Biobank for Translational Medicine: Standard Operating Procedures for Optimal Sample Management
Summary
Biobanks are crucial resources for biomedical research and the Biobank for Translational and Digital Medicine Unit at the European Institute of Oncology is a model in this field. Here, we provide a detailed description of biobanks’ standard operating procedures for the management of different types of human biological samples.
Abstract
Biobanks are key research infrastructures aimed at the collection, storage, processing, and sharing of high-quality human biological samples and associated data for research, diagnosis, and personalized medicine. The Biobank for Translational and Digital Medicine Unit at the European Institute of Oncology (IEO) is a landmark in this field. Biobanks collaborate with clinical divisions, internal and external research groups, and industry, supporting patients’ treatment and scientific progress, including innovative diagnostics, biomarker discovery, and clinical trial design. Given the central role of biobanks in modern research, biobanking standard operating procedures (SOPs) should be extremely precise. SOPs and controls by certified specialists ensure the highest quality of samples for the implementation of science-based, diagnostic, prognostic, and therapeutic personalized strategies. However, despite numerous efforts to standardize and harmonize biobanks, these protocols, which follow a strict set of rules, quality controls, and guidelines based on ethical and legal principles, are not easily accessible. This paper presents the biobank standard operating procedures of a large cancer center.
Introduction
Biobanks are biorepositories aimed at the collection, storage, processing, and sharing of human biological samples and associated data for research and diagnosis. Their role is crucial not only for biomarker discovery and validation but also for the development of new drugs1. Hence, the vast majority of translational and clinical research programs rely on access to high-quality biospecimens. In this respect, biobanks are considered a bridge between academic research and the pharmaceutical/biotechnology industry2,3,4,5. Owing to the unprecedented opportunities provided by big data collection and artificial intelligence, the role of biobanks in cancer research is continuously evolving6.
The broad spectrum of biomaterials handled by biobanks is coupled with clinicopathologic information, including demographic and environmental data, tumor type, histologic grade, stage, presence of lymphovascular invasion, and biomarkers status7,8. The more high-quality specimens and data are available, the faster research will advance and impact healthcare delivery9. There is a strict regulatory framework based on ethical and legal principles that should follow widely adopted SOPs, quality controls, and guidelines (e.g., the U.S. National Cancer Institute, the U.K. Confederation of Cancer Biobanks, and the E.U. International Society for Biological and Environmental Repositories)10,11.
The development of SOPs for all major aspects of biobanks brings several advantages in terms of quality, traceability, consistency, reproducibility, and turnaround times12,13. Another important aspect of SOP implementation is represented by the optimization of biobank management, which allows for better problem-solving and alternative procedures for biobank employees and researchers14. All of these facets are part of the biobank workflow (Figure 1).
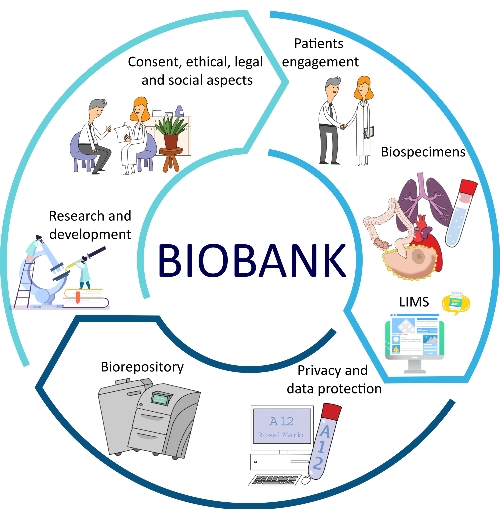
Figure 1: Different factors that contribute to the optimization of biobanking. Abbreviation: LIMS = laboratory information management system. Please click here to view a larger version of this figure.
These highly specific and sensitive data require stringent managerial standard procedures in biobanking. A detailed and validated project form should be made available to all researchers who need to get access to the biobank samples and data. The information provided in the request should include the study methodology and design, goals, objectives, and budget. A biobank Technical Scientific Committee should be established with the capital role of the evaluation of applications for research projects. This body should include members from the biobank unit, clinical divisions, research groups, data protection, legal office, and technology transfer office.
The Biobank for Translational and Digital Medicine Unit of the European Institute of Oncology (IEO) is a worldwide reference for biobanks in terms of the quality and quantity of services provided, as well as innovation. This fully certified facility (UNI EN ISO 9001:2015-Certiquality) is an integral part of the BBMRI-ERIC Italian node (i.e., Biobanking and BioMolecular Resources Research Infrastructure) and interacts with both clinical units and research infrastructure.
There is great heterogeneity in the types of biospecimens stored by biobanks. These include tissue samples-either fresh-frozen or paraffin preserved-biofluids (e.g., plasma, serum, blood, urine, stool), cell cultures, and peripheral blood mononuclear cells (PBMCs). Our biobank operates synergistically with the European research infrastructure for biobanking (BBMRI-ERIC), which is one of the largest biobank networks in Europe and provides a portal for access to biobanks and biomolecular resources coordinated by national nodes15. In addition to BBMRI-ERIC, the International Society for Biological and Environmental Repositories (ISBER) has also played an important role in the standardization of operating procedures for biobanking16.
