Summary
The genome is organized in the nuclear space into different structures that can be revealed through chromosome conformation capture technologies. The in-nucleus Hi-C method provides a genome-wide collection of chromatin interactions in Drosophila cell lines, which generates contact maps that can be explored at megabase resolution at restriction fragment level.
Abstract
The genome is organized into topologically associating domains (TADs) delimited by boundaries that isolate interactions between domains. In Drosophila, the mechanisms underlying TAD formation and boundaries are still under investigation. The application of the in-nucleus Hi-C method described here helped to dissect the function of architectural protein (AP)-binding sites at TAD boundaries isolating the Notch gene. Genetic modification of domain boundaries that cause loss of APs results in TAD fusion, transcriptional defects, and long-range topological alterations. These results provided evidence demonstrating the contribution of genetic elements to domain boundary formation and gene expression control in Drosophila. Here, the in-nucleus Hi-C method has been described in detail, which provides important checkpoints to assess the quality of the experiment along with the protocol. Also shown are the required numbers of sequencing reads and valid Hi-C pairs to analyze genomic interactions at different genomic scales. CRISPR/Cas9-mediated genetic editing of regulatory elements and high-resolution profiling of genomic interactions using this in-nucleus Hi-C protocol could be a powerful combination for the investigation of the structural function of genetic elements.
Introduction
In eukaryotes, the genome is partitioned into chromosomes that occupy specific territories in the nuclear space during interphase1. The chromatin forming the chromosomes can be divided into two main states: one of accessible chromatin that is transcriptionally permissive, and the other of compact chromatin that is transcriptionally repressive. These chromatin states segregate and rarely mix in the nuclear space, forming two distinct compartments in the nucleus2. At the sub-megabase scale, boundaries separate domains of high-frequency chromatin interactions, called TADs, that mark chromosomal organization3,4,5. In mammals, TAD boundaries are occupied by cohesin and CCCTC-binding factor (CTCF)6,7,8. The cohesin complex extrudes chromatin and halts at CTCF-binding sites that are disposed in a convergent orientation in the genomic sequence to form stable chromatin loops9,10,13,14. Genetic disruption of the CTCF DNA-binding site at the boundaries or reduction in CTCF and cohesin protein abundance results in abnormal interactions between regulatory elements, loss of TAD formation, and gene expression deregulation9,10,11,13,14.
In Drosophila, the boundaries between TADs are occupied by several APs, including boundary element-associated factor 32 kDa (BEAF-32), Motif 1 binding protein (M1BP), centrosomal protein 190 (CP190), suppressor of hairy-wing (SuHW), and CTCF, and are enriched in active histone modifications and Polymerase II16,17,18. It has been suggested that in Drosophila, TADs appear as a consequence of transcription13,17,19, and the exact role of independent APs in boundary formation and insulation properties is still under investigation. Thus, whether domains in Drosophila are a sole consequence of the aggregation of regions of similar transcriptional states or whether APs, including CTCF, contribute to boundary formation remains to be fully characterized. Exploration of genomic contacts at high resolution has been possible through the development of chromosome conformation capture technologies coupled with next-generation sequencing. The Hi-C protocol was first described with the ligation step performed "in solution"2 in an attempt to avoid spurious ligation products between chromatin fragments. However, several studies pointed to the realization that the useful signal in the data came from ligation products formed at partially lysed nuclei that were not in solution20,21.
The protocol was then modified to perform the ligation inside the nucleus as part of the single-cell Hi-C experiment22. The in-nucleus Hi-C protocol was subsequently incorporated into cell population Hi-C to yield a more consistent coverage over the full range of genomic distances and produce data with less technical noise23,24. The protocol, described in detail here, is based on the population in-nucleus Hi-C protocol23,24 and was used to investigate the consequences of genetically removing DNA-binding motifs for CTCF and M1BP from a domain boundary at the Notch gene locus in Drosophila25. The results show that altering the DNA-binding motifs for APs at the boundary has drastic consequences for Notch domain formation, larger topological defects in the regions surrounding the Notch locus, and gene expression deregulation. This indicates that genetic elements at domain boundaries are important for the maintenance of genome topology and gene expression in Drosophila25.
Protocol
1. Fixation
- Start with 10 million Schneider's line 2 plus (S2R+) cells to prepare 17.5 mL of a cell suspension in Schneider medium containing 10% fetal bovine serum (FBS) at room temperature (RT).
- Add methanol-free formaldehyde to obtain a final concentration of 2%. Mix and incubate for 10 min at RT, taking care to mix every minute.
NOTE: Formaldehyde is a hazardous chemical. Follow the appropriate health and safety regulations, and work in the fume hood. - Quench the reaction by adding glycine to achieve a final concentration of 0.125 M and mix. Incubate for 5 min at RT, followed by 15 min on ice.
- Centrifuge for 400 × g at RT for 5 min and then for 10 min at 4 °C; discard the supernatant. Resuspend the pellet carefully in 25 mL of cold 1x phosphate-buffered saline.
