概要
Leucine Rich Repeat Kinase 2 is a large multidomain kinase, mutations in which are the most common genetic cause of Parkinson’s disease. Analysis of the kinase activity of this protein has proven to be a crucial tool in understanding the biology and dysfunction of this protein. In this paper, in vitro assaying of the kinase activity of LRRK2 and a selection of its mutants is described, providing an experimental system to examine phosphorylation of putative substrates and potential dysfunction of LRRK2 in disease.
Abstract
Leucine Rich Repeat Kinase 2 (LRRK2) is a 2527 amino acid member of the ROCO family of proteins, possessing a complex, multidomain structure including a GTPase domain (termed ROC, for Ras of Complex proteins) and a kinase domain1. The discovery in 2004 of mutations in LRRK2 that cause Parkinson’s disease (PD) resulted in LRRK2 being the focus of a huge volume of research into its normal function and how the protein goes awry in the disease state2,3. Initial investigations into the function of LRRK2 focused on its enzymatic activities4-6. Although a clear picture has yet to emerge of a consistent alteration in these due to mutations, data from a number of groups has highlighted the importance of the kinase activity of LRRK2 in cell death linked to mutations7,8. Recent publications have reported inhibitors targeting the kinase activity of LRRK2, providing a key experimental tool9-11. In light of these data, it is likely that the enzymatic properties of LRRK2 afford us an important window into the biology of this protein, although whether they are potential drug targets for Parkinson’s is open to debate.
A number of different approaches have been used to assay the kinase activity of LRRK2. Initially, assays were carried out using epitope tagged protein overexpressed in mammalian cell lines and immunoprecipitated, with the assays carried out using this protein immobilised on agarose beads4,5,7. Subsequently, purified recombinant fragments of LRRK2 in solution have also been used, for example a GST tagged fragment purified from insect cells containing residues 970 to 2527 of LRRK212. Recently, Daniëls et al. reported the isolation of full length LRRK2 in solution from human embryonic kidney cells, however this protein is not widely available13. In contrast, the GST fusion truncated form of LRRK2 is commercially available (from Invitrogen, see table 1 for details), and provides a convenient tool for demonstrating an assay for LRRK2 kinase activity. Several different outputs for LRRK2 kinase activity have been reported. Autophosphorylation of LRRK2 itself, phosphorylation of Myelin Basic Protein (MBP) as a generic kinase substrate and phosphorylation of an artificial substrate – dubbed LRRKtide, based upon phosphorylation of threonine 558 in Moesin – have all been used, as have a series of putative physiological substrates including α-synuclein, Moesin and 4-EBP14-17. The status of these proteins as substrates for LRRK2 remains unclear, and as such the protocol described below will focus on using MBP as a generic substrate, noting the utility of this system to assay LRRK2 kinase activity directed against a range of potential substrates.
Protocol
Safety
The protocol described below utilizes ATP labelled with radioactive 32P at the γ phosphate position to follow the kinase activity of LRRK2. It is based on the standard protocols used in our laboratory, and so with regard to many of the steps in the process such as running the gels, western blotting et cetera the materials and precise conditions should be taken as a guide as the equipment and protocols used for these processes varies from laboratory to laboratory. Compounds containing isotopes that emit ionising radiation are potentially harmful to human health and strict licensing and regulations at an institutional and national level control their use. The experiments in this protocol were carried out following training in open source radiation use at University College London and following the good laboratory practise guidelines provided by the safety services at the college (guidelines available at http://www.ucl.ac.uk/estates/safetynet/training/). Use of open source radiation should not be attempted prior to appropriate training and regulatory approval. The regulatory body responsible for open source radiation in laboratory research varies from country to country. Examples of these are: in the United Kingdom, the Health and Safety Executive (http://www.hse.gov.uk/radiation/ionising/index.htm), in the United States the Nuclear Regulatory Commission ( http://www.nrc.gov/materials/miau/regs-guides-comm.html), in Canada the Canadian Nuclear Safety Commission (http://nuclearsafety.gc.ca/eng/), and in Germany Das Bundesamt für Strahlenschutz (http://www.bfs.de/de/bfs). Users in other countries should confirm local rules, regulations and licensing authorities with their radiation safety officer. Safety precautions relevant to this protocol have been noted in the text, highlighted with the radioactive trefoil symbol ( ).
).
1. Preparing the kinase reactions
 All reaction mixtures prepared in 1.5ml sample tubes with screw caps containing an O ring to prevent spread of radioactivity.
