Induction of Cellular Differentiation and Single Cell Imaging of Vibrio parahaemolyticus Swimmer and Swarmer Cells
概要
This protocol enables single cell microscopy of the differentially distinct Vibrio parahaemolyticus swimmer and swarmer cells. The method produces a population of swarmer cells easily available for single cell analysis and covers preparation of cell cultures, induction of swarmer differentiation, sample preparation, and image analysis.
Abstract
The ability to study the intracellular localization of proteins is essential for the understanding of many cellular processes. In turn, this requires the ability to obtain single cells for fluorescence microscopy, which can be particularly challenging when imaging cells that exist within bacterial communities. For example, the human pathogen Vibrio parahaemolyticus exists as short rod-shaped swimmer cells in liquid conditions that upon surface contact differentiate into a subpopulation of highly elongated swarmer cells specialized for growth on solid surfaces. This paper presents a method to perform single cell fluorescence microscopy analysis of V. parahaemolyticus in its two differential states. This protocol very reproducibly induces differentiation of V. parahaemolyticus into a swarmer cell life-cycle and facilitates their proliferation over solid surfaces. The method produces flares of differentiated swarmer cells extending from the edge of the swarm-colony. Notably, at the very tip of the swarm-flares, swarmer cells exist in a single layer of cells, which allows for their easy transfer to a microscope slide and subsequent fluorescence microscopy imaging of single cells. Additionally, the workflow of image analysis for demographic representation of bacterial societies is presented. As a proof of principle, the analysis of the intracellular localization of chemotaxis signaling arrays in swimmer and swarmer cells of V. parahaemolyticus is described.
Introduction
Bacteria constantly experience changes to their external environment and have developed several techniques to change and adapt their behavior accordingly. One such mechanism involves differentiation into distinct cell types that better complement the altered environment. Differentiation often involves major changes in the regulation of the cell cycle, cell morphology, and the spatiotemporal organization of the cells. One organism that can undergo differentiation is Vibrio parahaemolyticus. V. parahaemolyticus belongs to the Vibrionaceae, whichis a family of proteobacteria that usually inhabit fresh or salt water. Vibrionaceae are widely distributed in the environment and include several species that cause intestinal tract infections in humans, also including Vibrio cholerae.V. parahaemolyticus is a dimorphic organism and is able to differentiate into two distinct cell types as a response to accommodate changes to its external milieu. In aqueous environments, it exists as a short rod-shaped swimmer cell with a single polar flagellum positioned at the old cell pole. Upon surface contact, differentiation into a swarmer cell is triggered. Swarmer cell differentiation involves two major changes: swarmer cell morphogenesis through inhibition of cell division, and the induction of a second flagella system. This results in the formation of a peritrichous and highly elongated rod-shaped swarmer cell, which can either continue the swarmer life-style, where division events results in progeny swarmer cells, or alternatively differentiate back into swimmer cells.
Several factors have been reported to induce or influence swarmer differentiation. The primary stimulus appears to be driven by mechanosensing where V. parahaemolyticus uses the polar flagellum as a tactile sensor that detects inhibition of the rotation upon surface contact, but several other factors are involved as well1. In the lab, rotation of the polar flagellum can be artificially inhibited by addition of phenamil, which blocks the sodium-channel driven flagellar rotation, thereby inducing swarmer differentiation2. Furthermore, in an earlier study, Vibrio alginolyticus was induced to swarm on solid media when cells were propagated on growth medium in a Petri dish sealed with clear plastic tape. Alkali-saturated filter paper prevented swarming under these conditions, hence suggesting that one or more volatile acids might be involved in induction of swarming. Thus, sealing the Petri dish with plastic tape likely allowed for the accumulation of volatile acids, formed as by-products of cellular metabolism, within the head space of the plate. The same effect was achieved when H2O2 was added to the growth medium in un-sealed Petri dishes in order to artificially produce volatile acids by hydrolysing media components3,4,5. Nevertheless, the identity of such volatile acids remains unknown. Moreover, it has been shown that excess availability of calcium6 and iron-limitation7 both enhance swarmer differentiation and proliferation over solid surfaces. Cells can be starved for iron by adding the compound 2,2´-Bipyridyl to the growth medium, which has been shown to influence swarmer differentiation8. The factors known to regulate differentiation are now implemented in the design of a protocol that reproducibly induces V. parahaemolyticus differentiation into swarmer cells and their proliferation on solid agar surfaces.
