Assaying for Inorganic Polyphosphate in Bacteria
Summary
We describe a simple method for rapid quantification of inorganic polyphosphate in diverse bacteria, including Gram-negative, Gram-positive, and mycobacterial species.
Abstract
Inorganic polyphosphate (polyP) is a biological polymer found in cells from all domains of life, and is required for virulence and stress response in many bacteria. There are a variety of methods for quantifying polyP in biological materials, many of which are either labor-intensive or insensitive, limiting their usefulness. We present here a streamlined method for polyP quantification in bacteria, using a silica membrane column extraction optimized for rapid processing of multiple samples, digestion of polyP with the polyP-specific exopolyphosphatase ScPPX, and detection of the resulting free phosphate with a sensitive ascorbic acid-based colorimetric assay. This procedure is straightforward, inexpensive, and allows reliable polyP quantification in diverse bacterial species. We present representative polyP quantification from the Gram-negative bacterium (Escherichia coli), the Gram-positive lactic acid bacterium (Lactobacillus reuteri), and the mycobacterial species (Mycobacterium smegmatis). We also include a simple protocol for nickel affinity purification of mg quantities of ScPPX, which is not currently commercially available.
Introduction
Inorganic polyphosphate (polyP) is a linear biopolymer of phosphoanhydride-linked phosphate units that is found in all domains of life1,2,3. In diverse bacteria, polyP is essential for stress response, motility, biofilm formation, cell cycle control, antibiotic resistance, and virulence4,5,6,7,8,9,10,11. Studies of polyP metabolism in bacteria therefore have the potential to yield fundamental insights into the ability of bacteria to cause disease and thrive in diverse environments. In many cases, however, the methods available for quantifying polyP in bacterial cells are a limiting factor in these studies.
There are several methods currently used to measure polyP levels in biological materials. These methods typically involve two distinct steps: extracting polyP and quantifying the polyP present in those extracts. The current gold standard method, developed for the yeast Saccharomyces cerevisiae by Bru and colleagues12, extracts polyP along with DNA and RNA using phenol and chloroform, followed by ethanol precipitation, treatment with deoxyribonuclease (DNase) and ribonuclease (RNase), and digestion of the resulting purified polyP with the S. cerevisiae polyP-degrading enzyme exopolyphosphatase (ScPPX)13 to yield free phosphate, which is then quantified using a malachite green-based colorimetric assay. This procedure is highly quantitative but labor-intensive, limiting the number of samples that can be processed in a single experiment, and is not optimized for bacterial samples. Others have reported extracting polyP from a variety of cells and tissues using silica beads ("glassmilk") or silica membrane columns6,14,15,16,17,18. These methods do not efficiently extract short chain polyP (less than 60 phosphate units)12,14,15, although this is of less concern for bacteria, which are generally thought to synthesize primarily long-chain polyP3. Older methods of polyP extraction using strong acids19,20 are no longer widely used, since polyP is unstable under acidic conditions12.
There are also a variety of reported methods for quantifying polyP. Among the most common is 4′,6-diamidino-2-phenylindole (DAPI), a fluorescent dye more typically used to stain DNA. DAPI-polyP complexes have different fluorescence excitation and emission maxima than DAPI-DNA complexes21,22, but there is considerable interference from other cellular components, including RNA, nucleotides, and inositol phosphates12,15,16,23, reducing the specificity and sensitivity of polyP measurements made using this method. Alternatively, polyP and adenosine diphosphate (ADP) can be converted into adenosine triphosphate (ATP) using purified Escherichia coli polyP kinase (PPK) and the resulting ATP quantified using luciferase14,17,18. This allows the detection of very small amounts of polyP, but requires two enzymatic reaction steps and both luciferin and very pure ADP, which are expensive reagents. ScPPX specifically digests polyP into free phosphate6,12,13,24, which can be detected using simpler methods, but ScPPX is inhibited by DNA and RNA12, necessitating DNase and RNase treatment of polyP-containing extracts. Neither PPK nor ScPPX are commercially available, and PPK purification is relatively complex25,26.
PolyP in cell lysates or extracts can also be visualized on polyacrylamide gels by DAPI negative staining27,28,29,30, a method that does allow assessment of chain length, but is low-throughput and poorly quantitative.
We now report a fast, inexpensive, medium-throughput polyP assay that allows rapid quantification of polyP levels in diverse bacterial species. This method begins by lysing bacterial cells at 95 °C in 4 M guanidine isothiocyanate (GITC)14 to inactivate cellular phosphatases, followed by a silica membrane column extraction optimized for rapid processing of multiple samples. The resulting polyP-containing extract is then digested with a large excess of ScPPX, eliminating the need for DNase and RNase treatment. We include a protocol for straightforward nickel affinity purification of mg quantities of ScPPX. Finally, polyP-derived free phosphate is quantified with a simple, sensitive, ascorbic acid-based colorimetric assay24 and normalized to total cellular protein. This method streamlines the measurement of polyP in bacterial cells, and we demonstrate its use with representative species of Gram-negative bacteria, Gram-positive bacteria, and mycobacteria.
