C. elegans Positive Butanone Learning, Short-term, and Long-term Associative Memory Assays
Summary
Here we describe methods to test C. elegans associative learning and short- and long-term associative memory. These population assays employ the worms abilities to chemotax toward volatile odorants, and form positive associations upon pairing food with the chemoattractant butanone. Increasing the number of conditioning periods induces long-term memory.
Abstract
The memory of experiences and learned information is critical for organisms to make choices that aid their survival. C. elegans navigates its environment through neuron-specific detection of food and chemical odors1, 2, and can associate nutritive states with chemical odors3, temperature4, and the pathogenicity of a food source5.
Here, we describe assays of C. elegans associative learning and short- and long-term associative memory. We modified an aversive olfactory learning paradigm6 to instead produce a positive response; the assay involves starving ~400 worms, then feeding the worms in the presence of the AWC neuron-sensed volatile chemoattractant butanone at a concentration that elicits a low chemotactic index (similar to Toroyama et al.7). A standard population chemotaxis assay1 tests the worms’ attraction to the odorant immediately or minutes to hours after conditioning.
After conditioning, wild-type animals’ chemotaxis to butanone increases ~0.6 Chemotaxis Index units, its “Learning Index”. Associative learning is dependent on the presence of both food and butanone during training. Pairing food and butanone for a single conditioning period (“massed training”) produces short-term associative memory that lasts ~2 hours. Multiple conditioning periods with rest periods between (“spaced training”) yields long-term associative memory (<40 hours), and is dependent on the cAMP Response Element Binding protein (CREB),6 a transcription factor required for long-term memory across species.8
Our protocol also includes image analysis methods for quick and accurate determination of chemotaxis indices. High-contrast images of animals on chemotaxis assay plates are captured and analyzed by worm counting software in MatLab. The software corrects for uneven background using a morphological tophat transformation.9 Otsu’s method is then used to determine a threshold to separate worms from the background.10 Very small particles are removed automatically and larger non-worm regions (plate edges or agar punches) are removed by manual selection. The software then estimates the size of single worm by ignoring regions that are above a specified maximum size and taking the median size of the remaining regions. The number of worms is then estimated by dividing the total area identified as occupied by worms by the estimated size of a single worm.
We have found that learning and short- and long-term memory can be distinguished, and that these processes share similar key molecules with higher organisms.6,8 Our assays can quickly test novel candidate genes or molecules that affect learning and short- or long-term memory in C. elegans that are relevant across species.
Protocol
These C. elegans associative learning and memory assays require advance preparation. For a suggested schedule of preparation, see the timeline presented in Figure 1A. Also note that worms’ memories can be disturbed by physical agitation (rough pipetting, centrifugation, vortexing), and that naíve chemotaxis can be perturbed by excessive odors (perfume, etc.) in the environment.
1. Preparation of Animals for Assay
- Cultivate worms on 100 mm high growth media (HGM) plates (see Section 7.1 for recipe) seeded with 1 mL OP50 E. coli using standard methods.11
- Hypochlorite-treat at least two densely populated HGM plates of gravid adult worms to obtain a large synchronized population of eggs.
Note: For best yields, bleach the first generation to obtain a highly synchronized population, then bleach the second generation as Day 2 adults to obtain eggs for the subsequent step. - Evenly divide eggs onto 3-4 HGM plates seeded with 1 mL OP50 E. coli for each hypochlorite treated plate.
Note: It is important that the worms are cultivated with plenty of fresh food as starvation can affect the outcome of olfactory assays. - Incubate worms at 20oC for about 72 hours, the time it takes for animals reach the young adult stage.
2. Pre-conditioning Starve
- Double check your HGM plates with young adults to make sure the worms still have plenty of food, and that there are enough worms for the assay (≥ 200 worms per assay plate).
- Wash worms off of HGM plates with M9 buffer11 into a 15 mL conical tube. For the least agitation during washes, gently apply the M9 buffer by pouring a few milliliters onto the HGM plate, and gently swirl the plate to free worms from the sticky OP50 bacteria. Next, tilt the plate, pull up the M9 buffer/worm mixture from the corner of the plate using a P1000 pipetman, and transfer the worms to the 15 mL conical tube. If worms are still left on the plate, repeat this step 1-2X.