All reaction mixtures prepared in 1.5ml sample tubes with screw caps containing an O ring to prevent spread of radioactivity.
- Thaw protein on ice – LRRK2 wild type, D1994A, G2019S.
- make up reaction on ice – 10nM LRRK2, 0.5μg/μl MBP, 5ul of 10x kinase buffer, made up to 50μl with water.
2. Running the assay
 All steps utilizing 32P ATP should take place in designated radiation areas.
All steps utilizing 32P ATP should take place in designated radiation areas.
 Suitable personal protective equipment should be worn – under standard operating procedure in our laboratory these include lab coat, double gloves and protective goggles.
Suitable personal protective equipment should be worn – under standard operating procedure in our laboratory these include lab coat, double gloves and protective goggles.
 Samples containing 32P ATP should be shielded from users by 6mm Perspex screens to minimize exposure.
Samples containing 32P ATP should be shielded from users by 6mm Perspex screens to minimize exposure.
 Where applicable, personal monitoring devices should be used – within UCL, any certified open source radiation user must have a film badge to monitor radiation exposure during experiments.
Where applicable, personal monitoring devices should be used – within UCL, any certified open source radiation user must have a film badge to monitor radiation exposure during experiments.
 All experimental surfaces should be assessed for radioactive contamination before and after use using a Geiger counter.
All experimental surfaces should be assessed for radioactive contamination before and after use using a Geiger counter.
 All potentially contaminated consumables should be disposed of in strict adherence to institutional guidelines for radioactive waste disposal.
All potentially contaminated consumables should be disposed of in strict adherence to institutional guidelines for radioactive waste disposal.
- Prior to starting assay, set heating blocks to 30°C and 100°C respectively.
- Remove 32P ATP from -20 freezer (note that storage conditions for 32P ATP may vary depending on supplier or type of radionucleotides used).
 scan outside of container prior to use, thaw behind perspex screen.
scan outside of container prior to use, thaw behind perspex screen. - With reactions on ice, add 1μl of 32P ATP to each along with 10μM of cold ATP.
- Mix well with pipette.
 Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination.
Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination. - Remove 15μl aliquot for zero time point and terminate reaction in aliquot by addition of 5μl of 4x SDS sample buffer and denaturation at 100°C for 10 minutes.
 Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination.
Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination. - Remaining sample placed in heating block and incubated at 30°C for 60 minutes.
- 15μl removed at 60 minute time point and reaction terminated by addition of 5μl of 4x SDS sample buffer and denaturation at 100°C for 10 minutes.
 Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination.
Pulse centrifuge to bring liquid to bottom of tube, minimizing risk of contamination.
3. Immunoblotting the samples and analysing results
- Samples run on SDS-PAGE
- 10 well 4-12% Bis-tris polyacrilamide gel prepared for electrophoresis using MOPS running buffer.
- 20μl of each sample loaded onto gel along with 7μl of sharpstain protein standard ladder.
- Gel run at 160v for 90 minutes, or until dye front has reached the end of the gel.
 All liquid in contact with radioisotopes should be treated as radioactive waste and disposed of as per institutional guidelines. UCL regulations state that liquid radioactive waste should be discarded by pouring down designated radioactive disposal sinks with copious water.
All liquid in contact with radioisotopes should be treated as radioactive waste and disposed of as per institutional guidelines. UCL regulations state that liquid radioactive waste should be discarded by pouring down designated radioactive disposal sinks with copious water.
- Protein transferred to PVDF membrane via western blot.
- Transfer buffer prepared, 1x Tris Glycine plus 20% Methanol. PVDF Membrane and filter paper cut to correct size for gel and activated with glacial methanol in the case of the membrane or pre-wet with transfer buffer in the case of the filter paper. The membrane should be pre-labelled with ballpoint ink to allow identification of orientation.
 Gel removed from plastic casing and excess acrylamide removed and disposed of as radioactive waste.
Gel removed from plastic casing and excess acrylamide removed and disposed of as radioactive waste. - The gel should be formed into a sandwich with the membrane and filter paper, ensuring that no bubbles exist between the membrane and gel, and then arranged in the western blot apparatus with the membrane between the gel and the anode.
- Transfer carried out at 25V for 16 hours.
- Transfer buffer prepared, 1x Tris Glycine plus 20% Methanol. PVDF Membrane and filter paper cut to correct size for gel and activated with glacial methanol in the case of the membrane or pre-wet with transfer buffer in the case of the filter paper. The membrane should be pre-labelled with ballpoint ink to allow identification of orientation.