The Biobank Unit, which is part of the Division of Pathology, is committed to the centrality of the patient, support for the development of clinical research, continuous improvement, the enhancement of human resources, international collaboration, support for training events, safety in the workplace, and scientific and technological growth. The common vision is to set up the national and European landmarks for biobanks in terms of the quality and quantity of services and innovation. The collected biological samples are used to identify new biomarkers and new drugs (e.g., to develop increasingly personalized therapies) and to ensure the best available treatment for patients through excellence in research.
Each biological specimen is collected and handled after prior verification for the presence of the Scientific Research Participation Agreement expressed by the patient15. Biological collected specimens are used to conduct research projects or clinical trials and include excess (i.e., not needed for diagnostic purposes) pathologic and non-pathologic surgical samples, liquid biopsies (e.g., blood, serum, plasma, and urine), and other biological specimens. These biomaterials are stored according to dedicated cryopreservation protocols. This paper provides the biobank protocols of a large cancer center.
Protocol
This protocol focuses on the SOPs used for breast, ovarian, prostate, lung, and colon cancer. All the procedures described here were approved by the Scientific Technical Committee, Ethics Committee (EC), and the Directors of Breast, Gynecology, Urology, Thoracic surgery, and Digestive System Surgery Programs. The study procedures follow the Declaration of Helsinki of 1964, the General Data Protection Regulation (GDPR) act of 2018, and the subsequent amendments. An institutional Research Participation Agreement (RPA), derived from the GDPR act, represents the informed consent obtained from all patients to collect biological samples and personal, clinical, and genetic data. The RPA was obtained from all patients for storing, processing, and using the obtained data for scientific purposes.
1. Prerequisites of biological samples
- Check whether a patient meets the conditions for enrollment based on the RPA and projects protocol and provide a detailed RPA description to all patients.
- Increase patient engagement, e.g., broadcast an educational cartoon video in waiting rooms to inform the patients about the importance and the impact of RPA. Provide gadget bookmarks to all the patients (see the freely accessible short cartoon animation at https://www.ieo.it/it/PER-I-PAZIENTI/I-diritti-del-paziente/Consensi-informati/ and https://vimeo.com/679070846).
- Train the personnel to carry out the consent administration during each hospitalization phase and provide additional information if the patients participate in a specific study.
NOTE: If the RPA is not signed, biological samples are not collected. - Avoid including cases that have presented infection by SARS-CoV-2 (COVID-19), hepatitis B (HBV), hepatitis C (HCV), and human immunodeficiency virus (HIV).
2. Biobank software
- Use laboratory information management system (LIMS) software to track all biological samples. Ensure the availability of a good operating system that automatically obtains personal and clinical information at patient registration and can be integrated with medical records, administrative cases, RPA, and patient pathological data, as shown in Figure 2.
- Ensure the recording of the biological samples using the biobank software.
- Identify patients using codes. Assign a unique code to each individual, which matches the medical record number (individual visit, type of patient's service).
NOTE: During registration, the Scientific RPA is electronically updated in the operating system.
- Identify patients using codes. Assign a unique code to each individual, which matches the medical record number (individual visit, type of patient's service).
- Generate an aliquot ID
- Indicate the year and the anatomical site or biofluid type (Table 1) from which the sample originates (Table 2), and add a progressive unique number per sample.
- For bilateral organs, add a sequential number to distinguish the origin of the biological sample from the right or left organ. Assign the abbreviation with the suffix 1 (for the left) or 2 (for the right)-two digits.
NOTE: For instance, a detailed ID looks like "12-B-00100-01", where 12 indicates the year, and B indicates the organ = breast.
- Register with an ID for each aliquot
- Track two macro types of biological samples: solids and liquids.
- Divide into subcategories: fresh samples and frozen samples.
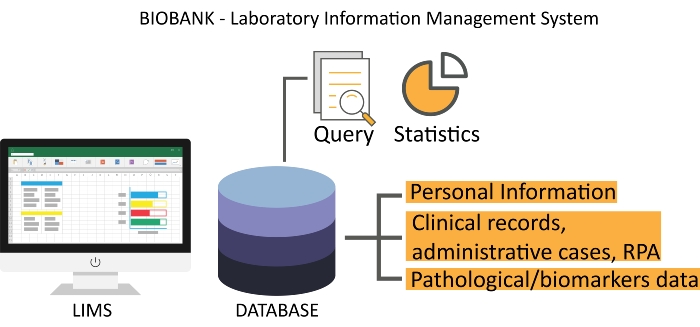
Figure 2: A representative interface of biobank LIMS. The Scientific RPA is electronically updated from the clinical record server. Abbreviations: LIMS = laboratory information management system; RPA = Research Participation Agreement. Please click here to view a larger version of this figure.
Table 1: Biofluid types and corresponding codes. Please click here to download this Table.
Table 2: Tissue sample types and corresponding codes. Please click here to download this Table.