Note: Rough washing dislodges bacteria off of the plate that can make it into the tube with the worms. This bacteria is impossible to get rid of and can interfere with the assays. DO NOT pipette worms against the side of the tube, as this can damage the worms and interfere with the assays. - Let worms settle by gravity (DO NOT centrifuge). Remove supernatant by vacuum and wash with at least 3-4 mL of M9. Repeat 2X for a total of 3 washes.
Note: Centrifuging during washes will pull down any remaining bacteria into the worm population, which can interfere with the assays. Instead, gently add M9 buffer for subsequent washes by pouring it directly into the 15 mL conical tube. Worms settled at the bottom of the tube should become mixed in the M9 buffer upon its addition. If worms remained settled, gently invert the tube to mix. - After the third wash, use some of the worm population for naíve chemotaxis assays (Section 5).
Note: It is critical to work fairly quickly at all wash steps, as worms will begin to starve if they sit too long in M9 buffer, which can increase the naíve chemotaxis toward butanone. - Add 3-4 mL of M9 buffer to the 15 mL conical tube, let worms starve in M9 buffer at room temperature for 1 hour (Figure 1B, C).
3. Short-term Associative Memory (massed) Training
See Figure 1B for short-term associative memory assay workflow.
- At the end of the starve period in M9 buffer (Section 2), streak 2 μL of 10% butanone (in 95% EtOH) on the inside of the lids of 60 mm nematode growth media (NGM)11 plates that have been seeded with 500 μL OP50 E. coli.
Note: A good starting population for short- and long-term memory assays is ≥ 500 μL of worms. Multiple conditioning plates are needed per genotype to support the entire population during training. When working with volatile chemicals, only open lids of plates when necessary, leaving them closed at all other times. - Remove the M9 buffer supernatant from the 15 mL conical tube of starved worms by vacuum and use a P1000 pipetman to apply 100-200 μL of worms to the conditioning plates.
Note: Try to remove as much M9 liquid as possible, so that worms can quickly get to the OP50 food source when transferred to the conditioning plates. Worms will stick to the inside of the pipette tip and can be conserved by pipetting smaller volumes. - Incubate conditioning plates at room temperature for 1 hour (Figure 1B).
- Wash worms off of plates with ~1 mL M9 buffer into a 15 mL conical tube. Let worms settle by gravity. Wash again 1-2X until M9 buffer in the tube is clear.
Note: Use the gentle washing methods described in Section 2. The same 15 mL conical tube can be used from Section 2. - After the final wash, remove the M9 buffer supernatant by vacuum, and use some of the worm population for chemotaxis assays (Section 5) to test for 1x (massed) associative learning of the food-butanone association immediately after conditioning (0 minute time point).
Learning Index (LI) = CI0 hr – CINaive. - Transfer the remaining population of worms to 60 mm NGM plates seeded with 500 μL of OP50 (hold plates). Again, apply 100-200 μL of worms per plate to ensure there are enough bacteria to support the population.
Note: The number of hold plates used per genotype is at least the number of short-term associative memory time points to be tested. - Incubate post-conditioning plates for 0.5, 1, or 2 hours (short-term associative memory time points) at room temperature (Figure 1B).
Note: Short-term associative memory of wild-type animals ends by 2 hrs after training. Time points may need to be adjusted for different genotypes or conditions. - To test for short-term associative memory of the food-butanone association, after each time point, gently wash worms off plates into a 15 mL conical tube and again 1-2X with M9 buffer. Use worms for chemotaxis assays (Section 5). Learning Index (LI) = CITime point – CINaive.
Note: Use the gentle washing methods described in Section 2. For each time point, discard any extra worms that are not used in chemotaxis assays.
4. Long-term Associative Memory (Spaced) Training
See Figure 1C for long-term associative memory assay workflow.
- Follow steps 3.1-3.2 in Section 3.
- Incubate conditioning plates at room temperature for 30 minutes (Figure 1C).
- Follow step 3.4 in Section 3.
Note: To remain inside a 30-minute timeframe, one can begin all washes during training at ~25 minutes. - After the final wash, remove M9 supernatant by vacuum and transfer worms to unseeded 60 mm NGM plates.
Note: The use of multiple plates per genotype is not required at this step since the worms are being starved. - Starve on 60 mm NGM plates at room temperature for 30 minutes (Figure 1C).
- Wash worms with M9 buffer into the 15 mL conical tube. Let worms settle by gravity (DO NOT centrifuge).