- Centrifuge at 400 × g for 10 min at 4 °C, then discard the supernatant.
NOTE: If continuing with the protocol, go to step 2.1 for lysis; otherwise, flash-freeze the pellet in liquid N2 and store the pellet at -80 °C.
2. Lysis
- Resuspend the cells in 1 mL of ice-cold lysis buffer (10 mM Tris-HCl, pH 8; 0.2% of non-ionic surfactant (see the Table of Materials); 10 mM NaCl; 1x protease inhibitors), and adjust the volume to 10 mL with ice-cold lysis buffer. Adjust the volume to obtain a concentration of 1 × 106 cells/mL. Incubate on ice for 30 min, mixing every 2 min by inverting the tubes.
- Centrifuge the nuclei at 300 × g for 5 min at 4 °C, and then carefully discard the supernatant. Wash the pellet 1x with 1 mL of cold lysis buffer, and transfer it to a microcentrifuge tube. Wash the pellet 1x with 1 mL of cold 1.25x restriction buffer, and resuspend each cell pellet in 360 µL of 1.25x restriction buffer.
- Add 11 µL of 10% sodium dodecyl sulfate (SDS) per tube (0.3% final concentration), mix carefully by pipetting, and incubate at 37 °C for 45 min, shaking at 700-950 rpm. Pipet up and down to disrupt clumps a few times during incubation.
- Quench the SDS by adding 75 µL of non-ionic surfactant (10% solution, see the Table of Materials) per tube (1.6% final concentration), and incubate at 37 °C for 45 min, shaking at 950 rpm. Pipet up and down a few times to disrupt clumps during incubation.
NOTE: If clumps are large and difficult to disrupt, decrease the rotating speed to 400 rpm during SDS and surfactant treatments. If the clumps are difficult to disaggregate by pipetting, split the sample into two; adjust the volumes of restriction buffer, SDS, and surfactant; and proceed with the permeabilization. Next, spin the nuclei at minimum speed (200 × g), carefully discard the supernatant, pool the samples together in 1X restriction buffer, and proceed with digestion. Take a 10 µL aliquot as the undigested sample (UD).
3. Enzymatic digestion
- Digest the chromatin by adding 200 units (U) of Mbo I per tube, and incubate at 37 °C for a period ranging from 4 h to overnight while rotating (950 rpm).
- On the next day, add an additional 50 U of Mbo I per tube, and incubate at 37 °C for 2 h while rotating (950 rpm).
- Inactivate the enzyme by incubating the tubes at 60 °C for 20 min. Place the tubes on ice.
NOTE: Take a 10 µL aliquot as the digested sample (D).
4. Biotinylation of DNA ends
- To fill in the restriction fragment overhangs and label the DNA ends with biotin, add 1.5 µL each of 10 mM dCTP, dGTP, dTTP, 20 µL of 0.4 mM biotin dATP, 17.5 µL of Tris low-EDTA (TLE) buffer [10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0], and 10 µL of 5 U/µL of Klenow (DNA polymerase I large fragment) to all the tubes. Mix carefully and incubate for 75 min at 37 °C. Shake at 700 rpm every 10 s for 30 s. Place all the tubes on ice while preparing the ligation mix.
5. Ligation
- Transfer each digested chromatin mixture to a separate 1 mL tube with ligation mix (100 µL of 10x ligation buffer, 10 µL of 10 mg/mL bovine serum albumin, 15 U of T4 DNA ligase, and 425 µL of double-distilled water [ddH2O]). Mix thoroughly by gentle pipetting, and incubate overnight at 16 °C.
6. Crosslink reversal and DNA purification
- Degrade proteins by adding 50 µL of 10 mg/mL Proteinase K per tube, and incubate at 37 °C for 2 h. Reverse crosslinks by increasing the temperature to 65 °C and incubate overnight.
- Degrade RNA by adding 20 µL of 10 mg/mL RNase A, and incubate at 37 °C for 1 h.
- Perform phenol:chloroform extraction followed by ethanol precipitation.
- Add 1 volume of phenol-chloroform, and mix thoroughly by inversion to obtain a homogeneous white phase.
NOTE: Phenol is a hazardous chemical. Follow the appropriate health and safety regulations. Work in the fume hood. - Centrifuge at 15,000 × g for 15 min. Transfer the aqueous phase into a fresh 2 mL microcentrifuge tube. Perform a back-extraction of the lower layer with 100 µL of TLE buffer, and transfer the aqueous phase into the same 2 mL tube.
- Precipitate DNA by adding 2 volumes of 100% ethyl alcohol (EtOH), 0.1 volumes of 3 M sodium acetate, and 2 µL of 20 mg/mL glycogen. Incubate at -20 °C for a period ranging from 2 h to overnight.
- Spin at 15,000 × g for 30 min at 4 °C, and wash the pellets 2x with ice-cold 70% EtOH. Dry the pellets at RT, and resuspend in 100 µL of TLE buffer. Quantify the DNA using a fluorogenic dye that binds selectively to DNA and a fluorometer according to the manufacturer's instructions (Table of Materials).