3. Sample collection
- Daily surgery plan preparation
- Determine whether the patient has signed the RPA or not and if they are eligible.
- Check the following conditions:
- Verify patient's compliance with the inclusion criteria: report any positive case of HIV, HBV, HCV, and COVID-19 in the appropriate infective risk "RI" field to avoid collecting biological specimens.
- Verify whether the patient is enrolled in a clinical trial or specific approved research project by correctly completing the Study/Project field in each patient profile.
- Inform technicians if the risk of infection is unknown; discard specimens with positive results or unknown risks.
- Process and store samples in the biobank, as presented in Figure 3.
- Collect fresh and frozen tissue samples related to patients who underwent surgery.
- Collect cytological samples, either by aspirative or esfoliative techniques, for surgically removed small lesions.
- Collect biological fluids (e.g., blood, serum, plasma, PBMC, buccal swab, urine, feces, and ascites) of patients in the prehospitalization stage, patients enrolled in clinical trials, and any other subject involved in approved screening projects.
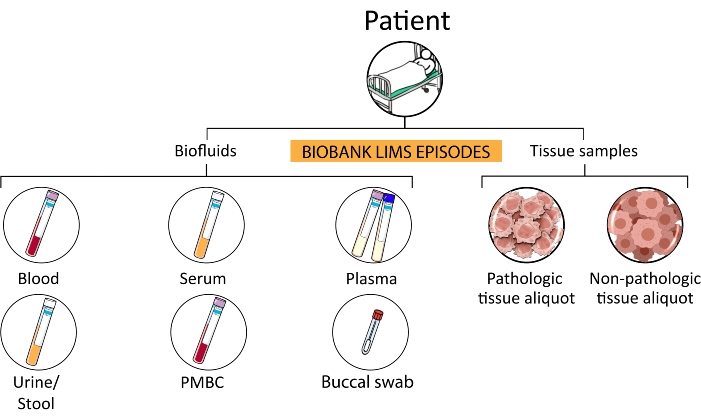
Figure 3: Sample hierarchy. From a single patient, several subcategories of episodes are processed and stored in the biobank. Abbreviations: PMBCs = peripheral blood mononuclear cells; LIMS = laboratory information management system. Please click here to view a larger version of this figure.
4. Blood sample collection
- Collect the patient's blood in 6 mL vacutainers containing the anticoagulant Na2 EDTA (7.2 mg, spray-dried). Register the vacutainers labeled with the number of the medical record and the episode code and process them in two different ways: fresh or frozen.
- For fresh samples, register the blood samples in the biobank software. Label the vacutainers with the biobank ID code and deliver them to the authorized users.
- For frozen samples stored in the biobank, prepare two Cryobank 2D coded tubes, each of 900 µL of blood (Figure 4). Register the aliquots in the biobank software, place them on a specific barcode plate, and store them in freezers (−80 °C) to ensure a constant temperature.
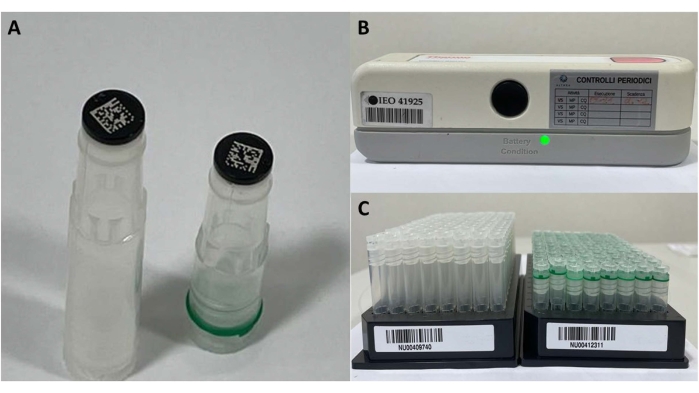
Figure 4: Frozen sample materials. (A) 2D barcode tubes, (B) barcode reader for single tube, and (C) tubes' plate for registration and storage. Please click here to view a larger version of this figure.
5. Serum sample collection
- Collect patient's blood in 6 mL vacutainer "plastic serum tubes" containing spray-coated silica to induce clotting. Leave the vacutainers for 3 h at room temperature (RT) to induce clotting, and then centrifuge at 828 x g for 10 min at 20 °C.
- Store serum (450 µL) in 0.5 mL Cryobank 2D coded tubes with a total of 3-4 aliquots for each sample. If the serum volume of the last aliquot is less than 450 µL, specify it as "LEFTOVER" during registration to track the correct amount of serum frozen.
- Register the aliquots in the biobank software, place them on a specific barcode plate, and store them in freezers (−80 °C) to ensure a constant temperature.
NOTE: The number of aliquots depends on the initial amount of whole blood taken and the amount of serum obtained.
6. Peripheral blood mononuclear cell isolation
- Open the laminar flow hood and clean it with 70% ethanol. Prepare the bag for biological waste, sterile 1x phosphate-buffered saline (PBS), and one empty sterile 50 mL conical tube.