Note: While there should be no bacteria in the buffer at this point, centrifuging can be a potentially damaging treatment of the worms, and may disrupt long-term memory formation. - Repeat steps 4.1-4.6 several times until a total of 7 conditioning blocks and 6 starve blocks have been completed (Figure 1C). (See Table 1 for a representative time sheet.)
- Gently wash worms off plates into the 15 mL conical tube and again 1-2X with M9 buffer.
- After the final wash, use some of the worm population for chemotaxis assays (Section 5) to test for 7x (spaced) associative learning of the food-butanone association immediately after conditioning (0 hour time point). Learning Index (LI) = CI0 hr – CINaive.
- Transfer the remaining population of worms to 100 mm HGM plates seeded with 1 mL of OP50 (hold plates). Again, apply 100-200 μL of worms per plate to ensure there are enough bacteria to support the population.
Note: The number of hold plates used per genotype is at least the number of long-term associative memory time points to be tested. 100 mm NGM plates can also be used as post-conditioning plates, but one must be very careful that there is enough OP50 to support the population of worms for at least 16 hours. - Incubate post-conditioning plates for 16, 24, and 40 hours (long-term associative memory time points) at 20oC (Figure 1C).
Note: Long-term associative memory of wild-type animals ends by 40 hrs after training. Time points may need to be adjusted for different genotypes or conditions. - To test for long-term associative memory of the food-butanone association, gently wash worms off plates into a 15 mL conical tube and again 1-2X with M9 buffer after each time point. Use worms for chemotaxis assays (Section 5). Learning Index (LI) = CITime point – CINaive.
Note: Use the gentle washing methods described in Section 2. For each time point, discard any extra worms not used in the chemotaxis assays.
5. Chemotaxis Assay
- Prepare the chemotaxis assay plates. Mark the bottom of unseeded 100 mm NGM plates with spots on the bottom and each side of the plate (Figure 2A).
Note: At least 3 replicates per genotype must be run to obtain statistically significant results. - Spot 1 μL of 1 M NaN3 at the odorant and control spot (Figure 2B).
Note: Do not spot your plates with this paralyzing agent more than 15 minutes before starting the assay, or the NaN3 will diffuse away from the spots, and animals will become paralyzed before reaching the odorant or control spots. Be careful not to puncture the agar when applying NaN3 since worms will burrow in punctured areas. - While the worms are settling in the conical tube after several M9 buffer washes (Section 2), spot 1 μL each of 95% EtOH and 10% butanone (see Section 7.2) at the appropriate spots on the marked assay plate (Figure 2C) on top of the previously spotted NaN 3.
Note: Chemicals must be spotted prior to adding the worms (Section 5.4). Make a note of which spot has EtOH or butanone. When working with these volatile chemicals, be careful to only open the lids of plates when necessary, and keep them closed at all other times. - Remove as much of the M9 buffer as possible from the tube of settled worms. Use a P20 pipetman with a pre-cut tip (see Section 7.3) to deliver 3X 5 μL worms (200-400 animals) to the origin of the marked assay plate (Figure 2D).
Note: Keep remaining worms in the 15 mL conical tube for use in Pre-conditioning Starve and memory assays (Sections 2-4). Worms will stick inside pipette tips, so adding worms to the plate in smaller quantities conserves your worm population and reduces the amount of liquid that gets released with the worms onto the assay plate. - Twist the corner of a KimWipe to a small point and use it to blot up the excess M9 buffer. This will release the worms onto the assay plate (Figure 2E).
Note: Be careful not to puncture the agar with the KimWipe, otherwise worms will burrow at the origin. Some worms may be lost in this step as they are pulled into the KimWipe with the M9 buffer when blotting. If using imaging and analysis to count worms (Section 6), take plates to imaging station before releasing worms from the origin. An image of the worms at the origin immediately after release will give the total number of animals for an assay plate. - Incubate the chemotaxis assay plate for 1 hour at room temperature.
- Count the number of worms at origin, EtOH, and butanone spots (Figure 2), as well as the total number of worms on the assay plate.
Note: Generally, worms paralyzed by NaN3 will be within a ~1 cm radius of the spots, and will appear stick-straight with many animals stacked against each other. If using imaging and analysis software to count worms (Section 6), take images of the origin, EtOH, and butanone spots. An image of the total amount of worms should have been taken in Section 5.5. Worms can be also be “hand-counted.” - Calculate the “Chemotaxis Index” (CI). CI = ([(nButanone)-(nEtOH)]/[(Total-nOrigin)].
