutions to final working concentration (100 nM MitoTracker Green or LysoTracker Green; henceforth mitochondria- or lysosome-specific dye, respectively) in pre-warmed 37 °C serum-free culture medium. This is the working solution for lysosome- or mitochondria-specific dye.
NOTE: Test multiple concentrations including the range of working concentrations suggested by manufacturer (20 – 200 nM for the mitochondria-specific dye and 50 – 75 nM for the lysosome-specific dye used here) in order to obtain the best staining efficiency. Choose a cell population that is known to have good expression of staining target (e.g. lysosomes) as positive control (e.g. human CD14+ monocytes). Choose the working concentration based on good separation between negative and positive staining, as well as good cell viability. - Resuspend the cells in 100 µL of lysosome- or mitochondria-specific dye working solution and incubate in 5% CO2 incubator at 37 °C for 15 min or 30 min, respectively.
- Add 1 mL of ice-cold FACS buffer to stop the reaction. Pellet cells by centrifugation (300 x g, for 5 min at 4 °C) and discard supernatant. Cells are now ready for surface marker staining.
3. Surface Marker Staining
- Block Fc receptors with 100 µL of pre-titrated 2.4G2 hybridoma supernatant on ice for 10 min. Wash cells with 1 mL of FACS buffer and centrifuge (300 x g, for 5 min at 4 °C). Discard supernatant.
- Incubate the cells with 50 µL pre-titrated fluorescence-conjugated antibody combination in FACS buffer on ice for 20 min. Avoid light.
NOTE: Prepare single color-stained cells that include all the fluorescent antibodies present in the antibody combination and cells treated only with organelle-specific dye for setting voltages of each fluorescence detector and compensation adjustment on flow cytometer. Likewise, prepare the Fluorescence Minus One (FMO) as background controls by using cells stained with all the antibodies, but without staining with organelle-specific dyes. - Wash the cells by adding 1 mL of FACS buffer and pellet by centrifugation (300 x g, for 5 min at 4 °C). Discard supernatant.
- Resuspend the cells in 300 µL of FACS buffer containing 1 µg/mL propidium iodide (PI) and immediately analyze the samples on flow cytometer.
4. Sample Collection on Flow Cytometer
- Turn on the computer, flow cytometer, FACS acquisition software and any other accessory devices depending on the machine platform. Run PBS for around 1 min to ensure the flow line is filled with PBS and sheath buffer. Make sure the flow stream runs smoothly.
- Open new experiment and select cytometer parameters (fluorescent detectors) according to manufacturer's instruction. Filter cells through 70-µm cell strainer and vortex prior to the acquisition of each sample to avoid clumps and doublet formation.
- Make dot plots of forward scatter (FSC) and side scatter (SSC), which represent cell size and granularity, respectively. Optimize the voltages of FSC and SSC to make sure the cells of interest appear in the plots. Make dot plots of FSC-A vs. FSC-W (or FSC-H) and draw a gate for the single cells.
- Make dot plots of each fluorescent parameter. Use unstained cells to determine background signal and use single color-stained controls to adjust voltages of each fluorescent parameter, so that positive and negative cell populations are clearly distinguishable and within the dynamic range of acquisition.
- After all the voltages are set, use auto-compensation if available, or manually adjust the compensation with unstained and single color-stained controls to correct the spectral spillover. Apply the compensation to samples.
- Make a dot plot of each parameter and draw gates according to your experimental set up. For example, FSC-A vs. FSC-H (or FSC-W) to exclude doublets, followed by FSC-A vs. SSC-A for lymphocyte gating; SSC-A vs. PI (live cells) and CD4 vs. CD8 (Figure 1).
- Collect enough events (total cell number) for meaningful comparison between populations. Typically, at least 1,000 events in the population of interest need to be obtained20.
- After all the samples are acquired, export flow data to be analyzed by flow cytometry analysis software. Save the experiment as template to preserve the cytometer setting and parameters for applying to similar experiments in the future.
5. Data Analysis
NOTE: There are many tools to analyze flow cytometric data, both commercial and free software. The acquisition software of the flow cytometers can also work for data analysis. Here we used FlowJo as an example.