- Pour the blood (from EDTA collection tubes) into the empty sterile 50 mL conical tube and dilute it 1:1 using sterile 1x PBS (e.g., 15 mL of blood + 15 mL of 1x PBS). Use the PBS to rinse the blood tube.
- Centrifuge the tubes at 400 x g for 30 min at 20 °C and process the tubes under a biohazard hood. Recover the middle white layer containing PBMCs using a Pasteur pipette and place it in a new sterile 50 mL conical tube. Add up to 45 mL of PBS to wash the PBMCs, mix, and centrifuge at 400 x g for 10 min at 4 °C. Recover the pellet, resuspend it in PBS, and count the cells using disposable Burker chambers.
NOTE: The volume of PBS depends on the pellet. From 15 mL of blood, ensure a resuspension volume of 2-3 mL and then a dilution of 1:10 to count. Use equation (1):
Mean of 3 squares × 10,000 × dilution factors × mL of resuspension volume = number of total cells (1) - Wash the PBMCs again with PBS, mix, and centrifuge at 400 x g for 10 min at 4 °C. Dilute the PBMCs at 2-3 x 106 cells/mL in freezer medium (fetal bovine serum (FBS) + 10% dimethyl sulfoxide [DMSO]). Prepare Cryobank 2D coded tubes by transferring 1 mL of resuspended cells into each cryotube, place them in a specific cryo box, and store them at −80 °C as soon as possible.
NOTE: Freezer medium is composed of FBS with sterile 10% DMSO and stored in aliquots at −20 °C for up to 6 months. Once slowly thawed at 4 °C, it must be used within 2 weeks.
7. Plasma sample collection
- Collect the patient's blood in 6 mL vacutainers containing the anticoagulant Na2 EDTA (7.2 mg, spray-dried). Centrifuge the vacutainer containing the whole blood at 2,000 x g for 10 min at 4 °C to separate the plasma.
- Remove the upper layer of the plasma using a 3 mL Pasteur pipette, transfer to a sterile 15 mL conical sterile tube, and centrifuge at 16,000 x g for 10 min at 4 °C to eliminate contaminating blood cells. Transfer the plasma into 1 mL Cryobank 2D coded tubes.
- Register the aliquots in the biobank software, place them on a specific barcode plate, and store them in a freezer (−80 °C) to ensure a constant temperature.
8. Stool and buccal swab sample collection
- Collect feces and buccal swabs in 15 mL tubes containing the following specific solution: 50 mM Tris-HCl, 10 mM NaCl, and 10 mM EDTA, pH 7.5.
- Register the tubes in the biobank software. Label the tubes with the biobank codes.
- Store the feces and buccal swabs at 4 °C in the biobank fridge.
9. Tissue processing
- Have tissue samples examined by a pathologist to determine whether the material that is not necessary for diagnostic procedures is sufficient for research purposes.
NOTE: When the volume of tissue is less than 1.5 mm3, it is either stored in OCT or provided as a fresh sample (A) linked to a specific research request. - Whenever possible, collect even the non-pathological counterpart (NP) of the pathological tissue (P). Place the samples in sterile cell culture Petri dishes labeled as P and NP (Figure 5). Keep the tissue on the ice at 4 °C and divide it into three parts each (A, B, and C) if enough material is available.
- Fresh tissue samples (A): place fresh aliquots of P and NP tissue in tubes with the appropriate culture medium defined in each specific protocol and send them to external research units (e.g., research laboratories).
- OCT tissue samples (B): place fresh aliquots of P and NP tissue in cryomolds, fill them with OCT resin (10.24% polyvinyl alcohol, 4.26% polyethylene glycol, and 85.5% non-reactive ingredients), and immediately place them in a flash-freeze apparatus at −80 °C.
NOTE: At −80 °C, OCT-embedded tissue takes from 60-150 s to freeze. - Tissue samples (C): place the remaining tissue samples in Cryobank 2D coded tubes in the flash-freeze apparatus.
- Store the plates at −80 °C.
NOTE: For each frozen OCT aliquot, perform a quality check test before distribution and its usage to have a histologic evaluation, as described in section 11.
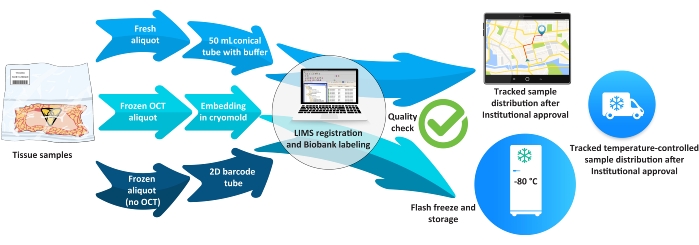
Figure 5: Biobank workflow for tissue samples. Abbreviations: LIMS = laboratory information management system; OCT = optimal cutting temperature. Please click here to view a larger version of this figure.
10. Tissue freezing
- Freeze tissue quickly in isopentane vapors using a flash-freeze apparatus. Put the tissue samples in isopentane at −120 °C for 3 min and store them in the cryopreservation rooms at −80 °C.