6. Worm Counting by Image Analysis
- Take high-contrast black-and-white images of worms on chemotaxis assay plates (Figure 3A, see Section 7.4 for description of camera setup). To calculate a chemotaxis index, images are needed at the butanone, EtOH, and origin spots of a chemotaxis assay plate at the end of an assay, as well as the total number of worms (picture of worms at origin immediately after they are released from M9 buffer at the beginning of the assay, Section 5.5).
Note: Images of the chemotaxis assay plates covering about a 2 cm radius around each spot generally capture all of the animals for each spot. - Make sure the files for all trials of one time point (example Naive, 0 hr, etc.) are contained in one folder, named appropriately.
- Files for each chemotaxis plate must be named as such: but#.png (butanone spot, trial #), ori#.png (origin, trial #), eth#.png (ethanol spot, trial #), tot#.png (total, trial #). (For example, if two trials were run for a time point, in the same folder the following files would be present: but1.png, but2.png, eth1.png, eth2.png, ori1.png, ori2.png, tot1.png, tot2.png)
- Open Matlab (see Section 7.5).
- In the “Current Directory” line at the top of the Matlab window, browse for folders, and choose the “count_worms_v0.5.3” folder (M-files available as Supplementary Information).
- In the “Command Window”, type “count_worms_directory().” Hit Enter.
Note: The default worm size when for this command is min 10, max 80 pixels. To adjust this based on a specific genotype or developmental stage, type “count_worms_directory (‘minsize’,10,’maxsize’, 80)” and change the pixel numbers accordingly. - When prompted, choose the folder of images to be analyzed.
Note: Only one folder of images can be analyzed at a time. - Each image in the folder being analyzed will pop up with a threshold already set, and particles selected (Figure 3A). Drag the rectangle tool over selected particles that are not worms to delete them. When finished with the image, hit the Esc key.
- After all images in the folder are checked, 4 columns will appear in the Command Window: (1) image name (example but1); (2) always “[1];” (3) average pixel size for one worm calculated by the program the particular image; and (4) the number of worms calculated to be in the image.
- Once this program has run, it will deposit two .csv files (can be opened as Excel spreadsheets) into the folder of images it has just analyzed. The “worm_counts_stats” file provides the output columns found in the Command Window (Section 6.9). The “worm_counts_summary” file provides 5 columns: (1) trial number; number of worms in (2) but, (3) eth, and (4) ori spots; and (5) total number of worms.
7. Materials Notes
- To prepare 100 mm high growth media (HGM) plates (Section 1): Dissolve 3 g NaCl, 20 g Bactopeptone, and 30 g Bacto-agar in 700 mL distilled water. Bring volume to 1 L with distilled water. After autoclaving, cool agar to 65oC and add 4 mL of 5 mg/mL cholesterol in ethanol, 1 mL each of 1 M CaCl2, 1 M MgSO4, and 25 mL of 1 M KPO4 (pH = 6.0).
- 95% EtOH is used, as higher percentage EtOH tends to contain impurities from the purification process that can affect results. It is a good idea to make the 10% butanone (in 95% EtOH) stock up fresh every few times the assay is run.
- P20 tips must have a few millimeters cut off to create a hole large enough for worms to fit through without being damaged. P1000 tips already have holes large enough for the worms to fit through.
- The chemotaxis assay plate imaging station (Figure 3B) in the Murphy lab contains a Firewire camera with a variable magnification lens attached to a boom stand. A backlight is placed at the bottom of the stand underneath a custom imaging platform with a glass top (Figure 3C), which allows chemotaxis assay plates to be illuminated from the bottom. “Measurement and Automation” software (National Instruments) is used to capture images. Materials for this imaging station set up are similar to those previously published,12 and can be found in the Table of specific reagents and equipment.
8. Representative Results:
A typical naïve chemotaxis index (CI) for wild-type worms for 10% butanone is around 0.2 (Figure 4A).6 Massed (1x) or spaced (7x) training generally increases the chemotaxis index to 0.7-0.8 at time 0 (Figure 4A, C-D) to give a learning index (LI = trained CI – naíve CI) of ~0.6.6 The learning that occurs with massed or spaced training is very robust. A LI of less than 0.5 typically arises when there are problems with the naíve chemotaxis assay, and the animals have a naíve CI of 0.3 or higher. Usually, this occurs because worms are starving or too crowded on cultivation plates between bleaching and the start of the assay, or worms sit too long in M9 buffer between washes and begin to starve; environmental odors can also increase the naíve CI. Starved worms have a much higher naíve CI for 10% butanone (Figure 4B).6 We find that problems with the naíve chemotaxis are generally resolved with improved worm care and culturing.