NOTE: The protocol can be paused here. Store ligation products at 4 °C for the short term or at -20 °C for the long term. Use an aliquot (100 ng) of the material as the ligated sample (L) for quality control.
- Add 1 volume of phenol-chloroform, and mix thoroughly by inversion to obtain a homogeneous white phase.
7. Assess Hi-C template quality
- Digestion and ligation qualitative controls
- Purify DNA from the UD and D aliquots by reversing the crosslinks, and perform phenol:chloroform extraction and ethanol precipitation as described above.
- Load 100 ng of UD, D, and L samples in a 1.5% agarose gel. Look for a smear centered around 500 bp in the D sample versus a high molecular weight band for the L sample (see representative results).
- Digestion efficiency quantitative control
NOTE: To assess the digestion efficiency more accurately, use the UD and D samples as templates to perform quantitative polymerase chain reactions (qPCR) using primers designed as follows.- Design a primer pair that amplifies a DNA fragment containing the DNA restriction site for the enzyme used for digestion (Mbo I in the present protocol), called R in the formula in step 7.2.3.
- Design a primer pair that amplifies a control DNA fragment that does not contain the restriction site for the enzyme used for digestion (Mbo I for the present protocol), called C in the formula in step 7.2.3.
- Use the cycle threshold values (Ct values) of the amplification to calculate restriction efficiency according to the formula shown below:
% Restriction = 100 – 100/2{(CtR – CtC)D – (CtR – CtC)UD}
Where CtR refers to the Ct value of fragment R, and CtC refers to the Ct value of the fragment C for sample D and sample UD.
NOTE: The restriction percentage reflects the efficiency of the restriction enzyme cleaving the restricted (R) DNA fragment compared to a control (C) DNA fragment that does not contain the restriction DNA site. A restriction efficiency of ≥ 80% is recommended.
- Detection of known interactions
- Perform PCR to amplify an internal ligation control to examine short-range and/or medium- or long-range interactions (see representative results).
- Alternatively, design primers to amplify a ligation product in which the primers are in forward-forward or reverse-reverse orientation in adjacent restriction fragments.
- Fill-in and biotin-labeling control
- Verify Hi-C marking and ligation efficiency by amplification and digestion of a known interaction or a ligation product between adjacent restriction fragments in the genome, as described above.
NOTE: Successful fill-in and ligation of the Mbo I site (GATC) generates a new site for the restriction enzyme Cla I (ATCGAT) at the ligation junction and regenerates the Mbo I site. - Digest the PCR product with Mbo I, Cla I, or both. After running the samples on a 1.5-2% gel, estimate the relative number of 3C and Hi-C ligation junctions by quantifying the intensity of the cut and uncut bands26.
NOTE: An efficiency of > 70% is desired (see representative results).
- Verify Hi-C marking and ligation efficiency by amplification and digestion of a known interaction or a ligation product between adjacent restriction fragments in the genome, as described above.
8. Sonication
- Sonicate the samples to obtain 200-500 bp DNA fragments. For the instrument used in this protocol (see the Table of Materials), dilute the sample (from 5 to 10 µg) in 130 µL of ddH2O per tube, and set the instrument to sonicate to 400 bp: fill level: 10; duty factor: 10%; peak incident power (w): 140; cycles per burst: 200; time (s): 80.
9. Biotin removal/end repair
NOTE: The steps shown below are adjusted for 5 µg of Hi-C DNA.
- To perform biotin removal, transfer the sample (130 µL) into a fresh microcentrifuge tube. Add 16 µL of 10x ligation buffer, 2 µL of 10 mM dATP, 5 µL of T4 DNA Polymerase (15U), and 7 µL of ddH2O (160 µL of total volume). Incubate at 20 °C for 30 min.
- Add 5 µL of 10 mM dNTPs, 4 µL of 10x ligation buffer, 5 µL of T4 polynucleotide kinase (10 U/µL), 1 µL of Klenow, and 25 µL of ddH2O (200 µL of total volume). Incubate at 20 °C for 30 min.
10. Size selection
- To select fragments mostly in the 250-550 bp size range, perform sequential solid phase reversible immobilization (SPRI) size selection first with 0.6x, followed by 0.90x according to the manufacturer's instructions, and elute the DNA using 100 µL of TLE.
11. Biotin pulldown/A-tailing/adapter ligation
NOTE: Perform the washes by resuspending the magnetic beads by vortexing, rotate the samples for 3 min on a rotating wheel, and then briefly spin down the sample and place it on the magnetic stand. Allow the beads to stick to the magnet, discard the supernatant, and proceed with the following wash step. Perform the washes at 55 °C on a thermo-block with rotation instead of the rotating wheel.
- Make up the final volume to 300 µL per sample with TLE for pull-down. Prepare the bead-washing buffers: 1x Tween Buffer (TB) (TB: 5 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 1 M NaCl, 0.05% Tween), 0.5x TB, 1x No-Tween buffer (NTB) (5 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 1 M NaCl), 2x NTB.