V. parahaemolyticus differentiation involves major changes in the regulation of cell division, cellular morphology, and the positioning of macromolecular machines such as flagella and chemotaxis apparatuses – processes that all require the specific localization of proteins in accordance with the cell cycle. Thus, the ability to study the intracellular localization of such proteins is essential to the understanding of the aforementioned cellular processes. In order to perform such studies fluorescence microscopy on single cells is required. This can be particularly challenging when imaging cells that exist within dense bacterial populations, as is the case for swarmer cells. There have been attempts to induce swarmer differentiation in liquid media, which could potentially generate single cells for microscopy studies. And although on a transcriptional level these cells have partially induced the swarmer-differentiation program, they do not undergo the same distinct morphological changes as fully differentiated swarmer cells grown on solid medium8. This paper offers a robust and reproducible protocol of a method to induce swarmer cell differentiation on an agar surface. The protocol produces a population of easily accessible swarmer cells readily available for single cell microscopy and subsequent analysis. Furthermore, the protocol enables localization studies of fluorescently labeled proteins in both the swimmer and swarmer cell-types (sections 1, 2, 3, 4). Additionally, the protocol describes the subsequent workflow on how to process and analyze the generated data from fluorescence microscopy experiments, which allows for demographic analysis of bacterial societies (section 5).
Protocol
1. Preparation of Electro-competent V. parahaemolyticus Cells and Electroporation of Plasmid DNA into Swimmer Cells
NOTE: The following section of the protocol allows for the preparation of electro-competent cells and the subsequent electroporation of plasmid DNA into swimmer cells. This step is important if fluorescent proteins are being ectopically expressed from a plasmid.
- Streak out V. parahaemolyticus cells from a -80 °C glycerol stock onto a fresh LB agar plate containing 100 µg/mL ampicillin and incubate overnight at 37 °C.
- The following day, inoculate 200 mL of LB medium with a single colony of V. parahaemolyticus and incubate it at 37 °C under shaking conditions until an OD600 of 1.0 is reached. It usually takes 5-6 h of incubation for the cell culture to reach the desired optical density.
- Immediately transfer the cells onto ice and perform all further steps on ice and in 4 °C pre-cooled centrifuges.
- Harvest the cells at 4 °C for 10 min at 2,000 x g.
- Discard the supernatant by decantation and re-suspend the cell pellet in 25 mL of ice-cold 273 mM sucrose solution (pH 7.4, buffered with KOH).
- Harvest the cells again at 4 °C for 10 min at 2000 x g. Repeat two additional times.
- Re-suspend the washed cell pellet in 400 µL of ice-cold 273 mM sucrose solution. Subsequently, add glycerol to a final concentration of 15% vol/vol. The cells are now ready for electroporation, but can also be frozen in aliquots of 70 µL at -80 °C for later use.
- For the electroporation, mix a 70 µL aliquot of electro-competent cells with 100 – 1,000 ng of plasmid DNA and transfer the mixture into an ice-cold electroporation cuvette (0.2 cm electrode gap). Perform electroporation with the following settings: 25 µF, 2400 V and 200 Ω.
- Suspend the cells in the electroporation cuvette in 600 µL of LB broth, transfer to a 2 mL tube and incubate while shaking at 37 °C for 3 h.
- Spin down the cells at 2,000 x g for 5 min and re-suspend the pellet in 100 µL of LB broth. Spread the cell-suspension onto selective LB agar plates containing the necessary antibiotics and incubate at 37 °C overnight.
- The following day, check the plates for colony formation. Colonies that grow carry the correct antibiotic resistance gene encoded on the plasmid and can now be used for further experiments.
2. Induction of Swarmer Cell Differentiation
NOTE: The following steps describe how to induce differentiation of V. parahaemolyticus into its swarmer cell life-cycle and how to stimulate proliferation on solid surfaces.
- Suspend 4 g of Heart Infusion (HI) Agar in 100 mL ddH2O in order to prepare the swarming plates.
- Carefully boil the HI agar solution in the microwave and shake the bottle from time-to-time until the powder is dissolved. Autoclave at 121 °C for 20 min.
- After autoclaving, cool down the HI agar to 60-65 °C. Add 2,2´-Bipyridyl to a final concentration of 50 µM (stock solution of 50 mM in 100% ethanol). Add CaCl2 to a final concentration of 4 mM (stock solution of 4 M in ddH2O). If necessary, add antibiotics into the solution to maintain plasmids.
NOTE: If planning to use an inducible plasmid, add inducing agent to its final concentration.
CAUTION: 2,2´-Bipyridyl: Acute toxicity (oral, dermal, inhalation). Use gloves and safety goggles when handling this chemical. - Pour 30 – 35 mL of the final HI agar solution into a round 150 mm Petri dish and let the agar solidify.
- Right before spotting the cell culture, dry the agar plate at 37 °C for at least 10 min (or until any liquid residues have disappeared, but no longer) up-right with an open lid.
NOTE: Drying times highly depend on the incubator used and heavily dried plates will not permit swarming of V. parahaemolyticus. (This step is very important!) Plates must be used fresh and should not be older than 2 – 3 h. Old plates will be too dry to stimulate swarming. - Inoculate a small amount of cells from the edge of a single colony into 5 mL of LB broth. Incubate the culture shaking at 37 °C until OD600 = 0.8 is reached, it usually takes around 2-3 hours. After that, the cells are ready to be spotted onto HI swarming agar plates.