The duration of memory in these assays is the time it takes for chemotaxis index immediately after training to return to naíve levels, when the learning index = 0. In wild-type animals, short-term associative memory typically begins to decline by 1 hour after massed training, lasts about 2 hours, and is independent of the transcription factor CREB (Figure 4C).6 Long-term associative memory typically does not begin to significantly decline until 16 hours after spaced training, lasts up to 40 hours, and is CREB-dependent (Figure 4D).6 Long-term associative memory may not reach its full potential in several instances: (1) worms do not have enough OP50 E. coli during 30-minute conditioning periods; (2) bacteria remain in the M9 buffer during starvation periods; or (3) worms are damaged during the assay (e.g. worms are pipetted against the side of the tube).
Results are generally statistically significant when 4 or more chemotaxis assay trials are run per genotype per time point. Running six replicates per genotype typically yields very significant results without becoming too much to handle; however, beginners may want to start with only three replicates. Generally, working with 2-3 genotypes or conditions at a time is comfortable for those who are experienced with these learning and memory assays.
Hand-counting worms on chemotaxis assay plates is very time consuming, can add variability between experiments or lab mates, and may also introduce biases when analyzing and interpreting data. Worm counting by image analysis (Section 6) standardizes data collection and analysis across the lab, and on average, cuts data analysis time for experienced lab members to 1/5 of that needed for hand counting. When comparing chemotaxis indices calculated using manual worm counts to those determined by the Count_worms software, we find an average error of 3.07 (± 1.19)%.
| Day 1 | |
| Time | Step |
| 9:00 | 1 hr starve in M9 buffer Start naíve chemotaxis assay |
| 10:00 | Condition #1 (60 mm NGM plates with food and butanone), 30 min End naíve chemotaxis assay |
| 10:30 | Starve #1 (60 mm NGM plates), 30 min |
| 11:00 | Condition #2, 30 min |
| 11:30 | Starve #2, 30 min |
| 12:00 | Condition #3, 30 min |
| 12:30 | Starve #3, 30 min |
| 1:00 | Condition #4, 30 min |
| 1:30 | Starve #4, 30 min |
| 2:00 | Condition #5, 30 min |
| 2:30 | Starve #5, 30 min |
| 3:00 | Condition #6, 30 min |
| 3:30 | Starve #6, 30 min |
| 4:00 | Condition #7, 30 min |
| 4:30 | Begin 0-hr chemotaxis assay Transfer remaining worms to hold plates for 16-40 hrs |
| 5:30 | End 0-hr chemotaxis assay |
| Day 2 | |
| Time | Step |
| 8:30 | Begin 16-hr chemotaxis assay |
| 9:30 | End 16-hr chemotaxis assay |
Table 1. Representative schedule for long-term associative memory assay. In this example, the assay begins on Day 1 at 9 am with a 1-hr starve. 30-min condition and starve periods are alternated until the worms have been conditioned seven times. Chemotaxis assays are run at the beginning (naíve) and end (0 hr) of training. The total run-time from start to finish is 8.5 hours. Worms on hold plates are tested for chemotaxis to butanone starting on Day 2, 16-40 hours after training.
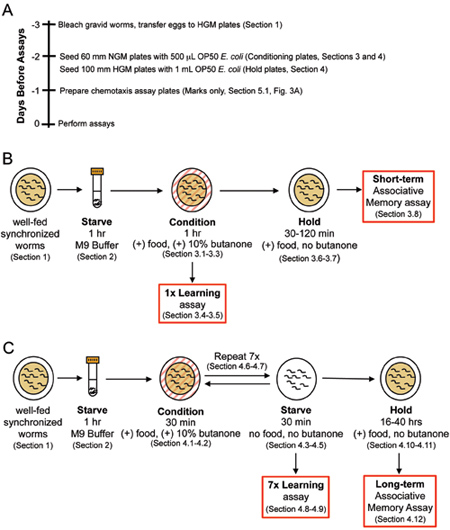
Figure 1. Short-term and long-term associative memory assay workflow. (A) Recommended timeline of preparation leading up to the day assays are performed. (B) Short-term associative memory assay: Starved worms are conditioned for 1 hour and tested immediately for 1x learning (0 min.) via chemotaxis assays, or transferred onto holding plates with food for 0.5, 1, or 2 hours after conditioning. (C) Long-term associative memory assay: Starved animals receive 7 training blocks of conditioning/starving before being tested for learning or LTAM 16-40 hours after training.
