Differential Radial Capillary Action of Ligand Assay: A High-Throughput Technique to Identify Bacterial Nucleotide Second Messenger-Binding Intracellular Proteins
Abstract
Source: Schicketanz, M. L., et al. Identifying the Binding Proteins of Small Ligands with the Differential Radial Capillary Action of Ligand Assay (DRaCALA). J. Vis. Exp. (2021)
This video demonstrates the differential radial capillary action of ligand assay — a technique for systematic high-throughput screening of intracellular nucleotide second messenger-binding proteins. This technique allows for direct binding of the messengers to the overexpressed target protein in the cell lysate — bypassing the need for protein purification.
Protocol
1. Preparation of whole cell lysates
- Inoculate the E. coli K-12 ASKA ORFeome collection strains into 1.5 mL Lysogeny broth (LB) containing 25 µg/mL chloramphenicol in 96-well deep well plates. Grow overnight (O/N) for 18 h at 30 °C with shaking at 160 rpm. On the next day, add isopropyl β-d-1-thiogalactopyranoside (IPTG) (final 0.5 mM) to the O/N cultures to induce protein expression at 30 °C for 6 h.
- Pellet cells at 500 x g for 10 min. Freeze the pellets at -80 °C until use. To lyse the cells, add 150 µL of lysis buffer L1 (40 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, supplemented with 2 mM phenylmethylsulfonyl fluoride (PMSF), 40 µg/mL DNase 1, and 0.5 mg/mL lysozyme) to resuspend the pellet.
- Freeze the cells at -80 °C for 30 min, and then thaw at 37 °C for 20 min. Repeat this cycle three times to lyse the cells. Store the lysates at -80 °C before use.
2. Purification of Relseq and GppA
NOTE: The recombinant proteins Relseq from Streptococcus equisimilis and GppA from E. coli K-12 are used to synthesize the radiolabeled pppGpp and ppGpp, respectively.
- Grow and collect cells overexpressing each protein.
- Grow the E. coli BL21 DE3 strain up to exponential phase (optical density (OD) ~0.3-0.4) in LB broth, and spin down 1 mL of culture at 6000 x g for 5 min. Decant the supernatant, and resuspend the cells with 100 µL of ice-cold TSB broth (LB broth supplemented with 0.1 g/mL PEG3350, 0.05 mL/mL dimethylsulfoxide, 20 mM MgCl2).
- Mix the plasmids bearing the histidine-tagged relseq and gppA, each 100 ng, with the above cell suspensions in TSB, and incubate on ice for 30 min. Heat-shock the cells at 42 °C for 40 s. Place the mixture on ice for 2 min, and add 1 mL of LB broth at room temperature to allow the cells to recover for 1 h at 37 °C with agitation at 160 rpm.
- Plate the recovered cells on LB agar plates supplemented with the corresponding antibiotics (Relseq:100 µg/mL ampicillin; GppA: 30 µg/mL kanamycin).On the next day, inoculate the colonies in LB broth to start O/N precultures of both strains at 37 °C.
- After 18 h, inoculate 500 mL of LB medium with 10 mL of the O/N cultures and the corresponding antibiotics. Grow the cultures by shaking at 160 rpm at 37 °C. When the OD600nm reaches 0.5-0.7, induce protein expression by adding 0.5 mM IPTG and growing for 3 h at 30 °C with shaking at 160 rpm.
- Collect the cells by spinning at 6084 x g for 10 min at 4 °C. Resuspend the pellet in 20 mL of ice-cold 1x phosphate-buffered saline (PBS), and re-centrifuge at 1912 x g for 20 min at 4 °C. Decant the supernatant, and freeze the pellets at -20 °C before use.
- Nickel-nitrilotriacetic acid (Ni-NTA) affinity purification
NOTE: From this point onwards, ensure that the samples are cold.- Add 40 mL of ice-cold lysis buffer L2 (50 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 10 mM imidazole, 10 mM β-mercaptoethanol supplemented with protease inhibitors (EDTA-free tablet; see the Table of Materials) to resuspend the pellet. Lyse the cells via sonication (60% amplitude, 2 s ON/ 4 s OFF for 8 min ON). Clear the lysate by spinning at 23,426 x g for 40 min at 4 °C, and continue with the supernatant for purification.