- Spot 1 µL of cell culture at the center of a HI agar swarming plate (Figure 1A).
- Let the spot dry (Figure 1B).
- Seal the plate with clear plastic tape and make sure that it is firmly attached, otherwise it might detach during overnight incubation (Figure 1C).
NOTE: It is very important that the seal is perfect. Avoid the formation of air-bubbles and folds to the tape when it is applied. - Incubate the plate at 24 °C overnight. This will result in the formation of a V. parahaemolyticus swarm-colony with swarm-flares extending from the periphery of the swarm-colony (Figure 2).
3. Preparation of V. parahaemolyticus Swarmer Cells for Fluorescence Microscopy
NOTE: The following protocol describes how to prepare microscopy agarose slides and the subsequent imprinting of swarming flares onto an agarose pad to obtain single swarmer cells for microscopy.
- To prepare agarose slides for microscopy, add 1 g of agarose to 100 mL of a 20% vol/vol PBS and 10% vol/vol LB broth solution. Carefully boil the solution in the microwave to dissolve the agarose and let it cool down to 65-70 °C while mixing with a magnetic stirrer.
- Place a clean microscope slide onto a perfectly level section of a working bench. Cover both ends of the slide with two layers of general-purpose laboratory labeling tape, fixing the slide to the working bench. The distance between the two pieces of tape should be around 3 cm.
NOTE: The two layers of tape create a very small elevation, which will determine the height of the agarose pad. It is important that the working space is level to make sure cells will be in the same plane for microscopy analysis. - Pipet ~ 250 µL of the previously prepared agarose solution onto the non-covered glass surface.
- Now place a second microscope slide in the same orientation as the bottom one on-top and slightly press it down, so that residual agarose can flow out to the sides.
- Let the agarose solidify 1-2 min and then slowly pull the top glass slide off the bottom one in a horizontal movement.
- Using a scalpel, cut off any residual agarose that is attached to the sides of the microscope slide and slowly remove the tape from the ends of the microscope slide.
NOTE: The agarose slide is now ready and should be used as soon as possible as it will slowly begin to dry out. - To perform microscopy on swarmer cells, cut out a ~3 mm x 10 mm piece of swarming agar from the most outer edge of a swarming colony (Figure 1D). It is very important not to break the tape seal of the swarm-plate until immediately before transfer of swarmer cells to the microscope slide. Once the tape is removed, swarming V. parahaemolyticus cells initiate differentiation into swimmer cells.
NOTE: Make sure to direct the cuts from the outside of the swarming colony edge to the inside of the colony (Figure 1D). This way you can prevent "swimmer contamination" into the edge of the swarming colony. - Transfer the piece of swarming agar onto a microscope agarose slide with the cells facing the agarose pad (Figure 1E). This imprints the swarming cells onto the agarose pad.
- After waiting ~30 s, remove the agar piece carefully from the agarose pad (Figure 1F).
- Place a glass coverslip onto the agarose pad where the cells were imprinted. The cells are now ready for microscopy.
NOTE: It is important to image swarmer cells immediately, since swarmer cells slowly initiate differentiation to the swimmer state.
4. Preparation of V. parahaemolyticus Swimmer Cell-cultures For Fluorescence Microscopy
NOTE: This section of the protocol describes how to prepare a culture of V. parahaemolyticus to perform fluorescence microscopy on the swimmer cell-type.
- Inoculate a small amount of cells from the edge of a single colony into 5 mL of LB broth. Incubate the culture shaking at 37 °C for 1 h or until slight cell-growth becomes visible.
- At this stage, if needed, add inducing agent for expression of proteins of interest. In this example, L-arabinose was added to a final concentration of 0.2% (w/vol) in order to induce expression of YFP-CheW9.
- Incubate the culture shaking for an additional 2 h at 37 °C.
- Follow steps 3.1-3.7 to prepare microscopy agarose slides.
- Spot 1 µL of cell culture at the center of the agarose pad and let the spot incubate until it is dry.
- Place a glass coverslip onto the agarose pad where the cells were spotted. The cells are now ready to be imaged under the fluorescence microscope.
5. Image Analysis
This part of the protocol describes a workflow for image analysis, particularly how to extract fluorescence data of single cells for demographic analysis.
- Before beginning the image analysis make sure that all microscopy images are saved in 16bit TIF format.
- Load the DIC (or phase contrast) and the corresponding fluorescent channel images into the software.
- Generate an overlay image of both channels by selecting "Display/Overlay images". Set the number of pictures ("# Images:") to two, and select the DIC image in the first channel and the fluorescent image in the second channel.