NOTE: Two methodologies are used for freezing samples of tumor and healthy tissue using flash freeze to maintain the integrity of the nucleic acids.
11. Tissue quality control
- House the cryostat instrument in a refrigerated container at a temperature between −20 ° C and −40 ° C, and cut a few cryosections from the OCT block17. Perform hematoxylin and eosin (H&E) staining on the tissue cryosection18. Analyze the morphology of the tissue under an optical microscope19.
NOTE: The sections have a suitable thickness from 3-12 µm.- Fill out a specific form (Table 3).
Table 3: Quality control form of OCT-embedded and frozen tissue sections. Abbreviation: OCT = optimal cutting temperature. Please click here to download this Table.
12. Request and retrieval of specimens for research purposes
- Request stored aliquots:
- Obtain a completed form with the research project name, principal investigator (PI), pickup date, and a brief description. Run a query on the software database (DB), select the aliquot, generate a pickup list for the technicians, and check the barcode for each retrieved aliquot.
NOTE: To obtain samples from the biobank, researchers from our institute or external collaborators (for-profit or not-for-profit) must apply using a specific form, and the project is evaluated by a technical-scientific committee and the Ethics Committee.
- Obtain a completed form with the research project name, principal investigator (PI), pickup date, and a brief description. Run a query on the software database (DB), select the aliquot, generate a pickup list for the technicians, and check the barcode for each retrieved aliquot.
Representative Results
Following the SOPs described above, we collected a total of 38,446 annotated biological liquid biopsies and a total of 10,205 tissue samples from April 2012 to December 2021 (Figure 6A). In addition, we analyzed in detail the collected samples from the Divisions of Urology, Gynecology, Senology, as well as the Divisions of Head and Neck, Abdominal-Pelvic, and Thoracic Surgery. The highest number of tissue samples we collected were from breast tumors (Figure 6B,C). Since 2019, we have also started collecting other biological specimens, such as urine, stool, and buccal swabs, following the significantly increased demand over the years (Figure 6D).
As shown in Figure 6A, the amount of collected samples, especially tissues, during 2020-2021 suffered due to the COVID-19 pandemic and the related reduction in oncologic procedures. Importantly, scientific work did not diminish during this period due to the use of properly stored and annotated biobank samples collected in the previous years. Proper collection of biological specimens and associated clinicopathological data allowed us to have a well-structured and competitive retrospective and prospective biobank. To this end, the selection of the surgical specimen must be performed by the pathologist, both to ensure a correct diagnosis and have the opportunity to conduct research with appropriate tissue specimens. In our biobank, specific work procedures are firmly established and followed, so that we apply standardized procedures that comply with the certification ISO 9001 in the context of biobanks for research.
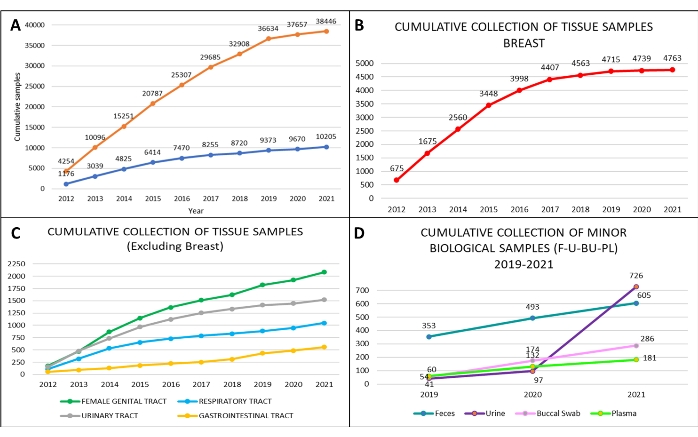
Figure 6: Biobank cumulative collection of biospecimens at the European Institute of Oncology, from 2012 to 2021. Cumulative collection of (A) tissue samples (orange curve) and blood with serum samples (blue curve); cumulative collection of (B) breast tissue samples (red curve); cumulative collection of (C) ovary tissue samples (green curve), prostate (grey curve), lung (light blue curve), and colon tissue samples (yellow curve). From 2019 to 2021, cumulative collection of (D) additional biological samples: feces (blue line), a buccal swab (pink line), plasma (light green line), and urine (violet line). Please click here to view a larger version of this figure.
Discussion
Although oncology has made tremendous advances, cancer remains a leading cause of morbidity and mortality worldwide20. Understanding tumor heterogeneity, its temporal evolution over time, and the outcomes of targeted treatment are strictly dependent on accurate data collection in the context of routine clinical care21. In this respect, the "multi-omics" approach is gaining momentum in oncologic predictive pathology22. The traditional tissue-based biomarker assessment is being integrated using multiple new bioanalytes, such as blood, plasma, urines, saliva, and stool23,24,25,26. Therefore, biobanks are now recognized as pivotal infrastructures to enhance clinical practice. Looking back at the history of cancer research, we realize that the most impressive and groundbreaking discoveries would never have been possible without direct examination of cancer tissue or liquid biopsies. Over time, the source of cancer tissue and the type of liquid biopsy to be examined has evolved from crude dissections, random "chance encounters", and in some cases, illicit trafficking to organized cancer collections and strategic modern oncology banks. Consideration of many ethical issues has changed considerably both in practice and in the main factors that distinguish modern oncology banks from the oncology collections of the past.
