순수 배양 및 줄무늬 평판배양: 혼합 검체에서 단일 박테리아 군집 분리
English
Diviser
Vue d'ensemble
출처: 틸드 앤더슨1,롤프 루드1
1 임상 과학 룬드학과, 감염 의학 학과, 생물 의학 센터, 룬드 대학, 221 00 룬드, 스웨덴
겉보기에는 불가능한 미생물 생물 다양성은 약 1조 종의 공존하는 종(1,2)으로 정말 놀랍습니다. 특히 가혹한 기후, 인간의 위 (3) 또는 남극의 빙하 호수 (4)의 산성 환경 처럼, 특정 종에 의해 지배 될 수 있습니다., 박테리아는 일반적으로 혼합 된 문화에서 발견. 각 균주는 다른 (5)의 성장에 영향을 미칠 수 있기 때문에, “순수”(한 가지 유형으로만 구성)를 분리하고 육성하는 능력은 모두 임상 및 학술 환경에서 필수적이되고있다. 순수한 배양은 추가 유전 (6) 및 proteomic 검사 (7), 샘플 순도의 분석 및, 아마도 더 주목할 만한, 임상 샘플에서 전염하는 에이전트의 식별 및 특성을 가능하게합니다.
박테리아는 성장 요구 사항의 넓은 범위를 가지고 있으며, 까다로운 종과 까다로운 종 (8)을 모두 유지하기 위해 설계된 영양소 미디어의 수많은 유형이있다. 성장 매체는 액체 형태(국물로서) 또는 전형적으로 한천계(적조류에서 유래한 겔화제) 고체 형태로 제조될 수 있다. 국물에 직접 접종은 유전적으로 다양하거나 심지어 혼합 된 세균 인구를 생성할 위험을 수반하지만, 도금및 재 줄무늬는 각 세포가 매우 유사한 유전 적 메이크업을 가지고 있는 순수한 문화를 만듭니다. 줄무늬 플레이트 기술은 개별 세포를 서로 분리하는 것을 목표로 시료(도1)의점진적 희석을 기반으로 합니다. 미디어 및 지정된 환경에 의해 유지되는 모든 실행 가능한 세포(이하 콜로니 형성 유닛, CFU라고 함)는 이후에 이진 분열을 통해 딸 세포의 고립된 식민지를 발견할 수 있다. 세균성 지역 사회 내의 급속한 돌연변이 비율에도 불구하고, 이 세포 단은 일반적으로 복제로 간주됩니다. 이 인구를 수확하고 다시 줄무늬하면 후속 작업이 단일 세균 유형만 포함되도록 합니다.
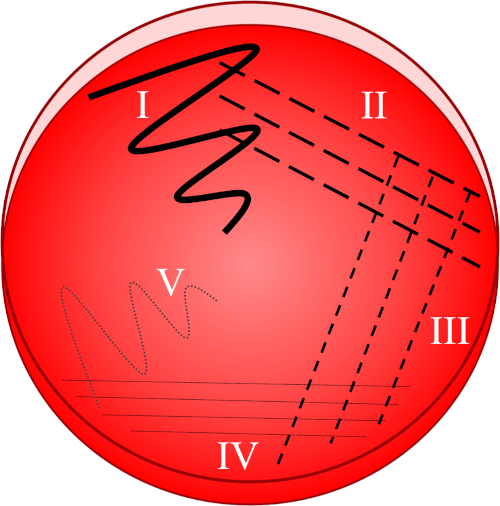
그림 1: 줄무늬 플레이트는 원래 샘플의 점진적 희석을 기반으로 합니다. I) 접종은 처음에 지그재그 운동을 사용하여 분산되어 상대적으로 조밀한 세균 인구가 있는 영역을 만듭니다. II-IV) 줄무늬는 네 번째 사분면에 도달 할 때까지, 매번 멸균 접종 루프를 사용하여, 이전 영역에서 그려집니다. V) 플레이트 의 중간을 향한 최종 지그재그 동작은 무접종이 현저하게 희석된 영역을 형성하여 식민지가 서로 분리된 것처럼 보일 수 있게 합니다.
줄무늬 플레이트 기술은 선택적 및/또는 차동 매체의 사용과 결합될 수도 있습니다. 선택적 배지는 특정 유기체의 성장을 억제합니다(예를 들어 항생제의 첨가를 통해) 차등 배지는 전적으로 다른 구별하는 데 도움이됩니다(예를 들어 혈액 천식판에 용혈을 통해).