- Use 150 µL of streptavidin-linked magnetic beads (see the Table of Materials) per library. Wash the beads 2x with 400 µL of 1x TB. Then resuspend beads in 300 µL 2x NTB.
- Mix the beads with the 300 µL Hi-C material and incubate at RT for 30 minutes on a rotating wheel to allow biotin binding to streptavidin beads.
- Wash the beads with 400 µL of 0.5x TB, and incubate at 55 °C for 3 min, rotating at 750 rpm. Wash the beads in 200 µL of 1x restriction buffer.
- Resuspend the beads in 100 µL of dATP tailing mix (5 µL of 10 mM dATP, 10 µL of 10x restriction buffer, 5 µL of Klenow exo-, and 80 µL of ddH2O). Incubate at 37 °C for 30 min.
- Remove the supernatant, wash the beads 2x with 400 µL of 0.5x TB by incubating at 55 °C for 3 min and rotating at 750 rpm.
- Wash the beads with 400 µL of 1x NTB and then with 100 µL of 1x ligation buffer.
- Resuspend the beads in 50 µL of 1x ligation buffer, and transfer the suspension to a new tube. Add 4 µL of pre-annealed PE adapters (15 µM stock) and 2 µL of T4 ligase (400 U/µL, i.e., 800 U/tube); incubate at RT for 2 h.
NOTE: Pre-anneal the adapters by adding equal volumes of both PE 1.0 and PE 2.0 adapters (30 µM stock) and incubating for 10 min at RT (see the Table of Materials). - Recapture the beads by removing the supernatant and washing the beads 2x with 400 µL of TB. Wash the beads with 200 µL of 1x NTB, then with 100 µL of 1x restriction buffer, and resuspend the beads in 40 µL of 1x restriction buffer.
12. PCR amplification
- Set up PCRs of 25 µL volume with 5, 6, 7, and 8 cycles. For each PCR, use:
Reaction Recipe Hi-C beads 2.5 µL 10 µM PE PCR primer 1 (Table of Materials) 0.75 µL 10 µM PE PCR primer 2 (Table of Materials) 0.75 µL 10 mM dNTP 0.6 µL 5x reaction buffer 5 µL DNA polymerase 0.3 µL ddH2O 14.65 µL Total 25 µL Cycles Temperature Time 1 98 °C 30 s n cycles 98 °C 10 s 65 °C 30 s 72 °C 30 s 1 72 °C 7 min
13. Final PCR amplification
- Perform final PCR amplification using the same conditions as described above and the number of cycles selected. Split the sample using 5 µL of Hi-C beads as template in 50 µL reactions.
- Collect all PCR reactions and transfer them to a fresh tube. Use the magnet to remove the streptavidin beads and recover the supernatant (PCR products). Transfer the beads to a fresh tube, wash the beads as indicated in step 11.2.9, and store in 1x restriction buffer at 4 °C as a backup.
- Purify the PCR products using 0.85x the volume of the SPRI beads according to the manufacturer´s instructions. Elute with 30 µL of TLE buffer.
- Quantify the Hi-C library using a fluorometric instrument, and confirm the quality of the library by chip-based capillary electrophoresis.
- As a last quality checkpoint, use 1 µL of the Hi-C library as a template to perform a PCR reaction using 10 cycles using the same conditions described in step 12.1. Divide the PCR product into two microcentrifuge tubes: digest one with Cla I and leave the other one undigested as a control. Run the products in a 1.5-2% agarose gel (see representative results).
- Proceed to 50 bp or 75 bp paired-end sequencing on a suitable sequencing platform.
Representative Results
Described below are the results of a successful Hi-C protocol (see a summary of the Hi-C protocol workflow in Figure 1A). There are several quality control checkpoints during the in-nucleus Hi-C experiment. Sample aliquots were collected before (UD) and after (D) the chromatin restriction step as well as after ligation (L). Crosslinks were reversed, and DNA was purified and run on an agarose gel. A smear of 200-1000 bp was observed when restriction with Mbo I was successful (Figure 1B). The expected size of the molecule depends on the restriction enzyme of choice. If the ligation was successful, a high molecular weight band was seen at the top of the gel (Figure 1B). Digestion efficiency can be also confirmed by qPCR as described in detail in the protocol. An acceptable digestion efficiency is 80% or higher (Figure 1C).
To assess Hi-C ligation efficiency in detail, primers can be designed to amplify an internal ligation product control in which the primers are in forward-forward or reverse-reverse orientation in adjacent restriction fragments. Alternatively, primers can be designed to amplify known interactions. Figure 2A shows the amplification of a known medium-range (300 kb) interaction in Drosophila25. Hi-C ligation products (in which the biotin marking, fill-in, and ligation occurred successfully) can be estimated by digestion of the PCR product recovered in the amplification. After fill-in and ligation, Hi-C amplicons will contain a new Cla I restriction site at the original Mbo I site, which is preserved upon blunt-end ligation. If restriction with Cla I is not complete, the fill-in reaction and biotin marking will be inefficient. A digestion efficiency of more than 70% is recommended to avoid having a large proportion of non-useful reads for the libraries after sequencing (Figure 2A, compare the Cla I digestion of the 3C versus the Hi-C template).