Due to the advances in cancer research and the vast amount of molecular information that is now provided by modern technologies, it is becoming more and more evident that biobanks, particularly those in cancer centers, may face several types of methodological issues. Among these, technology has become a universal challenge that still prevents SOP standardization and harmonization. Another critical aspect for maintaining core biobank activities is to have an integrated LIMS software able to receive and automatically maintain all hospital IDs and all the codified clinical data coming from the hospital's software. It is noteworthy that other valuable software used to manage biobanks and some freeware can be obtained for biobank managing27,28,29,30,31. Another critical step in biobanks is the implementation of the participation pact for all patients and the legal and ethical agreement necessary for the storage of clinical data and biospecimens10,32.
In this regard, this protocol has well-defined guidelines that do not permit the collection and storage of biospecimens in the absence of consent. This is also a critical issue since patients can withdraw their participation even after their samples have been stored; thus, methods to rapidly take such samples out of the biobanking system have been implemented. Biospecimens that arrive from patients recruited by our biobank follow strict protocols for collection and storage. In this regard, several important aspects have been evaluated to monitor this process and are being continuously improved. In particular, ISO9001 certification requires several indicators of performance, such as warm ischemic time, which has to be maintained for under 30 min or 60 min depending on the source of the tissue. Additionally, liquid biopsies and biological fluids are collected using standardized protocols following strict time procedures15,33,34,35,36.
Specific features are of great importance in biobanks' workflows. These include the presence of a certified pathologist, which guarantees the sampling of the tissue for diagnostic reasons, and the collection of tissue for biobanking in a time frame compatible with a high quality of samples (ischemic time is an important indication for some types of research, such as RNA-dependent assays, which require less warm ischemic time). Moreover, managing the space required for specimen storage is of great importance in biobanks. The number of collected liquid biopsies could be driven by the study design. Liquid biopsies may often be collected both during the preoperative and the follow-up period, as defined by each study design.
Owing to screening campaigns for cancer prevention and the early diagnosis of tumors, i.e., small-sized breast tumors at early stages of development, as well as the availability of minimally invasive surgical techniques, have reduced the number of tissues samples available for research (as most tissue samples are always used for diagnostic purposes). The capacity to collect and store biological specimens has improved considerably over the last few years. This could be observed for biological fluids, reflecting the increased capacity of this biobank to support this institute's research groups in the growing demand for patient-derived annotated material. Despite these improvements, we have experienced some limitations for multi-center studies that require coordination between biobanks from different parts of the world, which can be integrated only by implementing similar procedures.
Having ruled out most of the ethical and technical issues regarding biobanking, including the collection of all clinical and demographic information, the next goal is to implement the digitalization of all the histological preparations and staining used for diagnosis and research purposes. This is of fundamental importance for the next generation of studies that will greatly benefit from a fully integrated digital pathology and biobank, which is going to become the standard for the future. Only a large series of patients with integrated data and digital images may fuel multi-center, large artificial intelligence (AI) studies for the improvement of cancer patient care. In conclusion, we believe that good healthcare does not end with diagnosis and treatment. Best practices comprise finding the ways of continuous diagnosis and treatment improvement for any disease, in particular ones that severely affect life expectancy or quality.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank all the patients who actively participated during the last decade in our research programs through the donation of their biospecimens. Without them, this research would not be possible. We are also grateful to all the personnel working at IEO, nurses, technicians, biologists, doctors, and the directors of all the clinical and research units. The authors are grateful to Prof. Pier Paolo Di Fiore and Prof. Giancarlo Pruneri for their guidance. Finally, we dedicate this work to Prof. Umberto Veronesi, the founder of IEO, and his pioneering approach to integrating cancer research and patient care.