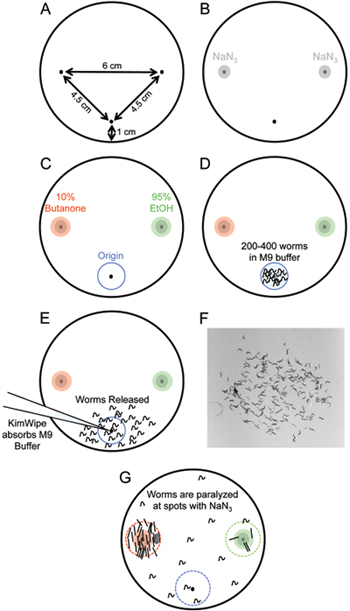
Figure 2. Chemotaxis assay setup. (A) Schematic of chemotaxis assay plate. Small spots are added to the plate with the help of a ruler or other guiding device to mark the origin (bottom) and butanone and EtOH (left and right) spots on the plate. (B) 1 μL NaN3 is added to the butanone and EtOH spots. (C) 1 μL each of 10% butanone and 95% EtOH are added the spots on the left or right side of the plate. (D) 200-400 worms suspended in M9 buffer are added to the origin of the plate. (E) A KimWipe twisted into a point is used to absorb M9 buffer and release worms onto the assay plate.
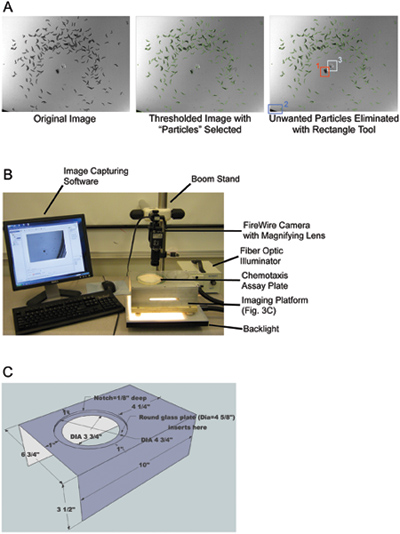
Figure 3. Worm counting with image analysis. (A) Example of “count_worms_v0.5.3” image particle selection window. The program opens original high-contrast black-and-white image (left) automatically pops up a window with a thresholded image (middle). The rectangle tool can be used to highlight and eliminate undesired non-worm “particles,” such as assay plate marks (1), plate edges (2), or agar punctures (3). (B) Image of the Murphy lab chemotaxis assay imaging station. Chemotaxis assay plates are illuminated from the bottom to create high contrast images required for analysis. (C) Schematic of the custom-made imaging platform used in the chemotaxis imaging station.
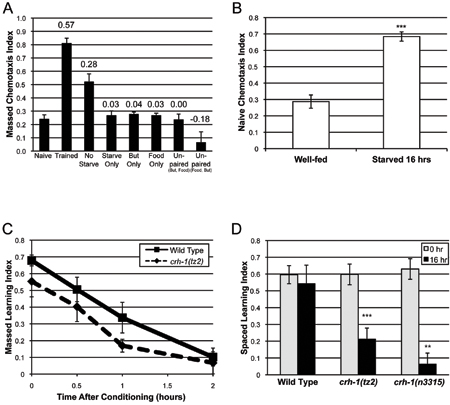
Figure 4. Typical associative learning and memory results. (A) The naíve chemotaxis index of wild-type worms to 10% butanone increases upon massed (1x) training. The learned association is requires the pairing of food (OP50 E. coli) and butanone. Numbers above bars represent the “Learning Index” (LI = CITrained – CINaive). (B) Wild type naíve chemotaxis to 10% butanone is greatly increased when worms are starved for 16 hours. Panels A-D are adapted from Kauffman et al., 2010.6 (C) Wild type short-term associative memory lasts about 2 hours, and is independent of the transcription factor CREB. (D) Wild type long-term associative memory lasts about 40 hours, and is CREB-dependent.