Protocol
1. Purifying Yeast Exopolyphosphatase (ScPPX)
- Transform the E. coli protein overexpression strain BL21(DE3)31 with plasmid pScPPX26 by electroporation32 or chemical transformation33.
- Inoculate 1 L of lysogeny broth (LB) containing 100 µg mL-1 ampicillin in a 2 L unbaffled flask with a single colony of BL21(DE3) containing pScPPX2 and incubate overnight at 37 °C without shaking, to an absorbance at 600 nm (A600) of approximately 0.3, as measured in a spectrophotometer.
- Start the culture shaking (180 rpm) and incubate for 30 min at 37 °C, during which time the cells will grow to an A600 of 0.4 – 0.5.
- Add isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and an additional 100 µg mL-1 ampicillin, then incubate for 4 h at 37 °C with shaking at 180 rpm.
NOTE: This overexpression protocol generates a large amount of soluble ScPPX. However, ScPPX overexpression is very forgiving, and a variety of other common protein overexpression protocols have been used to successfully purify ScPPX. - Transfer the cells to a 1 L centrifuge bottle and harvest them by centrifuging for 20 min at 5000 x g at 4 °C. Remove the supernatant and transfer the cell pellet to a 50 mL conical tube.
NOTE: Cell pellets can be stored indefinitely at -80 °C. - Thaw the pellet on ice, then resuspend it in 9 mL of 50 mM HEPES, 0.5 M NaCl, and 5 mM imidazole (pH 8) for a total volume of about 10 mL.
- Add (final concentrations) 1 mg mL-1 lysozyme, 2 mM MgCl2, and 50 units mL-1 of an RNA- and DNA-degrading endonuclease (see Table of Materials) and incubate for 30 min on ice.
NOTE: ScPPX binds nucleic acids (data not shown), so nuclease treatment is essential during purification. - Using a sonicator with a microtip (see Table of Materials), lyse the cells by 2 cycles of sonication on ice at 50% power, pulsing 5 s on and 5 s off, with a 2 min rest between cycles.
NOTE: Use appropriate hearing protection during sonication. Similarly to the case with overexpression, a variety of cell lysis methods are acceptable for ScPPX purification. - Remove insoluble debris by centrifuging the lysate for 20 min at 20,000 x g at 4 °C. Filter the resulting supernatant (approximately 10 mL) through a 0.8 µm pore size cellulose acetate syringe filter.
- Load the lysate onto a nickel-charged 5 mL chelating column (see Table of Materials) using either a peristaltic pump or a large syringe.
- Rinse the column with 50 mL of 50 mM HEPES, 0.5 M NaCl, 5 mM imidazole (pH 8).
- Rinse the column with 50 mL of 50 mM HEPES, 0.5 M NaCl, 20 mM imidazole (pH 8).
- Elute ScPPX with 50 mM HEPES, 0.5 M NaCl, and 0.5 M imidazole (pH 8), collecting 15 1-mL fractions. Test these elution fractions for protein content with the Bradford protein assay34 (see Table of Materials) and run an SDS-PAGE gel35 to confirm which fractions contain purified ScPPX (molecular weight = 45 kDa).
- Pool the fractions containing pure ScPPX and adjust the concentration of the pooled protein to about 2 mg mL-1 with 50 mM HEPES and 0.5 M NaCl. Higher protein concentrations may precipitate in the next step.
- Exchange the purified ScPPX into storage buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10% [v/v] glycerol) by dialysis. Place pooled ScPPX fractions in a sealed length of 12,000 – 14,000 Da molecular weight cutoff dialysis membrane tubing (see Table of Materials) and suspend in 1 L of storage buffer with continuous stirring at 4 °C for at least 4 h. Repeat this step with fresh storage buffer for a total of 3 buffer changes.
- Transfer the purified, dialyzed protein to a centrifuge tube or tubes and centrifuge for 20 min at 20,000 x g at 4 °C to remove any insoluble aggregates. Carefully transfer the supernatant to a clean 15 mL conical tube.
- Adjust the concentration of the purified ScPPX to 1 mg mL-1 with storage buffer, add 0.1% (w/v) protease-free bovine serum albumin (BSA; final concentration), and store for up to 6 months at 4 °C.
NOTE: ScPPX loses more than 90% of its activity when it is frozen13.
2. Harvesting Samples for Polyphosphate Extraction
- Grow bacteria under the conditions of interest for determining polyP content. For this protocol, grow Lactobacillus reuteri36 overnight at 37 °C without shaking in malic enzyme induction (MEI) medium37 without cysteine (MEI-C).
- Harvest enough cells by centrifugation in a 1.5 mL microcentrifuge tube to total 50 – 100 µg of cellular protein (see step 3 below). For E. coli, this is 1 mL of a log phase culture at an A600 of 0.2 – 0.4. For L. reuteri, this is 1 mL of an overnight culture. Adjust as necessary for other species of interest.