- Start the software and drag the FCS files into the workspace. Click on a sample to open a graphic window. Select parameters to be presented on X and Y axis by clicking on the X and Y axes. Use the toolbar to draw gates according to differentially expressed markers or defined phenotypes of cell populations.
- Add gates of each parameter as shown in Figure 1. Drag gates to all samples in the workspace, so that they get identical gating.
- Drag the plots to layout editor to create graphical reports.
- Display the mean fluorescence intensity (MFI) for each population of interest by clicking the statistics button (Σ) in a graphic window and choosing "Mean" and the appropriate parameter, for example, the channel used to detect organelle-specific dye. Then click "Add".
- Click the table editor icon and drag MFI to table editor to create statistical reports. Save as Excel, Text or CSV file to calculate delta MFI (ΔMFI) as MFI (organelle-specific dyes) – MFI (FMO).
NOTE: Do not calculate MFI between samples acquired with different cytometer settings (voltages, fluorescent channels or compensation). Comparisons of values derived from experiments with different settings are meaningless and biased. - Export all the values from the workspace and plots from layout editor for making tables and figures.
Representative Results
Identification of major T cell subsets in spleen and thymus
In brief, single cell suspensions from spleen and thymus were lysed of red blood cells, incubated with 2.4G2 supernatant, followed by organelle-specific dye and surface marker staining with fluorescence-conjugated antibodies (Table 1). The developmental progression of thymocytes can be described by using antibodies to the co-receptors CD8 and CD4. The most immature thymocytes do not express CD4 or CD8 and are called double negative (DN). Then, they up-regulate both CD4 and CD8 and become double positive (DP). Finally, they down-regulate CD4 or CD8 and become mature single positive (SP) cells ready to leave the organ. The earliest stages of T cell development, when the cells do not express the co-receptors CD4 and CD8, can be further classified on the basis of CD44 and CD25 expression. The earliest progenitors are CD44+ CD25– (DN1), and then they start expressing CD25 and become CD44+CD25+ (DN2) cells. After that, the cells down-regulate CD44 and become CD44–CD25+ (DN3) cells and, finally, they down-regulate CD25 as well to become CD44–CD25– (DN4) cells that are on the way to expressing CD4 and CD8 and becoming double-positive (DP) cells.
In the spleen, the T cells can be divided into cytotoxic CD8+ and helper CD4+ T cells. Both of these populations can be further divided into naive, effector and memory subsets based on CD62L and CD44 staining. The detailed gating strategy is shown in Figure 1.
Quantification of mitochondria mass in different T cell subsets
Mitochondria-specific dyes were used to measure the amounts of mitochondria in T cell populations. We found that the mitochondrial contents were the lowest in DN1, peaked at DN2-DN3, and then slightly decreased in DN4 and DP thymocytes and were even lower in CD4 and CD8 SP thymocytes (Figure 2A). However, CD4 or CD8 SP thymocytes had higher mitochondria staining MFI than splenic CD4 or CD8 T cells (Figure 2B). These observations suggest that mitochondrial content fluctuates during T cell development with immature thymocytes containing more mitochondria than their mature counterparts do, and the naive T cells that recirculate in oxygen-rich blood have even lower mitochondrial mass. Interestingly, this reduction of mitochondrial contents was more prominent in the CD8 than the CD4 T lineage (Figure 2B), and T cell activation further decreased it. Among activated T cells, CD4+ memory T cells had slightly more mitochondria compared to effectors; however, the opposite was the case for CD8+ T cells: CD8+ memory T cells had the lowest mitochondrial mass among all T cell subsets (Figure 2B).