- Following protein transfer to PVDF, the membrane should be dried at room temperature. Finally, the dry membrane should be isolated between cellulose acetate sheets and exposed to either a phosphor screen or x-ray film to allow detection of radiolabelled protein. Exposure time can run from several hours to over a week depending on the specific activity of the radioisotope used and the enzyme kinetics of the reaction.
- Phosphoscreen scanned/film processed. If using a phosphor screen, the image should be saved as a high-resolution TIFF or bitmap file, if using film then the resultant transparency should be scanned using a desktop scanner and saved as a high-resolution TIFF or bitmap file. The image can then be analyzed in ImageJ, a freeware program available on the National Institutes of Health website (http://rsbweb.nih.gov/ij/).
4. Representative Results
Figure 1 shows representative results for an assay carried out using wild type, G2019S and D1994A LRRK2 with Myelin Basic Protein as a generic phosphate acceptor substrate. Autophosphorylation of LRRK2 is visible at ≈ 200kDa, with multiple bands representing phosphorylated MBP visible from 20-40kDa. Note the absence of autophosphorylation in the D1994A (kinase dead) lane, and increased phosphorylation due to the G2019S mutation. Note also residual phosphorylation of MBP in the kinase dead lane. This may be due to incomplete ablation of the kinase activity of LRRK2 by the D1994A mutant or reflect the presence of trace contaminating kinases in the reaction.
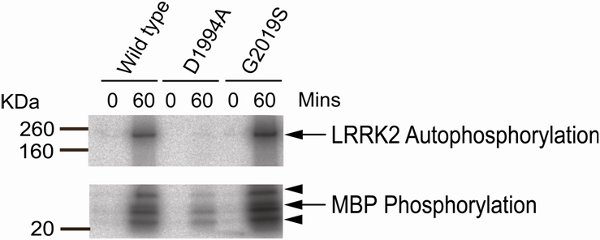
Figure 1.
Discussion
This paper describes a basic protocol for assaying the kinase activity of LRRK2 using an in vitro system. In the interests of brevity, this has been limited to a one-hour end point template using a generic substrate, but the general protocol is applicable to a range of potential substrates and amenable to more sophisticated analyses examining the kinetics of the kinase activity of LRRK2. This highlights one of the key advantages of using an in vitro system to examine the kinase behaviour of a protein such as this: because there is complete control over the concentrations of the enzyme and substrate, it is possible to generate kinetic data and calculate Km and Vmax values for the reaction. For more detailed kinetic evaluations or evaluating the impact of putative inhibitors, a higher throughput system (such as that afforded by using the LRRKtide substrate, available from Invitrogen) is a superior alternative to the approach described here.
It is important, however, to recognise that the reductionist model system provided by in vitro kinase assays is but one tool to examine the biology of a kinase and its relationships with potential substrates. Data from assays such as this should be used in conjunction with other approaches to gain a comprehensive picture of how a kinase behaves in vitro and in a cellular context. For example, a putative substrate phosphorylated in vitro can be analysed by mass spectrometry to identify possible phosphosites that can then be manipulated by targeted mutagenesis (e.g. converting serine or threonine phosphate acceptor residues to none-phosphorylatable residues such as alanine) to examine the functional consequences of phosphorylation ex vivo.
A key consideration in interpreting data from an in vitro assay system such as this is likelihood of either type I or type II (false positive or false negative) errors. The reductionist nature of this system unfortunately disposes it to both – in the case of the former, having a purified putative substrate and purified kinase in artificially close proximity and at high concentration (compared to the cellular milieu) can result in artefactual phosphorylation events. Conversely, many kinases function as part of a complex within the context of the cell, and require co-factors for phosphorylation of a given substrate to occur. As noted above, a positive result from phosphorylation of a putative substrate using an in vitro assay system should then be tested in a cellular system to validate the result, and a negative result should be interpreted with caution.
In light of this, it is critical where possible to have both positive and negative controls to allow a comparative study of the activity of your kinase of interest. Careful selection of a positive control that has a known activity towards your protein of interest, along with a negative control that is unlikely to phosphorylate your putative substrate, is extremely valuable. One candidate control for LRRK2 is Receptor interacting kinase 3, which has a closely related primary sequence to the kinase domain of LRRK2 but has a very different overall domain structure18. This is available as recombinant protein from Invitrogen and is used as a standard control in our laboratory.