- Completely remove the supernatant from the cell pellets.
- Resuspend the cell pellets in 250 µL of GITC lysis buffer (4 M guanidine isothiocyanate, 50 mM Tris-HCl, pH 7) and lyse by incubation at 95 °C for 10 min. Store lysates at -80 °C.
NOTE: Be consistent with lysis time, since extended incubation at high temperature may result in degradation of polyP by hydrolysis38. Lysates can be stored indefinitely at -80 °C.
CAUTION: Guanidine isothiocyanate is a chaotropic salt and should be handled with gloves and disposed of as hazardous waste.
3. Measuring the Protein Content of Cell Lysates
- Prepare BSA standards containing 0, 0.1, 0.2, and 0.4 mg mL-1 of BSA in GITC lysis buffer.
NOTE: It is important to make the BSA standards in GITC, since GITC influences the color development of the Bradford assay. BSA standards can be stored indefinitely at -20 °C. - Aliquot 5 µL of thawed, well-mixed cell lysates and of BSA standards to separate wells in a clear 96-well plate.
- Add 195 µL of Bradford reagent34 to each well and measure absorbance at 595 nm (A595) in a plate reader (see Table of Materials).
- Calculate the amount of protein in each well by comparison to the BSA standard curve, using the formula y = mx + b, where y is A595, x is µg of BSA, m is the slope of the standard curve, and b is the y-intercept of the standard curve. Multiply the resulting value by 0.05 to determine the total mg of protein in each sample.
4. Extracting Polyphosphate
- Add 250 µL of 95% ethanol to each GITC-lysed sample and vortex to mix.
- Apply that mixture to a silica membrane spin column and centrifuge for 30 s at 16,100 x g.
- Discard the flow-through, then add 750 µL of 5 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, 50% ethanol and centrifuge for 30 s at 16,100 x g.
- Discard the flow-through and centrifuge for 2 min at 16,100 x g.
- Place the column in a clean 1.5 mL microfuge tube and add 150 µL of 50 mM Tris-HCl (pH 8).
- Incubate at room temperature for 5 min, then elute polyP by centrifuging for 2 min at 8,000 x g.
NOTE: If desired, the eluates can be stored at -20 °C for at least 1 week.
5. Digesting Polyphosphate
- Prepare standards containing 0, 5, 50, or 200 µM potassium phosphate in 50 mM Tris-HCl (pH 8).
NOTE: Potassium phosphate standards can be stored indefinitely at room temperature. - Aliquot 100 µL of each phosphate standard and of extracted polyP samples into separate wells of a clear 96-well plate.
- Prepare a master mix containing (per sample): 30 µL of 5x ScPPX reaction buffer (100 mM Tris-HCl, 25 mM MgCl2, 250 mM ammonium acetate, pH 7.5)13, 19 µL of H2O, and 1 µL of purified ScPPX (1 mg mL-1).
- Add 50 µL of the master mix to each well of the 96-well plate. Incubate for 15 min at 37 °C.
NOTE: If desired, the digested polyP samples can be stored at -20 °C indefinitely.
6. Detecting Free Phosphate24
- Prepare detection solution base by dissolving 0.185 g of antimony potassium tartrate in 200 mL of water, adding 150 mL of 4 N H2SO4, then adding 1.49 g of ammonium heptamolybdate. Stir to dissolve and then bring to a final volume of 456 mL. Filter the solution to remove particulates and store protected from light at 4 °C for up to 1 month.
- Prepare a stock solution of 1 M ascorbic acid. Store protected from light at 4 °C for up to 1 month.
- Prepare a fresh working stock of detection solution each day by mixing 9.12 mL of detection solution base with 0.88 mL of 1 M ascorbic acid. Allow the detection solution to come to room temperature before use.
- Add 50 µL of detection solution to each sample and standard in the 96-well plate and incubate at room temperature for about 2 min to allow color development.
- Measure absorbance at 882 nm with a plate reader and calculate the phosphate concentration of each sample by comparison to the potassium phosphate standard curve.
CAUTION: The detection solution contains toxic salts and strong acids. Wear gloves and treat excess solution as toxic waste.
7. Calculating Cellular Polyphosphate Content
- Convert the phosphate concentrations determined in step 6.5 to nanomoles of polyP-derived phosphate in each entire cell lysate according to the following formula:
nmol polyP = 1.5 x (µM phosphate / 10)
NOTE: The standard curve-based method in step 6 determines the concentration of phosphate (in µM) in each 100 µL polyP sample aliquoted in step 5.2. To convert this concentration to a number of nmol, 100 µL is divided by 106 to give a volume in L, multiplied by the concentration (µmol L-1), then multiplied by 1,000 (the number of nmol in a µmol). This reduces to dividing the concentration by 10. The total extract volume prepared in step 4 is 150 µL, so it is necessary to multiply the resulting value by 1.5 to calculate the nmol of phosphate present in the entire extract. - Normalize cellular polyP content to total cellular protein by dividing nmol polyP by the mg total protein in each sample determined in step 3.4. PolyP levels are expressed in terms of the concentration of individual polyP-derived free phosphate.