Lysosomal content measurement in various subpopulations of T cells
Using lysosome-specific dye, we observed relatively low, but detectable, lysosomal contents in all T cell populations, with more prominent presence of lysosomes in DN1 thymocytes (Figure 3A). No significant difference was found among the thymocyte subsets from DN1 onward. In the periphery, a relatively high number of lysosomes was found in memory and effector CD8+ T cells. This phenomenon is in line with previous studies showing that activated CD8+ T cells increased their expression of lysosomal-associated membrane protein 1 (LAMP-1)21,22, reflecting their possession of lysosomal-cytotoxic granules (Figure 3B)
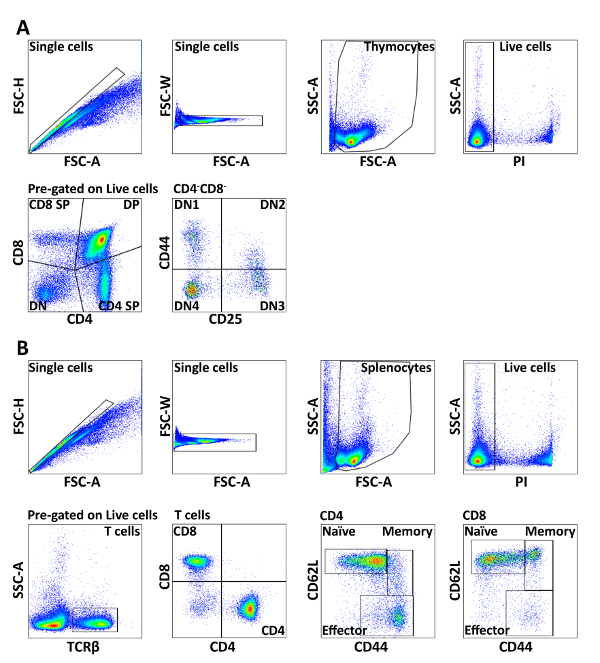
Figure 1: Gating strategy of splenic lymphocytes and thymocytes. Cells were pre-gated on FSC/SSC and PI– to obtain live singlets. (A) Total thymocytes from mice were stained with CD4, CD8, CD44, and CD25. CD4 and CD8 expressions were used to distinguish CD4+CD8– SP, CD4–CD8+ SP, CD4+CD8+ DP, and CD4–CD8– DN subsets. The CD4–CD8– DN subset was further divided into CD44+CD25– DN1, CD44+CD25+ DN2, CD44–CD25+ DN3, and CD44–CD25– DN4 subpopulations. (B) Total splenocytes from mice were stained with TCRβ, CD4, CD8, CD44, and CD62L. The TCRβ+ cells were separated into CD4+ and CD8+ T cells, which were further divided into naive (CD44–CD62L+), memory (CD44+CD62L+), and effector (CD44+CD62L–) populations. The results are representative of three independent experiments with n = 3-4. Please click here to view a larger version of this figure.
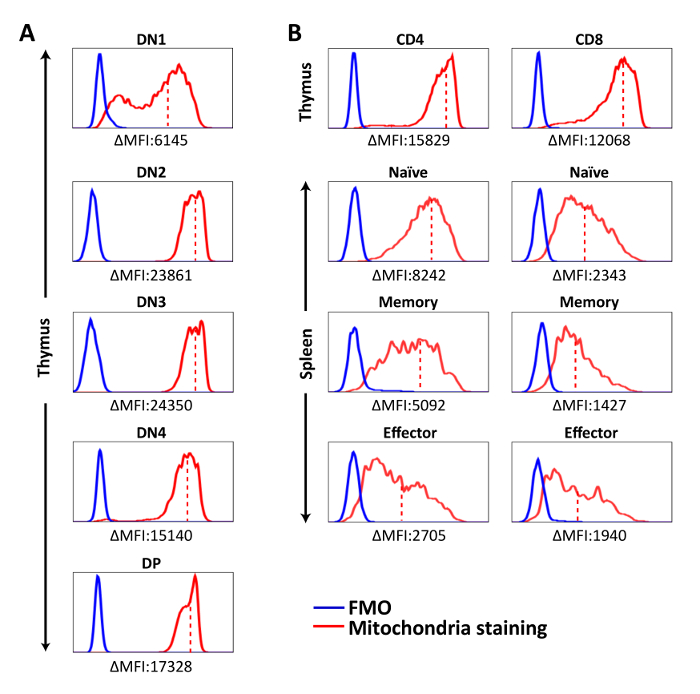
Figure 2: Representative histograms showing mitochondria staining in each cell population. Cells were stained with mitochondria-specific dye for 15 min at 37 ˚C, followed by surface marker staining. Fluorescent signal of stained cells was acquired by flow cytometer and analyzed on. (A) Mitochondria staining of DN and DP thymocytes. (B) Mitochondria staining of CD4SP and CD8SP thymocytes and splenic T cell populations. ΔMFI = MFI (Mitochondria staining) – MFI (FMO). The solid red line represents mitochondria staining, the solid blue line shows FMO control, and the dashed red line represents mean fluorescence intensity of mitochondria staining in each population. The results are representative of three independent experiments with n = 3-4. Please click here to view a larger version of this figure.