미생물학의 모든 작업의 기본은 무균 (멸균) 기술의 사용입니다. 위험한 긴장, 에어로졸 형성 및 장비/인력의 오염의 의도하지 않은 성장의 위험이 있기 때문에 모든 세균 문화는 잠재적으로 병원성으로 간주되어야 합니다. 이러한 위험을 최소화하기 위해 모든 미디어, 플라스틱, 금속 및 유리 제품은 일반적으로 사용 전후의 오토클레이브를 통해 멸균되어 약 121°C에서 고압 포화 증기를 적용하여 느린 세포를 효과적으로 제거합니다. 작업 공간은 일반적으로 에탄올을 사용하여 소독되어 사용 전과 이후에 모두 사용한다. 실험실 코트와 장갑은 항상 전염성 요원과 함께 작업하는 동안 착용됩니다.
Procédure
Résultats
The initial streak-plate may contain colonies originating from cells with different genetic makeup or (depending on sample purity) from different bacterial species (Figure 2A).
Through subsequent isolation of a single colony, where all units are derived from a common mother-cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth (Figure 2B).

Figure 2: A pure culture can be generated from a mixed sample through isolation of a single, secluded colony. A) Growth of a single bacterial cell (CFU) generated a clonal colony, separated from those of other species and strains. This CFU was used for subsequent streaking onto a new plate B) A second plate, where the bacterial population consists solely of cells derived from the initial CFU.
Applications and Summary
The ability to obtain and cultivate a pure bacterial colony is essential, both in clinical and academic settings. Streak plating enables the isolation of a relatively clonal cell population, originating from a shared CFU, that may be of particular interest during diagnosis or for additional characterization of the isolate. A sample is streaked onto a suitable agar-based nutrient medium and incubated until colonies become visible. An isolated colony is subsequently harvested and re-streaked onto a second plate.
References
- The Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 486:207-214. (2012)
- Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 113 (21) 5970-5975 (2016)
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infection and Immunity. 66:4517-21. (1998)
- Mikucki JA, Auken E, Tulaczyk S, Virginia RA, Schamper C, Sørensen KI, Doran PT, Dugan H, Foley N. Deep groundwater and potential subsurface habitats beneath an Antarctic dry valley. Nature Communications. 6:6831. (2015)
- Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nature Microbiology. 3:132-140. (2018)
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infectious Diseases. 7:711-23 (2007)
- Yao Z, Li W, Lin Y, Wu Q, Yu F, Lin W, Lin X. Proteomic Analysis Reveals That Metabolic Flows Affect the Susceptibility of Aeromonas hydrophila to Antibiotics. Scientific Reports. 6:39413 (2016)
- Medina D, Walke JB, Gajewski Z, Becker MH, Swartwout MC, Belden LK. Culture Media and Individual Hosts Affect the Recovery of Culturable Bacterial Diversity from Amphibian Skin. Frontiers in Microbiology. 8:1574 (2017)
Transcription
On a Petri dish, if a single bacterium undergoes multiple rounds of asexual reproduction, it will lead to the formation of a clonal colony. However, obtaining a single bacterium from a mixed sample, such as a soil suspension, can be difficult. If one loopful of this heterogeneous culture is taken, it can contain as many as one trillion individual bacteria. To spread this many bacteria out onto the surface of an agar plate and obtain a single colony, even using a zig-zag pattern, the loop would need to be dragged continuously over the surface of enough plates set side-by-side to encircle the entirety of Liberty Island. Obviously, scientists do not really use that many plates. Instead, they use a technique called streak plating.
The streak plate technique is based on progressive dilution of a bacterial sample, and it is performed over the solid media surface of a single Petri dish. To begin, the media surface is visually divided into five sections by assigning four fragments of the circumference as the first four sections, and the plate’s center as the fifth. This will effectively create five media plates out of a single Petri dish. Next, using a loopful of desired inoculum, the first section is streaked using a zig-zag pattern. Then, either a new disposable loop is used, or in the case of a wire loop, it is sterilized with a Bunsen burner, flaming it until it is red hot along the length of the wire. This use of a new loop, or flame sterilize loop, removes any remaining bacterial cells, assisting in the dilution of the bacteria. The hot loop is then cooled in the air for a few seconds before being dragged through the first section to create three to four separate lines, each carrying only a fraction of bacteria into the second section. The remaining sections are streaked in the same manner, using a sterile loop each time, and a single pass through the previous streak.