NOTE: In some cases, the amount of polyP-derived phosphate present in a sample may fall outside the linear range of the phosphate standard curve (step 6.5). If the levels of polyP are very high, the excess polyP-containing eluate from step 4.6 can be diluted 1:10 or 1:100, as necessary, and then measured again as described in steps 5 through 7.
Representative Results
The key steps of the protocol are diagrammed in simplified form in Figure 1.
To demonstrate the use of this protocol with Gram-negative bacteria, wild-type E. coli MG165539 was grown to mid-log phase in LB rich medium at 37 °C with shaking (200 rpm), then rinsed and incubated for an additional 2 h in morpholinopropanesulfonate-buffered (MOPS) minimal medium40 containing 4 g L-1 glucose and 0.1 mM K2HPO4, a condition known to induce production of polyP14,29. As shown in Figure 2A, we detected no polyP in wild-type E. coli grown in LB and 192 ± 14 (mean ± standard deviation [SD]) nmol polyP mg-1 total protein in MOPS, consistent with previous reports14,29. As expected, a ∆ppk mutant6, which lacks polyP kinase41, produced no polyP in either medium, a ∆ppx mutant6, which lacks exopolyphosphatase42, produced approximately the same amount of polyP as the wild-type, and a ∆phoB mutant29, which is defective in phosphate transport, produced significantly less polyP than the wild-type14,29,43.
To demonstrate the use of this protocol with Gram-positive bacteria, wild-type L. reuteri ATCC PTA 647536 and an isogenic ppk1 null mutant lacking polyP kinase, constructed using oligo-directed recombineering44, were grown overnight in MEI-C at 37 °C without shaking. As shown in Figure 2B, the wild-type accumulated 51 ± 6 nmol polyP mg-1 total protein under these conditions, while the ppk1 mutant contained less than half this amount. L. reuteri contains a second polyP kinase, encoded by the ppk2 gene45, which presumably accounts for the polyP present in the ppk1 null mutant.
To demonstrate the use of this protocol with mycobacteria, Mycobacterium smegmatis strain SMR546 was grown to mid-log phase in Hartmans-de Bont (HdB) medium47, then rinsed and diluted five-fold in HdB or HdB with 2% ethanol. These cultures were then incubated overnight at 37 °C with shaking (180 rpm). As shown in Figure 2C, in the absence of ethanol, M. smegmatis accumulated 141 ± 52 nmol polyP mg-1 total protein, while ethanol treatment resulted in a three-fold increase to 437 ± 102 nmol polyP mg-1 total protein. This result was expected, since ethanol has previously been reported to elevate polyP levels in M. smegmatis48.
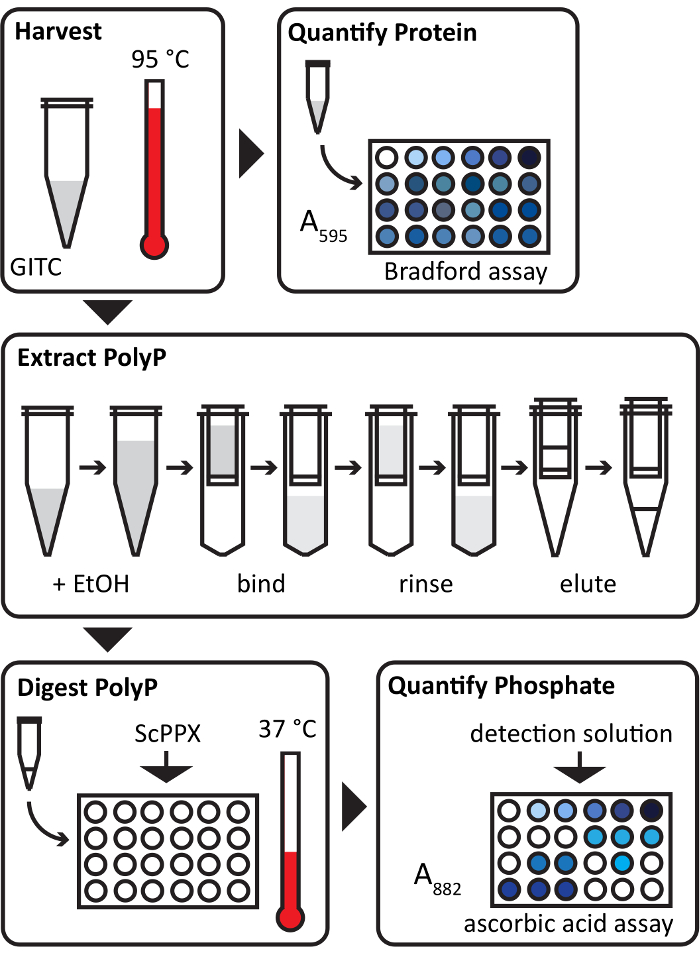
Figure 1: Diagram of polyphosphate extraction and measurement procedure. The essential steps of the polyP quantification protocol are illustrated. Bacterial cell pellets are lysed at 95 °C in GITC (guanidine isothiocyanate). Small aliquots of the lysates are assayed for protein content using the Bradford assay. PolyP is extracted using silica membrane columns and then digested with ScPPX. The resulting free phosphate is quantified using the ascorbic acid assay. Total polyP-derived phosphate is normalized to total protein for each sample. A595 = absorbance at 595 nm; A882 = absorbance at 882 nm. Please click here to view a larger version of this figure.