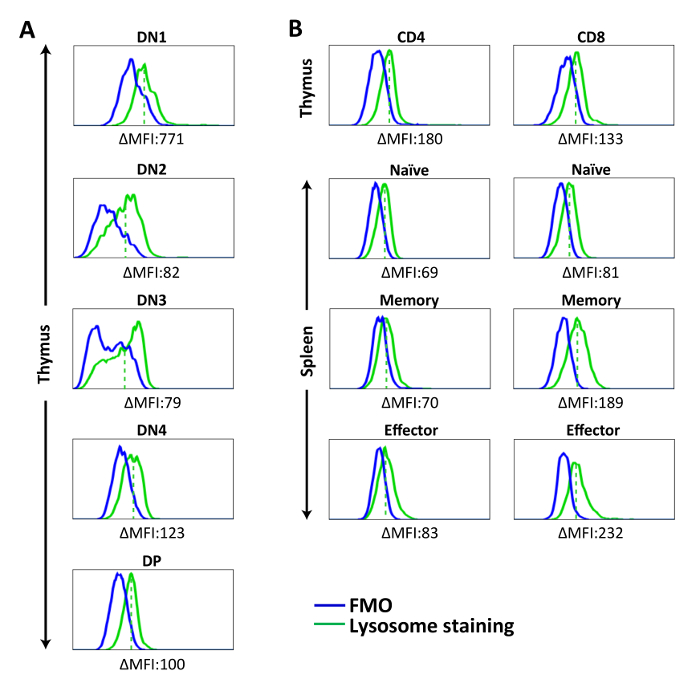
Figure 3: Representative histograms showing lysosome staining in splenic lymphocytes and thymocytes. Cells were stained with lysosome-specific dye for 30 min at 37 ˚C, followed by surface marker staining. Fluorescent signal of stained cells was acquired by flow cytometer and analyzed. (A) Lysosome staining of DN and DP thymocytes. (B) Lysosome staining of CD4SP and CD8SP thymocytes and splenic T cell populations. ΔMFI = MFI (Lysosome staining) – MFI (FMO). The solid green line represents lysosome staining, the solid blue line shows FMO, and the dashed red line represents mean fluorescence intensity of lysosome staining in each population. The results are representative of one experiment with n = 5. Please click here to view a larger version of this figure.
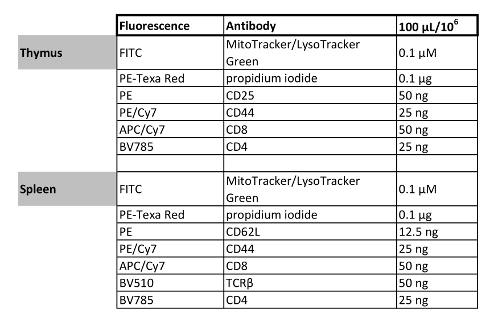
Table 1: Multicolor flow cytometry panel designs. The fluorochrome and concentration of antibodies/organelle dyes in the staining mixtures are listed here. It is strongly recommended to titrate and test each antibody when setting up new staining panels.
Discussion
This protocol combines organelle-specific dyes and surface marker staining to quantify the amount of mitochondria or lysosomes in different T cell populations. This method was developed to overcome the limitation of cell number and homogeneity requirements for traditional methods, such as electron microscopy and immunoblot analysis. It is especially useful in analyzing rare cell populations and simultaneously examining multiple cell types at the same time.
The duration of the procedure and the temperature of incubation are the most critical factors in this protocol. Proper organelle labeling requires accurate titration of organelle-specific dyes. Cells should also be strictly kept on ice to enhance the viability and avoid antibody capping and internalization. In addition, the fluorescence of these organelle-specific dyes is vulnerable to light exposure and temperature changes. Thus, immediate analysis of samples once the staining procedure is completed is very important. We have consistently been able to complete the whole experiment within 2 hours.