To determine an adequate number of PCR amplification cycles to amplify the final Hi-C library, PCR reactions were set up using 2.5 µL of a given library on beads, as described in the protocol. The number of PCR cycles for the final amplification is one cycle less than the number of cycles for which the smear is visible (Figure 2B). In this case, 4 cycles of PCR amplification were chosen. As a final quality checkpoint, an aliquot of the Hi-C library was re-amplified and digested with Cla I. The level of digestion of the library (a decrease in the smear size) indicates the abundance of valid Hi-C pairs and reflects the proportion of useful reads that will be obtained from the library (Figure 2C). A ratio of the upper size range (determined by the size present in the UD sample) and the bottom size range in both UD and D samples should produce a ratio > 1 for the UD and a ratio ≤ 1 for the digested sample if the Cla I digestion is efficient.
After paired-end sequencing, the FASTQ files (Table 1) were processed using HiCPro28 and the generated statistics plotted using MultiQC28. An alternative tool to HiCPro is the HiCUP30 pipeline that yields similar results (not shown). Figure 3 and Table 2 show the detailed statistical information of the sequenced reads. Full read alignment and alignment after trimming are reported. These two categories correspond to successfully aligned reads that will be used in subsequent analysis to find valid Hi-C pairs. The alignment-after-trimming category refers to reads spanning the ligation junction, which were not aligned in the first step and are trimmed at the ligation site to then realign their 5' extremity to the genome28 (Figure 3B,C and Table 2). The contact statistics show that the Hi-C library was of high quality with 82.2% valid pairs and 7.6% non-useful reads falling into the same-fragment self-circle, same-fragment dangling-ends, re-ligation, filtered pairs, and dumped pairs categories (Figure 3A, Figure 3D, and Table 2). Moreover, the number of PCR duplicates is very low, indicating that the library complexity is high, and that the PCR cycles introduced minimal artifacts (Figure 3E and Table 2).
Using the unique valid Hi-C pairs, basic analysis of the pair distribution was performed using HiCPro27. This experiment yielded 46.5% unique cis contacts ≤ 20 kbp, 47.1% unique cis contacts > 20 kbp, and 5.8% unique trans contacts (Figure 3D). The distribution of cis to trans valid pairs corresponded to the results expected for a successful Hi-C experiment with most of the interactions detected within the same chromosome. A high proportion of trans contacts indicates inefficient fixation. Using the Hi-C valid pairs from HiCPro27, matrices were normalized by iterative correction and eigenvector decomposition (ICE)30, and 1 kb and 5 kb resolution matrices were generated using HiCPlotter30,31,32. Normalized contact matrices at 1 kb and 5 kb resolution are presented for the Notch gene locus in Drosophila (Figure 4A, Figure 4C, and Figure 4D). In Figure 4A, the Notch gene locus can be seen along with the APs, domain l and II, as well as histone modifications along the locus (Figure 4A and Table 1). The design of the CRISPR-Cas9 deletion involved the motif of CTCF and M1BP (Figure 4B).
Upon deletion of the region containing both CTCF and M1BP DNA-binding sites at the 5' boundary of the Notch locus (5pN-delta343, Table 1), a dramatic change in chromatin contacts can be observed with loss of interactions inside the Notch locus and gain of contacts with the upstream TAD compared to the wild type (WT) (Figure 4C,D). Finally, Figure 4E shows a detailed panorama of WT and mutant interaction profiles at the restriction fragment level from the Notch gene 5' UTR, showing a decrease in the proportion of contacts made with the Notch gene locus and an increase in contacts with the upstream domain. The virtual 4C views of the Notch gene 5' UTR were obtained using HiC-Pro27. An alternative tool is the Hi-C other-ends quantification available in SeqMonk (SeqMonk (RRID:SCR_001913) http:www.bioinformatics.babraham.ac.uk/projects/seqmonk/. All the results presented in Figure 4 were obtained by applying the in-nucleus Hi-C protocol in WT and mutant S2R+ Drosophila cells, as described by Arzate-Mejía et al.25.