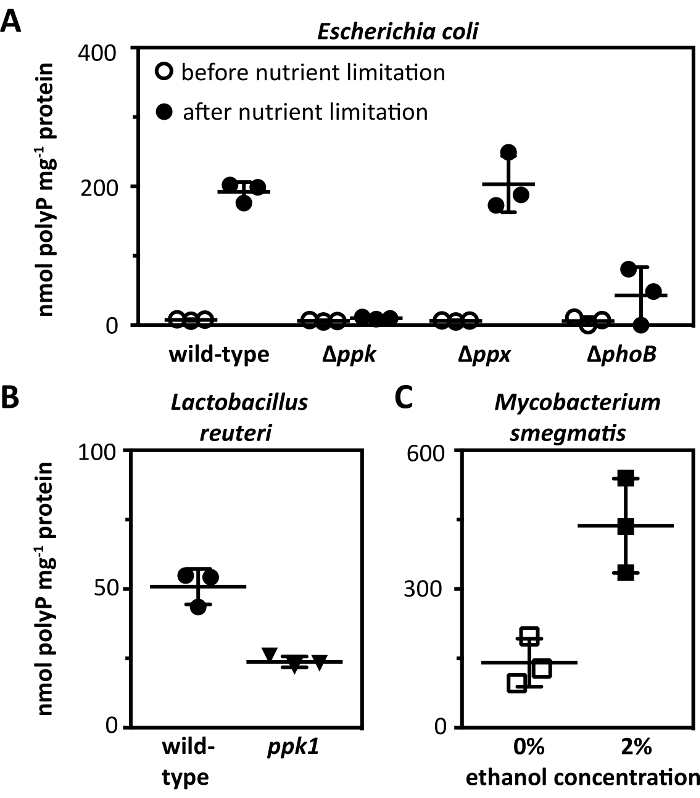
Figure 2: Representative results with diverse bacteria. (A) E. coli MG1655 wild-type and isogenic ∆ppk, ∆ppx, and ∆phoB mutants were grown at 37 °C with shaking (200 rpm) to A600 = 0.2 – 0.4 in rich LB medium (black circles) and then shifted to minimal medium (MOPS containing 4 g L-1 glucose and 0.1 mM K2HPO4) for 2 h (white circles; n = 3, ± SD). (B) L. reuteri ATCC PTA 6475 (circles) and an isogenic ppk1 null mutant (triangles) were grown overnight at 37 °C in MEI-C medium without shaking (n = 3, ± SD). (C) M. smegmatis SMR5 was grown at 37 °C with shaking (180 rpm) to A600 = 1 in HdB medium with (closed squares) or without (open squares) 2% ethanol (n = 3, ± SD). PolyP levels are expressed in terms of the concentration of individual polyP-derived free phosphate. Please click here to view a larger version of this figure.
Discussion
The protocol described here simplifies and accelerates quantification of polyP levels in diverse bacteria, with a typical set of 24 samples taking about 1.5 h to fully process. This permits rapid screening of samples and analysis of mutant libraries, and simplifies kinetic experiments measuring the accumulation of polyP over time. We have demonstrated that the protocol works effectively on representatives of three different phyla: proteobacteria, firmicutes, and actinobacteria, which are notorious for their resilient, difficult to lyse cell walls49.
Detection of polyP by digestion with ScPPX is more specific and more sensitive than fluorescence detection with DAPI, and is cheaper and more technically straightforward than conversion of polyP to ATP using PPK. Although ScPPX is inhibited by DNA and RNA12, we have overcome this by using a very large excess of enzyme (1 µg per sample) to achieve complete digestion even in bacteria containing more than 6,000 nmol polyP mg-1 protein29. Other published methods use 10 to 15 ng of ScPPX per reaction12,24. Our protocol for ScPPX purification typically yields more than 5 mg of pure ScPPX per liter of overexpression culture, which is enough for more than 5,000 assays. We have found that several different protocols for nickel-affinity purification of His-tagged proteins work well to purify ScPPX overexpressed from pScPPX2 (data not shown), and there are a wide variety of such protocols available to suit labs with varying technical capacities.