Supplementary Information
Discussion
The C. elegans olfactory associative learning and memory assays presented here distinguish between the processes of massed learning, spaced learning, short-term memory, and long-term memory. These assays rely on the worms’ abilities to sense food and chemical odors in their environment1,2 and to form positive associations between the two.6 Therefore, the assays are very sensitive to starvation in the starting population of animals, and great care must be taken to ensure that worms are well fed before and after training. Single massed training yields short-term associative memory. The increased number of conditioning periods and spaced training paradigm is critical for achieving long-term associative memory in C. elegans as it is in other organisms.6,8 Perhaps this assay could be modified to produce long-term memories of associations between different conditioned and unconditioned stimuli.
Our associative learning and memory assays can be used to compare the learning and memory capabilities of wild type to aging or genetic mutant animals. For example, while both associative learning and long-term associative memory abilities decline in aging wild-type worms, learning is maintained longer with age in animals with reduced insulin signaling, and learning and memory is maintained longer with age in calorically restricted animals.6 These assays are also amenable to RNAi treatment or pharmacological manipulation of C. elegans. For example, treatment of worms with daf-2 RNAi improves short- and long-term associative memory; conversely, blockage of gene transcription and protein synthesis with drugs (actinomycin D and cycloheximide, respectively) during spaced training disrupts long-term memory formation.6
Long-term memory is a process that is crucial to survival in all animals, and it appears that much of the molecular machinery involved in memory of a Pavlovian association is conserved from worms to mammals.6 The ability to distinguish and test associative learning, short-term and long-term memory using these olfactory assays makes C. elegans an excellent system for the further study of memory, and may provide insight into neurodegenerative processes that lead to loss of learning and memory in humans.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank W. Ryu for advice on our initial imaging set-up, Z. Gitai for the suggestion to use ImageJ13 to count worms, and J. Ashraf for help with Google SketchUp. CTM is a Pew Scholar in the Biomedical Sciences, a McKnight Scholar, and a Keck Scholar.
Materials
| Material Name | Tipo | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Sodium Azide | Fisher Scientific | S227 | Prepare 1 M concentration in distilled water | |
| Ethyl Alcohol 190 Proof | Pharmco-Aaper | DSP-CT-18 | >95% Ethanol may have impurities that interfere with chemotaxis assays | |
| 2-Butanone 99+% | Acros | 14967-0010 | ||
| Basler IEEE-1394/FireWire Area Scan CMOS Camera | Edmund Optics | NT56-683 | ||
| InfiniMite Video Lens – Alpha Industrial | Edmund Optics | NT56-202 | ||
| Dolan-Jenner MI-150 Fiber Optic Illuminator | Edmund Optics | NT55-718 | ||
| 8” x 8” Backlight | Schott Fostec | A08927 | ||
| Standard Boom Stand | Edmund Optics | NT54-120 | ||
| 3/4” Post Adaptor for Boom Stands | Edmund Optics | NT54-124 | ||
| Standard Fixed 1/4-20 Mounting Plate | Edmund Optics | NT54-122 |
Riferimenti
- Bargmann, C. I., Hartwieg, E., Horvitz, H. R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 74, 515-515 (1993).
- Ward, S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci U S A. 70, 817-817 (1973).
- Bono, M. d. e., Maricq, A. V. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 28, 451-451 (2005).
- Mori, I. Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. Annu Rev Genet. 33, 399-399 (1999).
- Zhang, Y., Lu, H., Bargmann, C. I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 438, 179-179 (2005).
- Kauffman, A. L. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 8, e1000372-e1000372 (2010).
- Torayama, I., Ishihara, T., Katsura, I. Caenorhabditis elegans integrates the signals of butanone and food to enhance chemotaxis to butanone. J Neurosci. 27, 741-741 (2007).
- Silva, A. J., Kogan, J. H., Frankland, P. W., Kida, S. CREB and memory. Annu Rev Neurosci. 21, 127-127 (1998).
- Meyer, F. Iterative image transformations for an automatic screening of cervical smears. J Histochem Cytochem. 27, 128-128 (1979).
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics. 9, 62-62 (1979).
- Brenner, S. The genetics of Caenorhabditis elegans. Genetica. 77, 71-71 (1974).
- Stephens, G. J., Johnson-Kerner, B., Bialek, W., Ryu, W. S. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol. 4, e1000028-e1000028 (2008).
- Abramoff, M. a. g. e. l. h. a. e. s., Ram, P. J., J, S. Image Processing with ImageJ. Biophotonics International. 11, 36-36 (2004).