- Select the "Multi-Line" tool from the toolbar and mark cells from one pole to the other through the middle of the cell. In order to get a list of all generated lines (line objects) open "Measure/Region Measurements". Configure the region measurements to only display the region label, distance, and average intensity.
- Calibrate the pixel size under "Measure/Calibrate Distances" to the pixel size that is specific for your microscope setup and click "Apply To All Open Images".
- Transfer all regions (line objects) to the fluorescent channel image. Under "Regions/Transfer Regions" select the source image (the overlay image) and the destination image (the fluorescent channel image). Select "All Regions" and press "OK".
NOTE: To be able to later re-produce the exact lines (line objects, regions, ROI), open "Regions/Save Regions" and save the regions. - Export all region measurements into a spreadsheet by opening "Measure/Region Measurements" and pressing "Open Log". Confirm to open and export the data. Once the spreadsheet is opened in the background, finally export the data by pressing "F9: Log Data".
- To determine the position of fluorescent foci along the cell, open "Measure/Linescan". Set "Linescan Width" to a value so that the whole width of a cell is covered by the region that you previously created. Right-click into the line-scan graph and select "Show Graph Data". By left-clicking and dragging the mouse-courser along the line-scan to the point of interest, the "Graph Data" window will display the corresponding pixel/distance between the two points. Select the blue highlighted row and then copy the content.
NOTE: The entire line-scan intensity profile can be logged to a spreadsheet by pressing "Log Data" in the "Linescan" window. The extracted data can now be used for visualization in graphs or further statistical analysis. Localization patterns over the cell cycle can be easily visualized in a demographic representation of the cell length and the fluorescent intensities along the cell body. These representations can be obtained using the same ROI's previously selected. The following steps will give an example of how to generate demographics similar to the ones shown in Figures 3C and 4C.- In Fiji/ImageJ load region markings (.rgn) previously created in the software (step 5.6) using the "Metamorph nd & ROI files importer". Alternatively, regions of interest (ROIs) of the cells to be analyzed can be created directly in Fiji/ImageJ using the build-in "Segmented Line" tool.
- Open "Analyze/Tools/ROI manager" and mark all active list entries".
- Afterward, click "More/Multi Plot" and then select "List" in the new "Multi Plot" window. Click on a table entry, then select and "copy all". Subsequently, paste into a new spreadsheet.
NOTE: Make sure that the data of the longest cell (two corresponding X and Y columns with the most number of rows) is the last one to the right in the spreadsheet. - Save the spreadsheet as a "CSV (Comma delimited) (*.csv)".
- Download the R scripts from here "https://github.com/ta-cameron/Cell-Profiles”10. The scripts sort cells by length and normalize the fluorescent intensity profiles of all cells as an average of each cell’s fluorescence.
- Run the entire script "cell profiles function.R" in R [version 3.0.1;11].
- Edit the "example plots.R" script in line 30 such that the file path refers to the folder where the previously generated .csv file is saved. Additionally, change the file name "profile.csv" in line 31 to the file name chosen for .csv file generated in step 5.9.4. Afterwards, run "example plots.R" and follow the documentation from "https://github.com/ta-cameron/Cell-Profiles” as well as the comments from the script file to generate demographic analysis.
Representative Results
Induction of differentiation and generation of swarming colonies
Figure 1 provides a schema of the important steps involved in producing swarming colonies of V. parahaemolyticus (Figure 1A-C). A swimmer culture was spotted on swarm-agar and incubated at 24 oC, inducing swarmer differentiation and proliferation over the solid agar surface. A representative stereo-microscopy image of a swarming colony is presented in Figure 2. Stereo-microscopy imaging clearly shows swarm-flares extending outward of the swarm-colony periphery. Higher magnification reveals that in swarm-flares cells are grouped in mono- or bi-layers only, whereas in the middle of the swarm-colony cells are stacked in multiple layers (Figure 2A). Swarm-flares can easily be transferred (as described in steps 3.8-3.11, Figure 1) to a microscope slide. Importantly, the flares only consist of a fully differentiated population of swarmer cells (Figure 2B). This is in contrast to cells from the middle of the swarm-colony, which are significantly shorter and likely represent a mixture of swimmer and swarmer cells (Figure 2B)9. Furthermore, careful transfer of the flare to the microscope slide leaves the overall structure and swarm-edge of the swarm-flare intact, resulting in a mono-layer of swarmer cells accessible for single cell microscopy. When imprinting the swarm-flares on the microscope slide, it is very important to do the transfer as gentle as possible and not to add too much pressure – otherwise cells from swarm-flares will spread over the slide mixing with non-swarmer cells originating from the middle of the swarm-colony.