Previous protocols using ScPPX digestion for polyP detection have often relied on malachite green-based colorimetric assays to quantify free phosphate6,12. These assays are sensitive, but typically require carefully timed sequential addition of two or three separate reagents to make accurate measurements24,50,51. The ascorbic acid-based detection system used here24 has a wider linear range of detection than malachite green with no loss of sensitivity, only involves addition of a single reagent, and does not require precise timing.
There are several steps that are critical to the success of this protocol. First, bacterial samples should be lysed in GITC immediately after harvesting, to ensure that polyP levels do not change after the time of sampling and to rapidly inactivate cellular phosphatases. Second, do not incubate GITC lysates at 95 °C for more than 10 min, and store them immediately afterwards at -80 °C to minimize possible polyP hydrolysis. Third, be careful when pipetting samples into 96-well plates for either protein or phosphate measurements not to splash or drip anything from one sample into another. The colorimetric assays, particularly for phosphate, are very sensitive, and small amounts of contamination can strongly skew the results. Finally, some reagents used in this protocol (i.e. ScPPX, detection solution) have limited shelf lives. Prepare fresh ScPPX every 6 months and fresh detection solution every month.
One limitation of our method is that silica columns do not bind polyP less than 60 phosphate units long very well12,14,15. Although bacteria typically synthesize mostly long-chain polyP3, we recommend preliminary experiments using polyacrylamide gel electrophoresis27,30 to assess chain length in a given bacterial species before using our procedure. For species where short-chain polyP makes up a substantial fraction of the polyP pool, our protocol is not appropriate and we recommend extracting polyP by a different method (e.g., phenol-chloroform extraction12), and would also recommend supplementing ScPPX digestion with commercially available S. cereviseae pyrophosphatase (PPA1)24, since ScPPX does not digest pyrophosphate13 and this has a proportionally higher impact on measurements of shorter-chain polyP. As an alternative to ScPPX, acid hydrolysis38 could be used to hydrolyze polyP to free phosphate, but this would require additional DNase, RNase, and purification steps to ensure that only polyP-derived phosphate was being measured. In some cases, it may be useful to use an alternative extraction method to test whether the efficiency of polyP extraction with GITC varies between strains or conditions, particularly with bacteria known to have thick cell walls, such as mycobacteria. If levels of polyP below the limit of detection of this assay are important for studies of a particular species, it may be necessary to use a different detection scheme, such as using PPK to convert polyP to ATP, which can be detected extremely sensitively using commercially available luciferase-based assays14,17,18.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This project was supported by University of Alabama at Birmingham Department of Microbiology startup funds and NIH grant R35GM124590 (to MJG), and NIH grant R01AI121364 (to FW).
Materials
| E. coli BL21(DE3) | Millipore Sigma | 69450 | |
| plasmid pScPPX2 | Addgene | 112877 | available to academic and other non-profit institutions |
| LB broth | Fisher Scientific | BP1427-2 | E. coli growth medium |
| ampicillin | Fisher Scientific | BP176025 | |
| isopropyl β-D-1-thiogalactopyranoside (IPTG) | Gold Biotechnology | I2481C | |
| HEPES buffer | Gold Biotechnology | H-400-1 | |
| potassium hydroxide (KOH) | Fisher Scientific | P250500 | for adjusting the pH of HEPES-buffered solutions |
| sodium chloride (NaCl) | Fisher Scientific | S27110 | |
| imidazole | Fisher Scientific | O3196500 | |
| lysozyme | Fisher Scientific | AAJ6070106 | |
| magnesium chloride (MgCl2) | Fisher Scientific | BP214-500 | |
| Pierce Universal Nuclease | Fisher Scientific | PI88700 | Benzonase (Sigma-Aldrich cat. # E1014) is an acceptable substitute |
| Model 120 Sonic Dismembrator | Fisher Scientific | FB-120 | other cell lysis methods (e.g. French Press) can also be effective |
| 5 mL HiTrap chelating HP column | GE Life Sciences | 17040901 | any nickel-affinity chromatography column or resin could be substituted |
| nickel(II) sulfate hexahydrate | Fisher Scientific | AC415611000 | for charging HiTrap column |
| 0.8 µm pore size cellulose acetate syringe filters | Fisher Scientific | 09-302-168 | |
| Bradford reagent | Bio-Rad | 5000205 | |
| Tris buffer | Fisher Scientific | BP1525 | |