Since this protocol relies on the capability of detecting multicolor fluorescence by flow cytometer to evaluate intracellular components, the acquisition settings (e.g. PMT voltages) and the compensation of spectra overlap (the spillover of fluorescence into neighboring channels) can heavily influence the intensity of the signal. This is especially relevant when the surface markers needed to identify the relevant cell types increase above six. In this situation, experience in designing multicolor flow cytometry experiments becomes very important for high quality data acquisition.
The immune system protects the body from pathogen insults and infections, and T cell activation has pivotal role in providing essential signals to other immune cells. Recent studies suggest that different T cell populations have unique metabolic programs, and mitochondrial and lysosomal contents are crucial indicators of metabolic changes. However, this phenomenon is not unique to T cells. The differentiation and activation of innate immune cells, such as macrophage and dendritic cells, are also closely linked to their metabolic status23,24. The advantage of this protocol is that it can be adapted to examine any cell populations of interest by using antibodies against lineage-specific markers. Moreover, in combination with other organelle-specific dyes, such as ER- or Cyto-ID, it has the potential to evaluate different organelles or cellular compartments in various types of cells.
The establishment of this mitochondria or lysosome quantification method is not only instrumental to study the metabolism in T cells, but also has great potential to benefit research that focuses on metabolic changes in disease settings, such as cancer or autoimmune diseases25,26.
開示
The authors have nothing to disclose.
Acknowledgements
The development of this protocol was supported by grants from the Taiwan Ministry of Science and Technology (MOST) NSC103-2320-B-010-002-MY2 and MOST104-2628-B-010-002-MY4 to C.L. Hsu. C.W. Wei is a recipient of Excellent Thesis Award of Institute of Microbiology and Immunology, National Yang-Ming University.
Materials
| 10 cm Graefe forceps (straight and serrated) | Dimeda | ||
| 11 cm Iris scissors (straight with sharp/sharp heads) | Dimeda | ||
| 15 mL centrifuge tube | Thermo Fisher Scientific | 339650 | |
| 5 mL syringe | TERUMO | ||
| 6 cm Petri dish | α-plus | ||
| 70 µm nylon cell strainer | SPL Lifesciences | 93070 | |
| Ammonium chloride (NH4Cl) | Sigma | A9434 | |
| Anti-mouse CD25 | Biolegend | 102007 | Clone:PC61 |
| Anti-mouse CD4 | Biolegend | 100453 | Clone:GK1.5 |
| Anti-mouse CD44 | Biolegend | 103029 | Clone:IM7 |
| Anti-mouse CD62L | Biolegend | 104407 | Clone:MEL-14 |
| Anti-mouse CD8 | Biolegend | 100713 | Clone:53-6.7 |
| Anti-mouse TCRβ | Biolegend | 109233 | Clone:H57-579 |
| BD LSRFortessa | BD Biosciences | ||
| Ethylenediaminetetraacetic acid (EDTA) | Bio basic | 6381-92-6 | |
| FALCON 5ml Polystyrene Round-Bottom Tube (FACS tube) | BD Biosciences | 352052 | |
| FcRgamma II (CD32) Hybridoma (2.4G2) | ATCC | HB-197 | |
| Fetal Bovine Serum (FBS) | Hyclone | ATD161145 | |
| Flowjo, LLC | BD Biosciences | ||
| Hydroxyethyl piperazineethanesulfonic acid (HEPES) | Sigma | H4034 | |
| L-glutamine (200 mM) | Gibco | A2916801 | |
| LysoTracker Green DND26 | Thermo Fisher Scientific | L7526 | |
| MEM Non-essential amino acids (100X) | Gibco | 11140050 | |
| MitoTracker Green FM | Thermo Fisher Scientific | M7514 | |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 | |
| Potassium bicarbonate (KHCO3) | J.T.baker | 298-14-6 | |
| Propidium iodide solution | Sigma | 25535-16-4 | |
| RPMI 1640 medium (powder) | Gibco | 31800089 | |
| Sodium bicarbonate (NaHCO3) | Sigma | S5761 | |
| Sodium pyruvate (100 mM) | Gibco | 11360070 |
参考文献
- Buck, M. D., O’Sullivan, D., Pearce, E. L. T cell metabolism drives immunity. Journal of Experimental Medicine. 212 (9), 1345-1360 (2015).