Materials
| Blue Max Con Tubes 15 mL | Falcon B.D | 352096 | |
| Blue Max Con Tubes 50 mL | Euroclone Spa | FLC352070 | |
| Box with 81 position for tissue storage | Ettore Pasquali Srl. | 06.0945.00 | |
| cf-DNA/cf-RNA Preservative Tubes | Norgen Biotek | 63950 | Preservation and isolation of both cf-DNA and cf-RNA from a single tube and in particular preserve cf-DNA/ct-DNA for 30 days at ambient temperature and for up to 8 days at 37 °C |
| Cryomold Standard (25 X 20 X 5 mm) | Olympus Italia S.r.l. | 4557 | Disposable plastic Cryomold molds create a uniformly shaped, flat-surface specimen block when used with O.C.T |
| Dimethyl Sulfoxide Plastic Bottle – 1 L | Vwr International S.R.L. | MFCD00002089 | It acts to preserve the reconstitution of the medium for the storage of frozen cells |
| Dpbs 1x W/o Ca And Mg – 500 mL | Microtech Srl | TL1006-500ML | Washing Buffer cell |
| Dualfilter T.I.P.S 1,000 µL | Euroclone Spa | 4809 | |
| Dualfilter T.I.P.S 200 µL | Euroclone Spa | 4823 | |
| Easytrack Barcode Reader for single tube datamatrix | Twin Helix Srl | TH-ETR4400 | 2D barcode tubes reader with USB connection |
| Fetal Bovine Serum Origin Brazileu S/fil | Microtech S.R.L | RM10532-500ML | Defrost at +4 °C, usually for two days, and once melted, start decomplementation at 56 °C for 45 min Let it cool down to room temperature, and aliquot it. Refroze them to -20 °C, and remember to defrost them every time the aliquots are needed |
| Ficoll Paque Plus (ge) 6 x 500 mL | Euroclone Spa | GEH17144003 | Ficoll is a medium for density gradient, It is sterile and ready for use. It alloes to get peripheral blood mononuclear cells, bone marrow and umbilical cord blood |
| Fixing solution Killik of 100 mL (OCT) | Bio-optica Milano S.p.a. | 05-9801 | Gel inclusion medium that solidifies at cold the water-soluble tissue (e.g., biopsies, frustules) |
| FLASH-FREEZE | Milestone | n.a. | Freezing appliance |
| Forma 8600 Series Chest Freezers (Temperature Range: -50 °C to -86 °C) 85 liters | Thermo Fisher Scientific Srl | 803CV | Orizzontal freezer |
| Isopentane 500 mL | Vwr International S.R.L. | 24872260 | Liquid included in theFLASH-FREEZE camera for freezing |
| Nautilus Lims Software | Thermo Scientific™ | n.a. | The software implementation is able to track all patients’ biological samples. Receives Personal and Clinical information automatically during registration due to the integration with IEO operating systems. Nautilus is integrated with the web service through three IEO operative systems: BAC – IEO central registry with personal information, wHospital – medical record |
| Pasteur pipette 10 mL | Euroclone Spa | CC4488 | |
| Pasteur pipette 3 mL | Euroclone Spa | APT1502 | |
| PATHOX | Dedalus ItaliTesi Elettronica e Sistemi Informativi S.p.A.a S.p.A. | n.a. | PATHOX – management system for the Pathology unit where several factors are registered for the Biobank, such as the histological samples, the related diagnoses, and biomarkers |
| Petri dishes, polystyrene – size 100 mm x 20 mm, slippable | Euroclone Spa | FLC353003 | |
| Set of 4 adapters 19 x 5/7 mL vac | Thermo Fisher Scientific Srl | 75003680 | |
| Set of 4 adapters 4 x 50 conical | Thermo Fisher Scientific Srl | 75003683 | |
| Set of 4 adapters 9 x 15 mL conical | Thermo Fisher Scientific Srl | 75003682 | |
| Single-use slide for counting cell | Biosigma S.P.A. | 347143/001 | Specifically used for individual cell count |
| Stamps Freezerbondz for tissue boxes, nitrogen-liquid proof , H 9,53 mm x L 25,40 mm | Twin Helix Srl | THT-152-492-3 | |
| Thermo Scientific TSX Series Ultra-Low Freezers (-50 °C to -86 °C) 949 liters | Thermo Fisher Scientific Srl | TSX70086V | Vertical freezer |
| Thermo Scientific Refrigerated Centrifuge SL16R | Thermo Fisher Scientific Srl | 75004030 | |
| Tissue box labels in Permanent | Twin Helix Srl | THT-199-482-3 | |
| Tuerks Solution | Merck Life Science S.R.L. | 1092770100 | In light microscopy, it is specifically used as stain for leukocyte |
| TX-400 Rotor TX-400 swinging bucket hol | Thermo Fisher Scientific Srl | 75003181 | |
| White box for storage | Bio Optica | 07-7300 | |
| wHospital Software | wHealth Lutech Group | n.a. | wHospital – medical record management system with personal information, administrative cases, and the informed consent of the patients |
References
- Pagni, F., et al. Targeting immune-related biological processes in solid tumors: We do need biomarkers. International Journal of Molecular Sciences. 20 (21), 5452 (2019).
- Braun, K. L., et al. Cancer patient perceptions about biobanking and preferred timing of consent. Biopreservation and Biobanking. 12 (2), 106-112 (2014).
- Bycroft, C., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 562 (7726), 203-209 (2018).
- Saifuddin, S. R., et al. King’s Health Partners’ Prostate Cancer Biobank (KHP PCaBB). BMC Cancer. 17 (1), 784 (2017).
- Lopez, G., et al. Molecular insights into the classification of luminal breast cancers: The genomic heterogeneity of progesterone-negative tumors. International Journal of Molecular Sciences. 20 (3), 510 (2019).
- Kinkorová, J. Biobanks in the era of personalized medicine: Objectives, challenges, and innovation: Overview. The EPMA Journal. 7 (1), 4 (2015).