| Spectrum Spectra/Por 4 RC Dialysis Membrane Tubing 12,000 to 14,000 Dalton MWCO | Fisher Scientific | 08-667B | other dialysis membranes with MWCO < 30,000 Da should also work |
| hydrochloric acid (HCl) | Fisher Scientific | A144-212 | for adjusting the pH of Tris-buffered solutions |
| potassium chloride (KCl) | Fisher Scientific | P217500 | |
| glycerol | Fisher Scientific | BP2294 | |
| 10x MOPS medium mixture | Teknova | M2101 | E. coli growth medium |
| glucose | Fisher Scientific | D161 | |
| monobasic potassium phosphate (KH2PO4) | Fisher Scientific | BP362-500 | |
| dibasic potassium phosphate (K2HPO4) | Fisher Scientific | BP363-500 | |
| dehydrated yeast extract | Fisher Scientific | DF0886-17-0 | |
| tryptone | Fisher Scientific | BP1421-500 | |
| magnesium sulfate heptahydrate | Fisher Scientific | M63-50 | |
| manganese sulfate monohydrate | Fisher Scientific | M113-500 | |
| guanidine isothiocyanate | Fisher Scientific | BP221-250 | |
| bovine serum albumin (protease-free) | Fisher Scientific | BP9703100 | |
| clear flat bottom 96-well plates | Sigma-Aldrich | M0812-100EA | any clear 96-well plate will work |
| Tecan M1000 Infinite plate reader | Tecan, Inc. | not applicable | any plate reader capable of measuring absorbance at 595 and 882 nm will work |
| ethanol | Fisher Scientific | 04-355-451 | |
| silica membrane spin columns | Epoch Life Science | 1910-050/250 | |
| ethylenediaminetetraacetic acid (EDTA) | Fisher Scientific | BP120500 | |
| 1.5 mL microfuge tubes | Fisher Scientific | NC9580154 | |
| ammonium acetate | Fisher Scientific | A637-500 | |
| antimony potassium tartrate | Fisher Scientific | AAA1088922 | |
| 4 N sulfuric acid (H2SO4) | Fisher Scientific | SA818-500 | |
| ammonium heptamolybdate | Fisher Scientific | AAA1376630 | |
| ascorbic acid | Fisher Scientific | AC401471000 |
Riferimenti
- Rao, N. N., Gomez-Garcia, M. R., Kornberg, A. Inorganic polyphosphate: essential for growth and survival. Annual Review of Biochemistry. 78, 605-647 (2009).
- Achbergerova, L., Nahalka, J. Polyphosphate–an ancient energy source and active metabolic regulator. Microbial Cell Factories. 10, 63 (2011).
- Kornberg, A., Rao, N. N., Ault-Riche, D. Inorganic polyphosphate: a molecule of many functions. Annual Review of Biochemistry. 68, 89-125 (1999).
- Albi, T., Serrano, A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World Journal of Microbiology and Biotechnology. 32 (2), 27 (2016).
- Gray, M. J., Jakob, U. Oxidative stress protection by polyphosphate–new roles for an old player. Current Opinion in Microbiology. 24, 1-6 (2015).
- Gray, M. J., et al. Polyphosphate is a primordial chaperone. Molecular Cell. 53 (5), 689-699 (2014).
- Racki, L. R., et al. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 114 (12), E2440-E2449 (2017).
- Rashid, M. H., et al. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 97 (17), 9636-9641 (2000).
- Candon, H. L., Allan, B. J., Fraley, C. D., Gaynor, E. C. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. Journal of Bacteriology. 189 (22), 8099-8108 (2007).
- Richards, M. I., Michell, S. L., Oyston, P. C. An intracellularly inducible gene involved in virulence and polyphosphate production in Francisella. Journal of Medical Microbiology. 57 (Pt 10), 1183-1192 (2008).
- Singh, R., et al. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. Journal of Bacteriology. 195 (12), 2839-2851 (2013).
- Bru, S., Jimenez, J., Canadell, D., Arino, J., Clotet, J. Improvement of biochemical methods of polyP quantification. Microbial Cell. 4 (1), 6-15 (2016).
- Wurst, H., Kornberg, A. A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. Journal of Biological Chemistry. 269 (15), 10996-11001 (1994).
- Ault-Riche, D., Fraley, C. D., Tzeng, C. M., Kornberg, A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. Journal of Bacteriology. 180 (7), 1841-1847 (1998).
- Lee, W. D., et al. Simple Silica Column-Based Method to Quantify Inorganic Polyphosphates in Cartilage and Other Tissues. Cartilage. , (2017).
- Martin, P., Van Mooy, B. A. Fluorometric quantification of polyphosphate in environmental plankton samples: extraction protocols, matrix effects, and nucleic acid interference. Applied and Environmental Microbiology. 79 (1), 273-281 (2013).
- Cremers, C. M., et al. Polyphosphate: A Conserved Modifier of Amyloidogenic Processes. Molecular Cell. 63 (5), 768-780 (2016).
- Dahl, J. U., et al. The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nature Microbiology. 2, 16267 (2017).
- Kulaev, I. S., Vagabov, V. M., Kulakovskaya, T. V. Ch. 2. The Biochemistry of Inorganic Polyphosphates. , 15-35 (2004).
- Werner, T. P., Amrhein, N., Freimoser, F. M. Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Archives of Microbiology. 184 (2), 129-136 (2005).