Fluorescence microscopy and image analysis
As proof of principle of single cell microscopy of differentially distinct V. parahaemolyticus swimmer and swarmer cells, a detailed analysis of ectopically expressed YFP-CheW was performed. CheW is an essential chemotaxis protein and can be used as a marker for the intracellular localization of chemotactic signaling arrays9,12. A plasmid encoding YFP-CheW under an L-arabinose inducible promoter was electroporated into V. parahaemolyticus as described in section 1 of the protocol. For microscopy, swimmer and swarmer cells were prepared as described in sections 4 and 3 respectively. As previously described YFP-CheW was localized in a uni-polar (Figure 3A, red arrows) and bi-polar manner (Figure 3A, yellow arrows)12,13. Accordingly, identification of the cell pole and the variation of YFP-protein localization at different lengths offer a great insight in how the localization pattern evolves as the cell cycle proceeds. These data can then be displayed in scatter plots correlating the variables "cell length" and "distance of the foci to the cell pole" (Figure 3B). To avoid imprecisions in contour detection, which is common when using automatic cell detection tools, cells are selected by means of manually drawing a line from one pole to the other using the "Multi-line" option in MetaMorph or "Segmented line" in the open-source platform ImageJ. Subsequently, the intensity profiles of the selected region of interest (ROI) can be collected, allowing the identification of the point in which YFP-protein displayed its highest intensity and also the overall length of the cell. The fluorescence profiles of single cells were analyzed as described in section 5 and were presented by plotting the distance of foci to the cell poles as a function of cell length (Figure 3B). This analysis clearly shows that short cells only display a uni-polar localization pattern of YFP-CheW, whereas in longer cells YFP-CheW is bi-polarly localized. Furthermore, localization patterns over the cell cycle can be easily visualized in a demographic representation of the cell length and the fluorescent intensities along the cell body. These representations can be obtained using the same ROIs previously selected when preparing the scatter plots. Hence, a demographic analysis of the whole cellular fluorescence profile as described in step 5.9 (Figure 3C) was performed. This analysis clearly shows a similar pattern of YFP-CheW localization, with YFP-CheW being uni-polarly localized in short cells and bi-polarly localized in longer cells. Thus, indicating a cell cycle-dependent localization pattern, where YFP-CheW is localized uni-polarly in recently divided cells (short), and then gets recruited to the opposite pole later in the cell cycle (long cells), resulting in a bi-polar localization. An analogous analysis was performed on swarmer cells (Figure 4). A population based analysis shows major differences in the localization of YFP-CheW in swarmer cells when compared to that of the swimmer cell-type. YFP-CheW is always bi-polarly localized (Figure 4A, yellow arrows) and importantly YFP-CheW also forms clusters positioned randomly along the cell length (Figure 4A, orange arrows). Thus, there are major changes in the intracellular localization of chemotaxis signaling arrays during differentiation of V. parahaemolyticus. This example shows that the method described allows for the production of a swarmer cell-population that is easily available for single cell fluorescence microscopy. Furthermore, it shows that the pipeline described for image analysis allows for demographic analysis of the intracellular organization of V. parahaemolyticus swarmer cells.
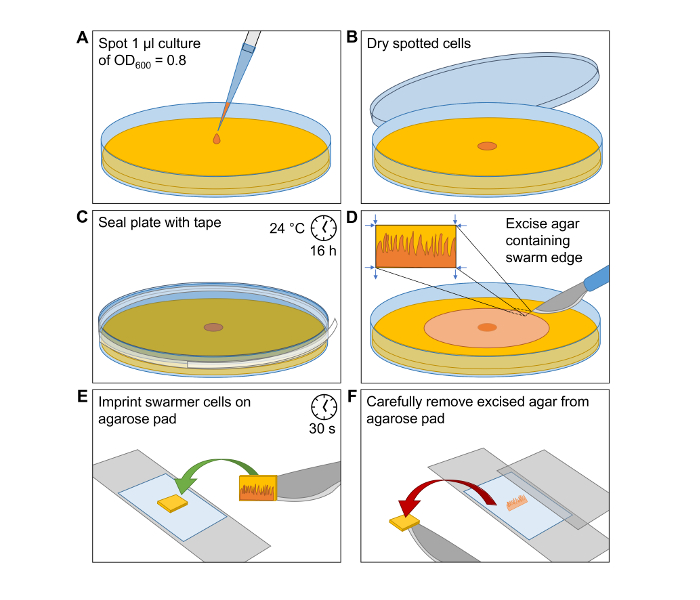
Figure 1: Induction of Swarmer Differentiation and Preparation of Cells for Microscopy Imaging. Simplified visualization of the workflow to produce V. parahaemolyticus swarmer cells and the preparation of microscope slides for microscopy analysis of single swarmer cells. (A) Spot 1 µL of an OD600 = 0.8 cell culture of V. parahaemolyticus at the center of a HI agar swarming plate. (B) Incubate the spot until it is dried and cells are attached to the agar surface. (C) Seal the Petri dish with clear tape. Incubate the plate at 24 °C for 16 h. (D) Carefully excise a small piece of HI agar from the outer edge of the swarming colony. (E) Transfer the excision onto a microscopy agarose pad with the swarmer cells facing the agarose pad. After a short incubation time of 30 s the swarmer cells are imprinted onto the agarose pad. (F) Carefully remove excised agar from agarose pad and place a microscopy coverslip on top. Please click here to view a larger version of this figure.