- During the above centrifugation, prepare the Ni-NTA resin.
- Transfer 500 µL of homogenized Ni-NTA resin into a standing polypropylene chromatography column, and let it settle for 15 min and the storage solution drain through. Wash the resin with 15 mL of ultrapure water twice, and then wash the column with 15 mL of the lysis buffer L2.
- Load the cleared supernatant of cell lysate from step 1 onto the column, and let it flow through. Wash the column with 30 mL of washing buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 20 mM imidazole).
- Elute the proteins with 400 µL of the elution buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 500 mM imidazole) three times. Then, repeat elution with another 300 µL of the elution buffer. Combine the eluted proteins to a final volume of 700 µL.
- Gel filtration
- Prepare gel filtration buffer (50 mM Tris, pH 7.5; 200 mM NaCl; 5% glycerol). Wash the size exclusion column with one column volume (25 mL) of the gel filtration buffer.
- Load the above 700 µL sample by using a 500 µL loop, run at 0.5 mL/min, and collect 2-3 fractions, each of 0.5 mL volume, containing the respective proteins.
- Combine and concentrate the fractions containing each of the proteins using a spin column, and measure the protein concentration using the Bradford assay.
3. Synthesis of 32P-labeled pppGpp and ppGpp
- Assemble a small-scale Relseq reaction in a screw cap tube (see Table 1).
NOTE: Work with radioactive reagents only in a licensed place and with personal protective equipment. - Incubate the tube at 37 °C in a thermomixer for 1 h, then at 95 °C for 5 min, and place on ice for 5 min. Spin down the precipitated protein at 15,700 x g for 5 min, and transfer the supernatant (synthesized 32P-pppGpp) to a new screw cap tube.
- To synthesize 32P-ppGpp from 32P-pppGpp, transfer half of the 32P-pppGpp product to a new screw cap tube, and add 1 µM GppA. Incubate the tube at 37 °C for 10 min, at 95 °C for 5 min, and then place on ice for 5 min.
- Spin down the precipitated protein at 15,700 x g for 5 min, and transfer the supernatant (synthesized 32P-ppGpp) to a new screw cap tube.
- Analyze the 32P-pppGpp and 32P-ppGpp by running 1 µL of the samples on a thin layer chromatography (TLC) plate (polyethyleneimine-modified cellulose TLC plates) using the 1.5 M KH2PO4, pH 3.4, as mobile phase.
NOTE: Use the α-32P-labeled guanosine 5'-triphosphate (32P-α-GTP) as control. - Dry the TLC plate completely, place it between a transparent plastic folder, and expose it to a storage phosphor screen for 5 min. Visualize and quantify the signals by using a phosphorimager.
NOTE: When the ratios of 32P-pppGpp and 32P-ppGpp are higher than 85%, a large-scale reaction (500 µL, sufficient for screening 20 96-well plates) could be assembled and synthesized by using Table 1.
4. DRaCALA screening of the target proteins of (p)ppGpp
- Thaw and transfer 20 µL of whole cell lysates to a 96-well V-bottom microtiter plate. Add 2.5 U/well of the endonuclease from Serratia marcescens, and incubate at 37 °C for 15 min to reduce lysate viscosity. Place the lysates on ice for 20 min.
- Mix the 32P-pppGpp and 32P-ppGpp in a 1:1 ratio, and add 1x lysis buffer L1 to make the final concentration of (p)ppGpp equal to 4 nM.
NOTE: Given the chemical similarity between pppGpp and ppGpp, a mix of both chemicals will simplify the screening process. - Use a multichannel pipette and filtered pipette tips to add and mix 10 µL of the (p)ppGpp mixture with the cell lysate. Incubate at room temperature (RT) for 5 min.
- Wash the 96 x pin tool by placing it in a 0.01% solution of non-ionic detergent for 30 s, and dry it on tissue paper for 30 s. Repeat the washing of the pin tool 3x.