- Marelli-Berg, F. M., Fu, H., Mauro, C. Molecular mechanisms of metabolic reprogramming in proliferating cells: implications for T-cell-mediated immunity. 免疫学. 136 (4), 363-369 (2012).
- Pua, H. H., Guo, J., Komatsu, M., He, Y. W. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. The Journal of Immunology. 182 (7), 4046-4055 (2009).
- Chao, T., Wang, H., Ho, P. C. Mitochondrial Control and Guidance of Cellular Activities of T Cells. Frontiers in Immunology. 8, 473 (2017).
- Youle, R. J., Narendra, D. P. Mechanisms of mitophagy. Nature Reviews Molecular Cell Biology. 12 (1), 9-14 (2011).
- Diogo, C. V., Yambire, K. F., Fernández Mosquera, L., Branco, F. T., Raimundo, N. Mitochondrial adventures at the organelle society. Biochemical and Biophysical Research Communications. 500 (1), 87-93 (2018).
- Todkar, K., Ilamathi, H. S., Germain, M. Mitochondria and Lysosomes: Discovering Bonds. Frontiers in Cell and Developmental Biology. 5, 106 (2017).
- McLelland, G. L., Soubannier, V., Chen, C. X., McBride, H. M., Fon, E. A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. The EMBO Journal. 33 (4), 282 (2014).
- Wrighton, K. H. Metabolism: Mitophagy turns beige adipocytes white. Nature Reviews Molecular Cell Biology. 17 (10), 607 (2016).
- Altshuler-Keylin, S., et al. Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metabolism. 24 (3), 402-419 (2016).
- Settembre, C., Fraldi, A., Medina, D. L., Ballabio, A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature Reviews Molecular Cell Biology. 14 (5), 283-296 (2013).
- Qu, P., Du, H., Wilkes, D. S., Yan, C. Critical roles of lysosomal acid lipase in T cell development and function. The American Journal of Pathology. 174 (3), 944-956 (2009).
- Wei, C. W., et al. Equilibrative Nucleoside Transporter 3 Regulates T Cell Homeostasis by Coordinating Lysosomal Function with Nucleoside Availability. Cell Reports. 23 (8), 2330-2341 (2018).
- Baixauli, F., et al. Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses. Cell Metabolism. 22 (3), 485-498 (2015).
- Piantadosi, C. A., Suliman, H. B. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. The Journal of Biological Chemistry. 281 (1), 324-333 (2006).
- Zhu, Y., et al. Constitutive association of the proapoptotic protein Bim with Bcl-2-related proteins on mitochondria in T cells. Proceedings of the National Academy of Sciences of the United States of America. 101 (20), 7681-7686 (2004).
- Ding, W. X., Yin, X. M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biological Chemistry. 393 (7), 547-564 (2012).
- van der Windt, G. J. W., et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences of the United States of America. 110 (35), 14336 (2013).
- Chikte, S., Panchal, N., Warnes, G. Use of Lyso dyes: A flow cytometric study of autophagy. Cytometry Part A. 85 (2), 169-178 (2013).
- Allan, A. L., Keeney, M. Circulating tumor cell analysis: technical and statistical considerations for application to the clinic. Journal of Oncology. 2010, 426218 (2010).
- Curtsinger, J. M., Lins, D. C., Johnson, C. M., Mescher, M. F. Signal 3 tolerant CD8 T cells degranulate in response to antigen but lack granzyme B to mediate cytolysis. The Journal of Immunology. 175 (7), 4392-4399 (2005).
- Wolint, P., Betts, M. R., Koup, R. A., Oxenius, A. Immediate cytotoxicity but not degranulation distinguishes effector and memory subsets of CD8+ T cells. Journal of Experimental Medicine. 199 (7), 925-936 (2004).
- Van den Bossche, J., et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Reports. 17 (3), 684-696 (2016).
- Pearce, E. J., Everts, B. Dendritic cell metabolism. Nature Reviews Immunology. 15 (1), 18-29 (2015).
- Dugnani, E., et al. Integrating T cell metabolism in cancer immunotherapy. Cancer Letters. 411, 12-18 (2017).
- Yang, Z., Matteson, E. L., Goronzy, J. J., Weyand, C. M. T-cell metabolism in autoimmune disease. Arthritis Research & Therapy. 17 (1), 29 (2015).