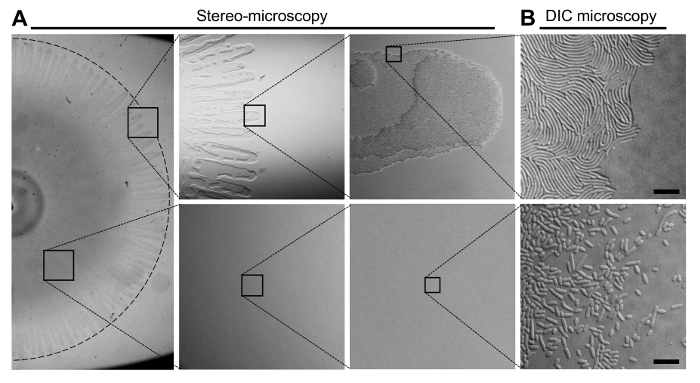
Figure 2: The Organization of Cells in a V. parahaemolyticus Swarming Colony. (A) Stereo-microscopy of a V. parahaemolyticus swarmer colony. The upper panels display the outer edge of a swarming colony in increasing magnification. The bottom panels display an area of the swarming colony between the center and the edge in increasing magnification. Swarm-flares extend from the periphery of the swarm-colony. (B) Representative DIC microscopy images of the swarmer colony edge (upper panel) and an area between the center and the periphery (bottom panel) after transfer of cells from the respective indicated locations of the swarming colony. The swarm-flares were imprinted directly onto a microscopy agarose pad as described in steps 3.8-3.11. Cells from the area between the center and the periphery were first scraped off the swarm-colony and re-suspended in LB broth, before being spotted onto a microscopy agarose pad. Scale bar = 7.5 µm. Please click here to view a larger version of this figure.
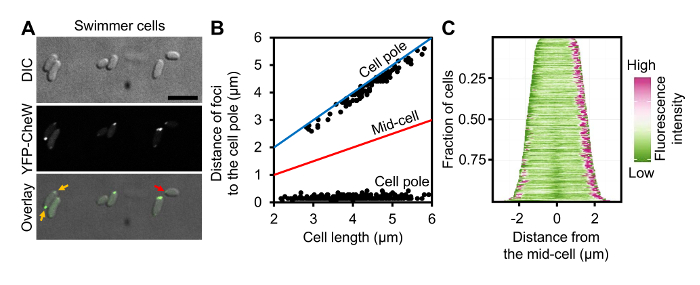
Figure 3: Localization of Chemotaxis Arrays in Swimmer Cells of V. parahaemolyticus. (A) DIC and fluorescent microscopy images of wild-type V. parahaemolyticus swimmer cells ectopically expressing YFP-CheW. (B) Graph displaying YFP-CheW foci distribution by plotting the distance of foci from the cell poles as a function of cell length in swimmer cells. (C) Demographic analysis of fluorescence intensity profiles of V. parahaemolyticus swimmer cells expressing YFP-CheW. Cells are sorted by cell length and displaying relative fluorescent intensity. Scale bar = 5 µm. Panel B and C are adapted from reference9. Please click here to view a larger version of this figure.
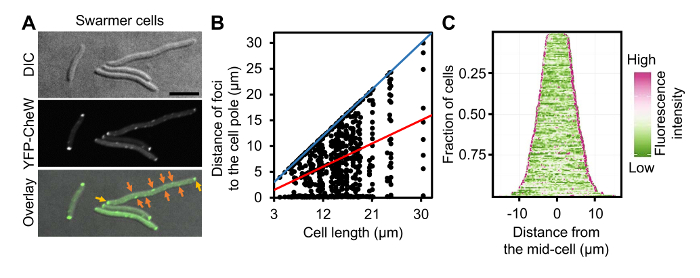
Figure 4: Localization of Chemotaxis Arrays in Swarmer Cells of V. parahaemolyticus. (A) DIC and fluorescent microscopy images of wild-type V. parahaemolyticus swarmer cells ectopically expressing YFP-CheW. (B) Graph displaying YFP-CheW foci distribution by plotting the distance of foci from the cell poles as a function of cell length in swarmer cells. (C) Demographic analysis of fluorescence intensity profiles of V. parahaemolyticus swarmer cells expressing YFP-CheW. Cells are sorted by cell length and display relative fluorescent intensity. Scale bar = 5 µm. Panel B and C are adapted fromreference9. Please click here to view a larger version of this figure.