- Place the pin tool in the above 96-well sample plates, and wait for 30 s. Lift the pin tool straight up, and place it straight down on a nitrocellulose membrane for 30 s.
NOTE: If a spot is missing, spot 2 µL of the corresponding samples with a pipette and filtered tips. It is advisable to make a duplicate spot of the same sample as indicated below. - Dry the membrane for 5 min at RT. Place the membrane between a transparent plastic folder, and expose it to a storage phosphor screen for 5 min. Visualize by using a phosphorimager.
5. Quantification and identification of potential target proteins
- Use the analysis software associated with the phosphorimager to open the .gel file of the visualized plates. Use the Array analysis function to define the 96 spots by setting up a grid of 12 columns x 8 rows.
- Define big circles to circumscribe the outer edge of the whole spots (see Figure 1). Export the Volumn+Background and Area of the defined big circles, and save in a spreadsheet.
NOTE: If required, reposition each individual circle to perfectly overlap with the spots, and resize each individual circle to make it slightly bigger than the actual spot. - Size down the defined circles to circumscribe the small inner dots. Export the Volumn+Background, and Area of the defined small circles, and save in a spreadsheet.
- Calculate the binding fractions in the spreadsheet by using the equation in Figure 1, and plot the data. Identify the potential binding proteins in the wells that show high binding fractions in comparison to the majority of other wells.
Table 1: Assembling information for the small- and large-scale synthesis reactions of 32P-labeled pppGpp. *10x Relseq buffer contains 250 mM Tris-HCl, pH 8.6; 1M NaCl; 80 mM MgCl2. Abbreviation: pppGpp = guanosine pentaphosphate.
| Volume (μL) | ||
| Small scale | Large scale | |
| Ultrapure water | ||
| 10x Relseq buffer* | 2 | 50 |
| ATP (8 mM final) | ||
| Relseq (4 μM final) | ||
| 32P-α-Guanosine triphosphate (GTP) (final 120 nM) (CAUTION) | 0.2 | 5 |
| total | 20 | 500 |
Representative Results
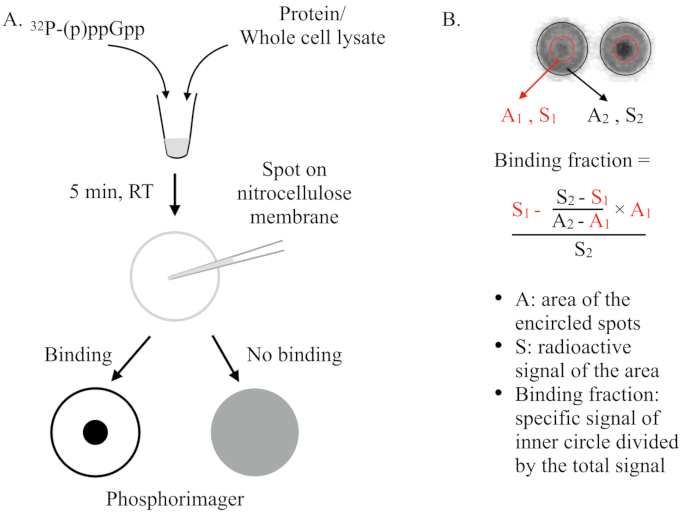
Figure 1: Quantification and calculation of the binding fraction. See the text for details. Briefly, the DRaCALA spots will be analyzed by drawing two circles that circumscribe the whole spot and the inner dark dot (i.e., the retained (p)ppGpp due to the binding of the tested protein). The specific binding signal is the radioactive signal of the inner circle (S1) after subtracting the non-specific background signal (calculated by A1 × ((S2-S1)/(A2-A1))). The binding fraction is the specific binding signal divided by the total radioactive signal (S2). Abbreviations: DRaCALA = Differential Radial Capillary Action of Ligand Assay; (p)ppGpp = guanosine penta- and tetraphosphates; RT = room temperature.
Divulgazioni
The authors have nothing to disclose.