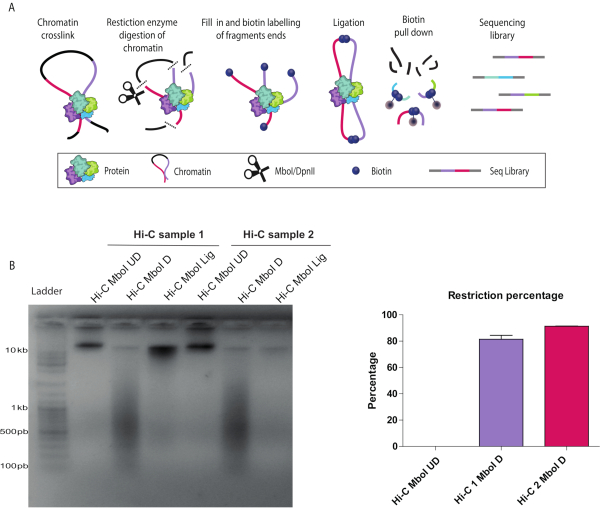
Figure 1: In-nucleus Hi-C digestion and ligation controls. (A) Hi-C protocol overview. Cells are cross-linked with formaldehyde, resulting in covalent links between chromatin segments (DNA fragments: pink, purple) and proteins. Chromatin is digested with a restriction enzyme (represented by scissors), in this example, Mbo I. The resulting sticky ends are filled in with nucleotides including a biotinylated dATP (Dark blue circles). DNA is purified, and the biotinylated junctions are enriched using streptavidin-coated magnetic beads (grey circles). Interacting fragments are identified by next-generation, paired-end sequencing. (B) Hi-C digestion and ligation quality controls for two biological replicates (Hi-C 1 and Hi-C 2). Hi-C libraries were resolved on a 1.5% agarose gel. Both digested, D, libraries run as a smear around 600 bp. Ligated samples, Lig, run as a rather tight band larger than 10 kb similar to the undigested UD samples. The differences in signal strength are due to uneven amounts of loaded DNA on the gel. (C) Hi-C digestion quantitative control by quantitative polymerase chain reaction for the same two biological replicates as in (B) (Hi-C 1 and Hi-C 2) using the cycle threshold values as detailed in the protocol. A successful digestion has ≥ 80% restriction. Please click here to view a larger version of this figure.
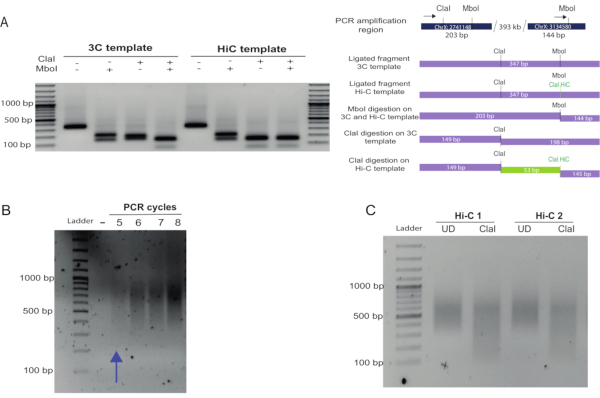
Figure 2: In-nucleus Hi-C fill-in and blunt-end ligation controls. (A) Fill-in and biotin labeling assessment. A known interaction between fragments located 300 kb apart in chromosome X was used as a control and amplified using the primers indicated with black arrows (see top of the scheme in (B), primer 1 (left), primer 2 (right), Table 1), generating a 347 bp amplicon. Hi-C ligation products can be distinguished from those produced in a 3C experiment by digestion of the ligation site. Hi-C junctions were digested by Cla I at the original Mbo I site, as this formed upon blunt-end ligation. Hi-C and 3C junctions were digested with Mbo I as the restriction site regenerates upon ligation (left of the gel). In contrast, 3C junctions were not digested by Cla I at the Mbo I site, but only by Mbo I. Compare the digestion profile of the Hi-C and 3C products using Cla I. A 53 bp fragment was obtained by digesting the Hi-C product (due to restriction of the Cla I site formed at the Mbo I site and restriction of a Cla I site already present in the region). This fragment was not observed in the 3C product digestion as the only Cla I site available was the one that was already present in the region. (B) After PCR amplification of the Hi-C library using different PCR cycles, the products were run on a 1.5% agarose gel. A smear of 400-1000 bp was expected and observed. The appropriate number cycles for the final amplification PCR should be taken as the number immediately lower than that at which a smear is just visible. (C) Final library Cla I digestion. An aliquot of the final library was re-amplified and digested with Cla I. The size reduction of the smear confirmed that a large proportion of the molecules in the library were valid Hi-C pairs. Densitometric analysis of this gel can be performed to obtain a ratio between the UN and D samples, as detailed in the representative results section. Please click here to view a larger version of this figure.