Discussion
This paper reports a method for microscopy imaging of V. parahaemolyticus swarmer cells, followed by downstream analyses intended to resolve and quantify the subcellular localization and other features of the studied fluorescently labeled proteins. Although several tools exist for microscopy image processing and analysis14,15,16,17,18, the analysis of V. parahaemolyticus swarming cells remains challenging mostly because they normally exist within a dense cell community and there is a high variation in cell length (Figure 2). Therefore, automated cell detection is sometimes faulty, and thus the results somewhat unreliable. This protocol outlines a pipeline designed to grow, image and analyze swarming cells of V. parahaemolyticus, which is tailored to the specific needs of these cells, such as a controlled growth environment on a surface and downstream image analyses that accommodate the presence of cells of variable lengths which grow in close proximity.
The ability to ectopically express a protein of interest from a plasmid can be important for several experiments, e.g. expression of fluorescently labeled proteins for microscopy studies or complementation of gene deletions. Here the process of preparing chemically competent cells and electroporation of plasmid DNA into V. parahaemolyticus is also described. The advantage of electroporation over conjugation, which is often used for V. parahaemolyticus, is that the plasmid backbone does not need to be modified to permit conjugation. Nevertheless, some precautions should be taken; as described in step 1.2, it is very important to reach a cell density of OD600 = 1.0. At lower optical densities cells will lyse upon electroporation and at higher densities the plasmid is not taken up by the cells. Furthermore, too harsh centrifugation of V. parahaemolyticus cells (steps 1.4-1.6) will drastically decrease electroporation efficiency. It is also important to carefully re-suspend the cell-pellet during the washing steps to generate electro-competent cells.
Although the preparation of a cell culture sounds trivial, culture density is very important for swarming performance. Generally, a cell culture in the early exponential growth phase (OD600 <0.5) will produce a larger swarming colony during overnight incubation than a culture in late exponential or early stationary phase. Depending on the experiment, a larger swarming colony might be beneficial. However, to compare colony expansion and morphology between different strains grown on the same HI agar swarming plate, it is important to have enough space available between individual swarming colonies. This can be achieved by spotting a cell culture of higher optical density which will usually result in smaller swarming colonies.
Induction of the swarming phenotype varies significantly among bacterial species and the reproducibility hereof is highly influenced by factors such as agar and nutrient composition, the ambient temperature, and the moisture content of both surroundings and growth medium. Consequently, minor changes in protocol and external environment significantly influence swarm assay results and induction of swarmer differentiation. Protocols for inducing swarming of other bacterial species such as Pseudomonas aeruginosa and Bacillus subtilis have been reported19. The steps described in this protocol will reliably produce the swarmer cell-type of V. parahaemolyticus and maintain them in the form of swarming colonies that produce single layered swarming flares at the edge of the colony. To perform stable and reproducible experiments on the swarmer cell-type of V. parahaemolyticus, preparation of the HI swarming agar plates is the most important part of this protocol. Particularly, the drying step might require optimization, as every incubator will perform differently. Another factor that can influence swarming behavior is the type of plastics used in the Petri dishes. The typical Petri dishes used in our laboratory to perform swarming experiments are made from polystyrene (PS). However, using Petri dishes made out of low-density polyethylene (PE LD) produced mixed results in swarming assays. It is crucial to follow the described steps, because only then reproducible swarming colonies that show distinct peripheral flare formation will form, which enable single swarmer cell microscopy.
Undoubtedly, different approaches can be taken to display the results rendered by fluorescent microscopy studies of V. parahaemolyticus swarming cells. For instance, the widely used program MicrobeTracker16 represents a plausible alternative. The recently available MicrobeTracker application Oufti20, which does not require MATLAB, is also another alternative. However, the use of differential interface contrast (DIC) images is not possible in Oufti, and phase contrast imaging is not suitable to discern cell segmentation in swarmer cells. Furthermore, not all microscopes are equipped for phase contrast imaging and in turn phase contrast imaging is not suited for all kinds of microscopy analysis. Another promising tool, which is free and ImageJ-based, is MicrobeJ17, but yet again only phase contrast and fluorescent images can be used. Furthermore, this protocol only allows for imaging and analysis of cells at specific time-points of their life-cycle. However, an excellent protocol describing time-lapse imaging and the subsequent image analysis, which allows for following single cell trajectories and protein dynamics21 can likely also be applied to V. parahaemolyticus.
Following the steps described in this protocol will enable scientists to perform reliable and re-producible single cell fluorescence microscopy on both the swimmer and swarmer cell-type of V. parahaemolyticus. This protocol details a robust way to transform V. parahaemolyticus from its swimmer into its swarmer cell-type. By imprinting the monolayer of swarmer cells from the peripheral flares of swarming colonies onto an agarose pad, the intracellular localization of proteins can be observed, the pipeline for image analysis can likely be applied to other bacterial species that exist within bacterial communities where cells grow in very close proximity and situations where automated image analysis might not be to an advantage.