Materials
| 32P-α-GTP | Perkinelmer | BLU006X250UC | |
| 96 x pin tool | V&P Scientific | VP 404 | 96 Bolt Replicator, on 9 mm centers, 4.2 mm Bolt Diameter, 24 mm long |
| 96-well V-bottom microtiter plate | Sterilin | MIC9004 | Sterilin Microplate V Well 611V96 |
| Agar | OXOID – Thermo Fisher | LP0011 | Agar no. 1 |
| ASKA collection strain | NBRP, SHIGEN, JAPAN | Ref: DNA Research, Volume 12, Issue 5, 2005, Pages 291–299. https://doi.org/10.1093/dnares/dsi012 | |
| Benzonase | SIGMA | E1014-25KU | genetically engineered endonuclease from Serratia marcescens |
| Bradford Protein Assay Dye | Bio-Rad | 5000006 | Reagent Concentrate |
| DMSO | SIGMA | D8418 | ≥99.9% |
| DNase 1 | SIGMA | DN25-1G | |
| gel filtration10x300 column | GE Healthcare | 28990944 | contains 20% ethanol as preservative |
| Glycerol | PanReac AppliChem | 122329.1214 | Glycerol 87% for analysis |
| Hypercassette | Amersham | RPN 11647 | 20 x 40 cm |
| Imidazole | SIGMA | 56750 | puriss. p.a., ≥ 99.5% (GC) |
| IP Storage Phosphor Screen | FUJIFILM | 28956474 | BAS-MS 2040 20x 40 cm |
| Isopropyl β-d-1-thiogalactopyranoside (IPTG) | SIGMA | I6758 | Isopropyl β-D-thiogalactoside |
| Lysogeny Broth (LB) | Invitrogen – Thermo Fisher | 12795027 | Miller's LB Broth Base |
| Lysozyme | SIGMA | L4949 | from chicken egg white; BioUltra, lyophilized powder, ≥98% |
| MgCl2(Magnesium chloride) | SIGMA | 208337 | |
| MilliQ water | ultrapure water | ||
| multichannel pipette | Thermo Scientific | 4661110 | F1 – Clip Tip; 1-10 ul, 8 x channels |
| NaCl | VWR Chemicals | 27810 | AnalaR NORMAPUR, ACS, Reag. Ph. Eur. |
| Ni-NTA Agarose | Qiagen | 30230 | |
| Nitrocellulose Blotting Membrane | Amersham Protran | 10600003 | Premium 0.45 um 300 mm x 4 m |
| PBS | OXOID – Thermo Fisher | BR0014G | Phosphate buffered saline (Dulbecco A), Tablets |
| PEG3350 (Polyethylene glycol 3350) | SIGMA | 202444 | |
| phenylmethylsulfonyl fluoride (PMSF) | SIGMA | 93482 | Phenylmethanesulfonyl fluoride solution – 0.1 M in ethanol (T) |
| Phosphor-imager | GE Healthcare | 28955809 | Typhoon FLA-7000 Phosphor-imager |
| Pipette Tips, filtered | Thermo Scientific | 94410040 | ClipTip 12.5 μl nonsterile |
| Poly-Prep Chromatography column | Bio-Rad | 7311550 | polypropylene chromatography column |
| Protease inhibitor Mini | Pierce | A32955 | Tablets, EDTA-free |
| screw cap tube | Thermo Scientific | 3488 | Microcentifuge Tubes, 2.0 ml with screw cap, nonsterile |
| SLS 96-deep Well plates | Greiner | 780285 | MASTERBLOCK, 2 ML, PP, V-Bottom, Natural |
| spin column | Millipore | UFC500396 | Amicon Ultra -0.5 ml Centrifugal Filters |
| Thermomixer | Eppendorf | 5382000015 | Thermomixer C |
| TLC plate (PEI-cellulose F TLC plates) | Merck Millipore | 105579 | DC PEI-cellulose F (20 x 20 cm) |
| Tris | SIGMA | BP152 | Tris Base for Molecular Biology |
| Tween 20 | SIGMA | P1379 | viscous non-ionic detergent |
| β-mercaptoethanol | SIGMA | M3148 | 99% (GC/titration) |