It should also be noted that the commercially available form of LRRK2 lacks the N-terminus of the protein and is tagged with Glutathione-s-transferase and this should be taken into consideration as a potential confounding factor when carrying out kinase assays with this protein, as it is not known what role the N-terminal of LRRK2 may play in the normal function of this protein. If, for example, the N-terminal portion of LRRK2 that is absent in the protein used in this protocol is critical for recruitment of a specific substrate to the kinase domain of LRRK2 then this will have a major impact on the observed phosphorylation of said substrate in the in vitro system described above.
Even with these caveats, however, the importance attached to biology of LRRK2 in Parkinson’s research highlights the utility of this protocol as a valuable tool for investigating the behaviour of this protein in an in vitro setting.
開示
The authors have nothing to disclose.
Acknowledgements
The author is a Parkinson’s UK Research Fellow (grant F1002). This work was supported in part by the Wellcome Trust/MRC Joint Call in Neurodegeneration award (WT089698) to the UK Parkinson’s Disease Consortium (UKPDC) whose members are from the UCL Institute of Neurology, the University of Sheffield and the MRC Protein Phosphorylation Unit at the University of Dundee.
Materials
| Reagent | Company | Catalogue Number | Comment |
| LRRK2 wild type | Invitrogen | PV4873 | |
| LRRK2 G2019S | Invitrogen | PV4881 | |
| LRRK2 D1994A | Invitrogen | PV6051 | |
| Kinase buffer | Cell Signalling | 9802S | |
| MBP | Sigma | M1891 | |
| 32P g ATP | Perkin Elmer | BLU002X500UC | 500µCi, 30Ci /mMole |
| 4x LDS sample buffer | Invitrogen | NP0007 | |
| 2-Mercaptoethanol | Sigma | M6250 | |
| Protein standards | Invitrogen | LC5800 | |
| MOPS running buffer | Invitrogen | NP0001 | |
| Bis-tris acrylamide gel | Invitrogen | NP0321 | 4-12% 1mm 10well |
| PVDF membrane | Millipore | IPVH00010 | |
| Transfer buffer | Invitrogen | NP0006 |
Table 1. Reagents
Distilled and de-ionized water was used for all dilution steps
| Equipment type | Company | Comment |
| Geiger counter | Mini Instruments | |
| SDS PAGE tank | Invitrogen | |
| Transfer tank | Invitrogen | |
| Heat blocks (2) | Eppendorf | |
| Phosphor screen | GE | X-ray film can be substituted |
| Phosphor imager | GE | |
| Exposure cassette | GE | |
| Centrifuge | Eppendorf | |
| Perspex shielding | ||
| 1.5ml tubes | VWR | Screw top with O ring |
Table 2. Equipment
参考文献
- Lewis, P. A. The function of ROCO proteins in health and disease. Biol. Cell. 101, 183-191 (2009).
- Paisan-Ruiz, C. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 44, 595-600 (2004).
- Zimprich, A. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 44, 601-607 (2004).
- West, A. B. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U. S. A. 102, 16842-16847 (2005).
- Gloeckner, C. J. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 15, 223-232 (2006).
- Lewis, P. A. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357, 668-671 (2007).
- Greggio, E. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 23, 329-341 (2006).
- Smith, W. W. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 9, 1231-1233 (2006).
- Deng, X. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat. Chem. Biol. 7, 203-205 (2011).
- Lee, B. D. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat. Med. 16, 998-1000 (2010).
- Nichols, R. J. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. The Biochemical Journal. 424, 47-60 (2009).
- Anand, V. S. Investigation of leucine-rich repeat kinase 2: enzymological properties and novel assays. FEBS. J. 276, 466-478 (2009).
- Daniels, V. Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. Journal of Neurochemistry. 116, 304-315 (2011).
- Jaleel, M. LRRK2 phosphorylates moesin at Thr558; characterisation of how Parkinson’s disease mutants affect kinase activity. Biochem. J. , (2007).
- Qing, H., Wong, W., McGeer, E. G., McGeer, P. L. Lrrk2 phosphorylates alpha synuclein at serine 129: Parkinson disease implications. Biochem. Biophys. Res. Commun. 387, 149-152 (2009).
- Imai, Y. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. Embo. J. 27, 2432-2443 (2008).
- Greggio, E. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 283, 16906-16914 (2008).
- Manning, G., Whyte, D. B., Martinez, R., Hunter, T., Sudarsanam, S. The protein kinase complement of the human genome. Science. 298, 1912-1934 (2002).