- Luo, J., et al. Intravital biobank and personalized cancer therapy: The correlation with omics. International Journal of Cancer. 135 (7), 1511-1516 (2014).
- Invernizzi, M., et al. Quality of life interventions in breast cancer survivors: State of the art in targeted rehabilitation strategies. Anticancer Agents in Medicinal Chemistry. 22 (4), 801-810 (2021).
- Roux, J., Zeghidi, M., Villar, S., Kozlakidis, Z. Biosafety and biobanking: Current understanding and knowledge gaps. Biosafety and Health. 3 (5), 244-248 (2021).
- Sanchini, V., et al. A trust-based pact in research biobanks. From theory to practice. Bioethics. 30 (4), 260-271 (2016).
- Vaught, J., Kelly, A., Hewitt, R. A review of international biobanks and networks: Success factors and key benchmarks. Biopreservation and Biobanking. 7 (3), 143-150 (2009).
- Ferrin, I., et al. Isolation, culture, and expansion of mesenchymal stem cells. Methods in Molecular Biology. 1590, 177-190 (2017).
- Hermansen, J. U., et al. The Norwegian childhood cancer biobank. Cancer Reports. , 1555 (2021).
- Schmelz, M., et al. A plan for emergency shutdown and reopening for a consortium of biobanks. Biopreservation and Biobanking. 19 (5), 394-398 (2021).
- Salvaterra, E., Corfield, J. . Advances in Biobanking Practice Through Public and Private Collaborations. , (2017).
- Snapes, E., Simeon-Dubach, D. ISBER best practices for repositories, moving toward the fifth edition. Biopreservation and Biobanking. 20 (1), 107-108 (2022).
- Fischer, A. H., Jacobson, K. A., Rose, J., Zeller, R. Cryosectioning tissues. Cold Spring Harbour Protocols. (8), 4991 (2008).
- Staining methods in frozen section: Best lab practices. Laboratory Best Practice Blog. UC Davis Health Available from: https://health.ucdavis.edu/blog/lab-best-practice/staining-methods-in-frozen-section-best-lab-practices/2020/03 (2020)
- Craciun, L., et al. Tumor banks: A quality control scheme proposal. Frontiers in Medicine. 6, 225 (2019).
- Ma, X., Yu, H. Global burden of cancer. The Yale Journal of Biology and Medicine. 79 (3-4), 85-94 (2006).
- Angerilli, V., et al. The role of the pathologist in the next-generation era of tumor molecular characterization. Diagnostics. 11 (2), 339 (2021).
- Correa-Aguila, R., Alonso-Pupo, N., Hernández-Rodríguez, E. W. Multi-omics data integration approaches for precision oncology. Molecular Omics. , (2022).
- Salati, M., et al. ctDNA analysis in the personalized clinical management of gastroesophageal adenocarcinoma: Turning hope into reality. Future Oncology. 17 (33), 4607-4618 (2021).
- Mirzayi, C., et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nature Medicine. 27 (11), 1885-1892 (2021).
- Cortvrindt, C., Speeckaert, R., Delanghe, J. R., Speeckaert, M. M. Urinary epidermal growth factor: A promising "next generation" biomarker in kidney disease. American Journal of Nephrology. , (2022).
- Fusco, N., Fumagalli, C., Guerini-Rocco, E. Looking for sputum biomarkers in lung cancer secondary prevention: Where are we now. Journal of Thoracic Disease. 9 (11), 4277-4279 (2017).
- Im, K., Gui, D., Yong, W. H. An introduction to hardware, software, and other information technology needs of biomedical biobanks. Methods in Molecular Biology. 1897, 17-29 (2019).
- Paul, S., Gade, A., Mallipeddi, S. The state of cloud-based biospecimen and biobank data management tools. Biopreservation and Biobanking. 15 (2), 169-172 (2017).
- Fthenou, E., et al. implementation, and integration of heterogenous information technology infrastructures in the Qatar biobank. Biopreservation and Biobanking. 17 (6), 494-505 (2019).
- Tukacs, E., et al. Model requirements for Biobank Software Systems. Bioinformation. 8 (6), 290-292 (2012).
- Willers, C., et al. A versatile, secure, and sustainable all-in-one biobank-registry data solution: The A3BC REDCap model. Biopreservation and Biobanking. , (2021).
- D’Abramo, F., Schildmann, J., Vollmann, J. Research participants’ perceptions and views on consent for biobank research: A review of empirical data and ethical analysis. BMC Medical Ethics. 16, 60 (2015).
- Policiuc, L., et al. The foundation of personalized medicine is the establishment of biobanks and their standardization. Journal of BUON. 23 (3), 550-560 (2018).
- Lygirou, V., Makridakis, M., Vlahou, A. Biological sample collection for clinical proteomics: Existing SOPs. Methods in Molecular Biology. 1243, 3-27 (2015).
- Pisapia, P., Malapelle, U., Troncone, G. Liquid biopsy and lung cancer. Acta Cytologica. 63 (6), 489-496 (2019).
- Spruessel, A., et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 36 (6), 1030-1037 (2004).