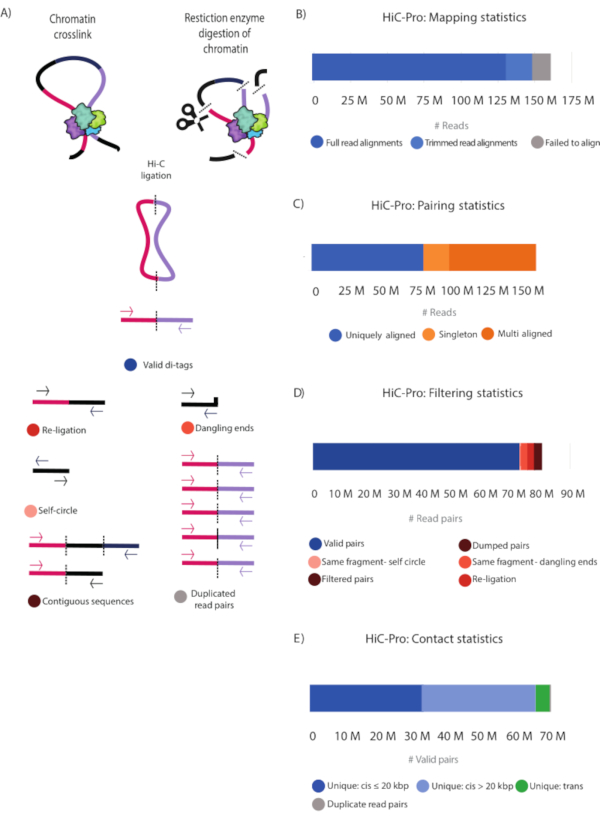
Figure 3: HiC-Pro statistics of the Hi-C library. (A) Schematic representation of valid Hi-C pairs and the different types of non-valid pairs that can be produced during the experiment and filtered out by HiCPro27 (Table 2). These include reads falling into contiguous sequences, dangling ends, same-fragment, self-circle, re-ligations, and PCR duplicates. (B) Mapping statistics. Reads that failed to align are shown (grey), and both fully aligned reads and reads aligned after trimming are shown in blue and light blue, respectively. These two categories represent the useful reads that are considered in subsequent analyses. (C) Pairing statistics. Multi Aligned reads (dark orange) represent reads that are aligned in multiple regions in the genome. Uniquely Aligned (dark blue) reads represent the read pairs that are aligned once in the genome, and singletons (light orange) represent read pairs in which just one genomic region was sequenced in both reads. (D) Filtering statistics. Valid read pairs (blue) represent successful Hi-C ligation products as described in (A). Self-fragment self-circles (light pink) are non-useful reads as they represent the same genomic fragment shown in (A). Same-fragment dangling ends (orange) represent reads in which a single restriction fragment was sequenced. Filtered and dumped pairs (brown) are also non-useful reads that have the wrong size or for which the ligation product could not be reconstructed. Finally, re-ligation reads (red) represent reads in which two adjacent fragments were re-ligated, thus producing non-useful information. (E) Valid read pairs contact distribution in the genome. Unique cis contacts (blue) are more frequent than unique trans contacts (green). Please click here to view a larger version of this figure.
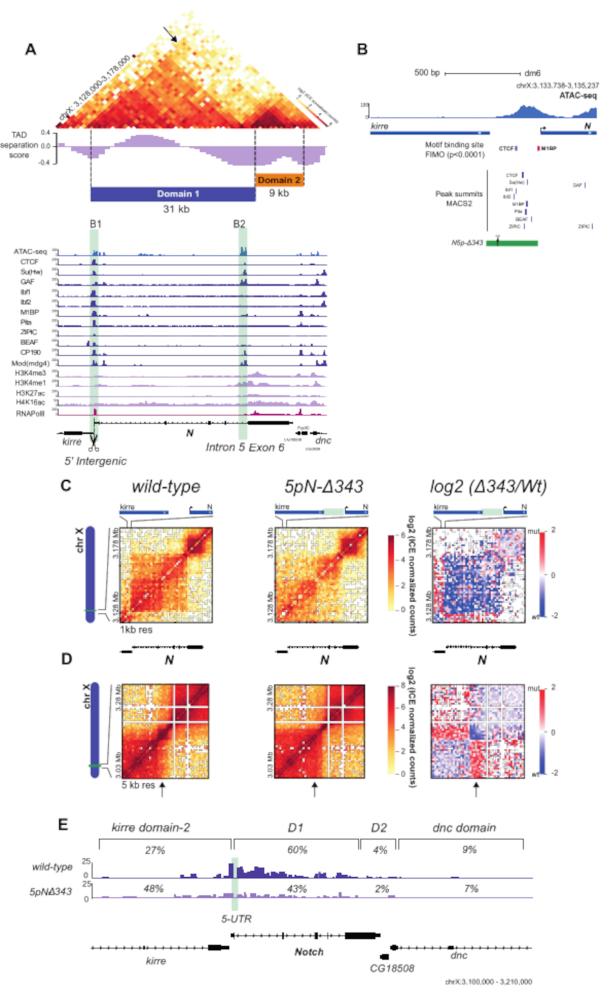
Figure 4: Hi-C contact matrices and virtual 4C analysis of WT and mutant S2R+ cells. (A) Hi-C normalized heatmap of a 50 kb region at 1-kb resolution centered in the Notch gene locus. TAD separation score32 for the locus is shown, along with the partitioning of the Notch locus into two topological domains (Domain 1 and Domain 2). ChIP-seq data for APs, RNA Pol II, and histone marks for S2/S2R+ cells34,35,36,37 are shown below the heatmap (Table 1). The positions of the Notch domain 1 boundaries are highlighted in light green. (B) Schematic representation of B1 boundary CRISPR mutant. The green rectangle indicates the deleted 343 bp region. Scissors indicate sgRNAs used for CRISPR-mediated genome editing. Motif-binding sites for APs are shown as boxes for CTCF and M1BP. Peak summits for DNA-binding APs shown in (A) are also indicated35. (C) Hi-C normalized heatmaps at 1 kb resolution covering a 50 kb region centered in Notch for the WT and the mutant cells. Left, Hi-C heatmaps of the log2 differences in interaction frequency between WT and mutant cells. (D) Hi-C normalized heatmaps covering a 250 kb region centered in Notch at 5 kb resolution for WT and mutant cells. Left, Hi-C heatmaps of the log2 differences in interaction frequency between WT and mutant cells. (E) Virtual-4C for WT and mutant cells using the 5' UTR of Notch as viewpoint. The percentages of interactions between the viewpoint and regions within the upstream kirredomain-2, Notch domain 1, Notch domain 2, and the downstream dnc domain for both WT and mutant cells are shown25. Abbreviations: WT = wild type; TAD = topologically associated domain; ChIP = chromatin immunoprecipitation; AP = architectural protein; RNA Pol = RNA polymerase; S2R+ = S2 receptor plus; sg RNA = single guide RNA; UTR = untranslated region. Please click here to view a larger version of this figure.