開示
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Max Planck Society.
Materials
| Media components | |||
| LB-medium | Roth | X968.3 | |
| Difco Agar, granulated | BD | 214510 | For preparation of LB agar with LB-medium |
| Difco Heart Infusion Agar | BD | 244400 | For preparation of HI agar swarming plates |
| Agarose NEEO, ultra quality | Roth | 2267.3 | For preparation of microscopy agarose pads |
| Name | Company | Catalog Number | コメント |
| Additives | |||
| L-arabinose | Roth | 5118.3 | Used to induce pBAD derivatives |
| 2,2`-Bipyridyl | Sigma Aldrich | D216305-25G | For preparation of HI agar swarming plates |
| Calcium chloride dihydrate | Roth | 5239.1 | For preparation of HI agar swarming plates |
| Ampicillin sodium salt | Roth | K029.3 | |
| Chloramphenicol | Roth | 3886.3 | |
| PBS buffer | – | – | Standard recipe for Phosphate-buffered saline |
| D(+) Sucrose | Roth | 4621.1 | For preparation of electrocompetent cells |
| Glycerol | Roth | 3783.5 | For preparation of electrocompetent cells |
| Name | Company | Catalog Number | コメント |
| Hardware | |||
| Petri dish 150x20mm with cams | Sarstedt | 82.1184.500 | For preparation of HI agar swarming plates |
| kelvitron t (B 6420) | Heraeus | – | Used for drying HI agar swarming plates |
| Gene Pulser Xcell Microbial System | BioRad | 1652662 | Used for elctroporation of plasmid DNA |
| Name | Company | Catalog Number | コメント |
| Software | |||
| MetaMorph Offline (version 7.7.5.0) | Molecular Devices | – | Used for microscopy image analysis |
| Excel | Microsoft | – | Used for microscopy data analsis |
参考文献
- McCarter, L., Hilmen, M., Silverman, M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 54 (3), 345-351 (1988).
- Kawagishi, I., Imagawa, M., Imae, Y., McCarter, L., Homma, M. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20 (4), 693-699 (1996).
- Ulitzur, S. Induction of swarming in Vibrio parahaemolyticus. Arch. Microbiol. 101 (4), 357-363 (1974).
- Ulitzur, S. Effect of temperature, salts, pH, and other factors on the development of peritrichous flagella in Vibrio alginolyticus. Arch. Microbiol. 104 (3), 285-288 (1975).
- Ulitzur, S. The mechanism of swarming of Vibrio alginolyticus. Arch. Microbiol. 104 (1), 67-71 (1975).
- Gode-Potratz, C. J., Chodur, D. M., McCarter, L. L. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J. Bacteriol. 192 (22), 6025-6038 (2010).
- McCarter, L., Silverman, M. Iron regulation of swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 171 (2), 731-736 (1989).
- Gode-Potratz, C. J., Kustusch, R. J., Breheny, P. J., Weiss, D. S., McCarter, L. L. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79 (1), 240-263 (2011).
- Heering, J., Ringgaard, S. Differential Localization of Chemotactic Signaling Arrays during the Lifecycle of Vibrio parahaemolyticus. Front Microbiol. 7 (November), 1-12 (2016).
- Cameron, T. A., Anderson-Furgeson, J., Zupan, J. R., Zik, J. J., Zambryski, P. C. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio. 5 (3), (2014).
- Development Core Team, R. F. F. S. C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 1, 2673 (2008).
- Ringgaard, S., et al. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc Natl Acad Sci U S A. 111 (2), E255-E264 (2014).
- Ringgaard, S., Schirner, K., Davis, B. M., Waldor, M. K. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 25, 1544-1555 (2011).
- Vischer, N. O. E., et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front Microbiol. 6 (JUN), (2015).
- Schneider, C. a., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9 (7), 671-675 (2012).
- Sliusarenko, O., Heinritz, J., Emonet, T., Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80 (3), 612-627 (2011).
- Ducret, A., Quardokus, E. M., Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
- Carpenter, A. E., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7 (10), R100 (2006).
- Morales-Soto, N., et al. Preparation, Imaging, and Quantification of Bacterial Surface Motility Assays. J. Vis. Exp. (98), e52338 (2015).
- Paintdakhi, A., et al. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 99 (4), 767-777 (2016).
- de Jong, I. G., Beilharz, K., Kuipers, O. P., Veening, J. -. W. Live Cell Imaging of Bacillus subtilis and Streptococcus pneumoniae using Automated Time-lapse Microscopy. J. Vis. Exp. (53), e1-e6 (2011).