Using this cycle of streaking and sterilizing, the bacterial concentration in every subsequent section should be diluted so that the final section contains only a few discretely located bacteria. Upon incubation, these discrete bacteria multiply to produce isolated clonal colonies of daughter cells, which are referred to as Colony Forming Units, or CFUs. These can be harvested and re-streaked to ensure that subsequent work involves only a single bacterial type, referred to as a pure culture. As well as isolating single colonies from a mixed-bacterial culture, the streak plating technique is also used to select media-specific strains, determine bacterial colony morphology, or identify different bacterial species. In this video, we will demonstrate how to isolate single-bacterial colonies from a mixed-bacterial sample suspension via streak plating technique.
To begin, put on laboratory gloves and a lab coat. Next, sterilize the workspace using 70% ethanol. Next, select a suitable medium that will sustain the utilized bacterial species or strain and begin preparing the media. Here, common LB agar is prepared by weighing out ten grams of pre-formulated, powdered media and 7.5 grams of agar. Add the weighed, dried components to a glass bottle which is able to hold twice the final volume to avoid overflow. Then, add 500 milliliters of water to the bottle, and cap it semi-tightly. Sterilize the media by placing the bottle in an autoclave set to 121 degrees Celsius for twenty minutes. After completion, use heat-proof gloves or a hot pad to remove the media from the machine and then immediately twist the bottle cap to close it tightly.
For the same-day use, let the media cool down by placing the bottle into a water bath heated to approximately 45 degrees Celsius, to preserve the media in a liquid state. Alternatively, the media can be left at room temperature to store at solid state. When needed, microwave the bottle with the lid slightly open to melt the media, and allow the media to cool using a 45 degree Celsius water bath.
Next, take a sleeve of sterile Petri dishes, and with a permanent marker, label them with the investigator and media names as well as the date. Then, transfer the required volume of media into a sterile vessel, and add antibiotics or other sensitive components if necessary. Here, 50 milliliters of media is mixed with 100 microliters of Kanamycin for a final concentration of 25 micrograms per milliliter. Swirl the tube to ensure even distribution of the added components throughout the media. Slowly, so as to avoid bubble formation, pour 20 to 25 milliliters of approximately 45 degree Celsius culture medium into each of the plates. If bubbles or foam appear, swiftly remove using a regular pipette and a sterile tip. Then, immediately replace all lids to prevent contamination. Allow the agar to solidify at room temperature for at least two hours or overnight. Once solidified, store the culture plates upside down at four degrees Celsius to minimize condensation on the medium’s surface.
To streak the culture of choice, first take a clean culture plate and remove the lid. Working quickly, submerge a disposable, sterile loop into the desired inoculum and then immediately swab the loop over the first quadrant of the plate using a zig-zag motion. Replace the lid of the dish, discard the used inoculation loop, and then select a new sterile loop. Using the new loop, make three to four strokes crossing the original swab line radiating from the first quadrant, which should contain a relatively dense bacteria population into the second quadrant. Close the lid once more, and discard the loop. With a new loop, repeat this action again, but this time streaking from the second into the third quadrant. Then, with a new loop again, make another streak from the third into the fourth section of the plate. Finally, with a fresh loop, make one last stroke in a zig-zag pattern from the fourth quadrant towards the center of the plate. The bacterial prevalence will be lower in this area, ideally allowing individual colonies to be established from a single viable mother cell.
Replace the plate lid, and if appropriate for the bacterial species, seal the plate with para film to prevent airflow. Turn the culture plate upside down to prevent condensation drips, and then place at a suitable temperature for growth. Here, an incubator is set to 37 degrees Celsius. Allow the plate to incubate until bacterial colonies are visible. To generate a clonal bacterial population, select one discrete colony from this plate. Now, with the sterile loop, touch the target colony, and as before, make a streak in the first quadrant of a new plate. Continue to alternately sterilize the loop and streak the remaining quadrants of the plate as previously demonstrated, ending with the zig-zag to the center. Close the plate, and place it to incubate until discrete colonies form. Once these colonies are grown, they will typically represent pure clonal strains.
The initial streak plate may contain colonies originating from cells from different bacterial species or cells with different genetic makeup, depending upon the sample purity. Through subsequent isolation of a single colony, where all units are derived from a common mother cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth.