- Aschar-Sobbi, R., et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. Journal of Fluorescence. 18 (5), 859-866 (2008).
- Kulakova, A. N., et al. Direct quantification of inorganic polyphosphate in microbial cells using 4′-6-diamidino-2-phenylindole (DAPI). Environmental Science and Technology. 45 (18), 7799-7803 (2011).
- Kolozsvari, B., Parisi, F., Saiardi, A. Inositol phosphates induce DAPI fluorescence shift. Biochemical Journal. 460 (3), 377-385 (2014).
- Christ, J. J., Blank, L. M. Enzymatic quantification and length determination of polyphosphate down to a chain length of two. Analytical Biochemistry. 548, 82-90 (2018).
- Ahn, K., Kornberg, A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. Journal of Biological Chemistry. 265 (20), 11734-11739 (1990).
- Zhu, Y., Lee, S. S., Xu, W. Crystallization and characterization of polyphosphate kinase from Escherichia coli. Biochemical and Biophysical Research Communications. 305 (4), 997-1001 (2003).
- Smith, S. A., Morrissey, J. H. Sensitive fluorescence detection of polyphosphate in polyacrylamide gels using 4′,6-diamidino-2-phenylindol. Electrophoresis. 28 (19), 3461-3465 (2007).
- Livermore, T. M., Chubb, J. R., Saiardi, A. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proceedings of the National Academy of Sciences of the United States of America. 113 (4), 996-1001 (2016).
- Rudat, A. K., Pokhrel, A., Green, T. J., Gray, M. J. Mutations in Escherichia coli Polyphosphate Kinase That Lead to Dramatically Increased In Vivo Polyphosphate Levels. Journal of Bacteriology. 200 (6), e00697-e00617 (2018).
- Smith, S. A., Wang, Y., Morrissey, J. H. DNA ladders can be used to size polyphosphate resolved by polyacrylamide gel electrophoresis. Electrophoresis. , (2018).
- Studier, F. W., Moffatt, B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Journal of Molecular Biology. 189 (1), 113-130 (1986).
- JoVE Science Education Database. . Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: Electroporation. , (2018).
- JoVE Science Education Database. . Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: The Heat Shock Method. , (2018).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72, 248-254 (1976).
- JoVE Science Education Database. . Basic Methods in Cellular and Molecular Biology. Separating Protein with SDS-PAGE. , (2018).
- Mu, Q., Tavella, V. J., Luo, X. M. Role of Lactobacillus reuteri in Human Health and Diseases. Frontiers in Microbiology. 9, 757 (2018).
- Alcantara, C., Blasco, A., Zuniga, M., Monedero, V. Accumulation of polyphosphate in Lactobacillus spp. and its involvement in stress resistance. Applied and Environmental Microbiology. 80 (5), 1650-1659 (2014).
- Kulaev, I. S., Vagabov, V. M., Kulakovskaya, T. V. Ch. 1. The Biochemistry of Inorganic Polyphosphates. , 3-13 (2004).
- Blattner, F. R., et al. The complete genome sequence of Escherichia coli K-12. Science. 277 (5331), 1453-1462 (1997).
- Neidhardt, F. C., Bloch, P. L., Smith, D. F. Culture medium for enterobacteria. Journal of Bacteriology. 119 (3), 736-747 (1974).
- Akiyama, M., Crooke, E., Kornberg, A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. Journal of Biological Chemistry. 267 (31), 22556-22561 (1992).
- Akiyama, M., Crooke, E., Kornberg, A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. Journal of Biological Chemistry. 268 (1), 633-639 (1993).
- Rao, N. N., Liu, S., Kornberg, A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. Journal of Bacteriology. 180 (8), 2186-2193 (1998).
- van Pijkeren, J. P., Britton, R. A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Research. 40 (10), e76 (2012).
- Zhang, H., Ishige, K., Kornberg, A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 99 (26), 16678-16683 (2002).
- Sander, P., Meier, A., Bottger, E. C. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Molecular Microbiology. 16 (5), 991-1000 (1995).
- Hartman, S., Bont, J. A. M. D., Balows, A. . The Prokaryotes, a handbook on the biology of bacteria: ecophysiology, isolation, application. , 1215-1237 (1992).
- Winder, F. G., Denneny, J. M. The metabolism of inorganic polyphosphate in mycobacteria. Journal of General Microbiology. 17 (3), 573-585 (1957).
- Jankute, M., Cox, J. A., Harrison, J., Besra, G. S. Assembly of the Mycobacterial Cell Wall. Annual Review of Microbiology. 69, 405-423 (2015).
- Carter, S. G., Karl, D. W. Inorganic phosphate assay with malachite green: an improvement and evaluation. Journal of Biochemical and Biophysical Methods. 7 (1), 7-13 (1982).
- Cogan, E. B., Birrell, G. B., Griffith, O. H. A robotics-based automated assay for inorganic and organic phosphates. Analalytical Biochemistry. 271 (1), 29-35 (1999).