| Experiment | Sample | GEO Accession number |
| ChIP | CP190 | GSM1015404 |
| ChIP | SuHW | GSM1015406 |
| ChIP | Mod(mdg4) | GSM1015408 |
| ChIP | CTCF | GSM1015410 |
| ChIP | Ibf1 | GSM1133264 |
| ChIP | Ibf2 | GSM1133265 |
| ChIP | BEAF32 | GSM1278639 |
| ChIP | Pita | GSM1313420 |
| ChIP | ZIPIC | GSM1313421 |
| ChIP | RNA PolII | GSM2259975 |
| ChIP | H3K4me1 | GSM2259983 |
| ChIP | H3K4me3 | GSM2259985 |
| ChIP | H3K27ac | GSM2259987 |
| ChIP | MSL2 | GSM2469507 |
| ChIP | H4K16ac | GSM2469508 |
| ChIP | M1BP | GSM2706055 |
| ChIP | GAF | GSM2860390 |
| ChIP | Input | GSM1015412 |
| Hi-C | S2R+ WT cells | GSE136137 |
| Hi-C | S2R+ 5pN-delta343 cells | GSE136137 |
Table 1: GEO accession numbers.
| HiC-Pro statistics | Reads | Percentage |
| Mapping Statistics | ||
| Full read Alignments | 131515921 | 82.20% |
| Trimmed read Alignments | 16408309 | 10.30% |
| Failed to align | 12110964 | 7.60% |
| Pairing Statistics | ||
| Uniquely Aligned | 79428455 | 50.90% |
| Singleton | 19063418 | 12.20% |
| Multi Aligned | 57700021 | 36.90% |
| Filtering Statistics | ||
| Valid Pairs | 71373989 | 90.12% |
| Same-Fragment: Self-circle | 2340697 | 2.90% |
| Same-Fragment: Dangling Ends | 2578783 | 3.20% |
| Filtered Pairs | 2773043 | 3.50% |
| Dumped pairs | 196565 | 0.20% |
| Contact Statistics | ||
| Unique: cis ≤ 20 kbp | 33108815 | 46.50% |
| Unique: cis > 20 kbp | 33539888 | 47.10% |
| Unique: trans | 4133602 | 5.80% |
| Duplicate read pairs | 398400 | 0.60% |
Table 2: HiC-Pro statistics.
Discussion
The in-nucleus Hi-C method presented here has allowed detailed exploration of Drosophila genome topology at high resolution, providing a view of genomic interactions at different genomic scales, from chromatin loops between regulatory elements such as promoters and enhancers to TADs and large compartment identification25. The same technology has also been efficiently applied to mammalian tissues with some modifications33. For example, when processing a tissue instead of a single-cell suspension, the tissue is sieved through a 70 µm filter, and the lysis step is performed while homogenizing the material using a Dounce homogenizer. In addition, as the mammalian genome is 25x larger than the Drosophila genome, the number of valid read pairs needed to build 1-5 kb resolution matrices is greater. The in-nucleus Hi-C method differs from the original Hi-C method2 in its avoidance of nuclear lysis with 1% SDS at 65 °C prior to ligation, thus preserving the nuclear integrity, and by ligating in 1 mL instead of 7 mL23,24.
The protocol has some key steps to assure high efficiency. The first step that can introduce digestion and fill-in inefficiencies is the formation of clumps during 0.3% SDS permeabilization and surfactant treatments. If the clumps are large and difficult to disrupt, the rotating speed should be decreased to 400 rpm during SDS and surfactant treatments. If the clumps remain difficult to disaggregate by pipetting, the sample should be split in two by adjusting the volumes with restriction buffer, SDS, and the non-ionic surfactant before proceeding with the permeabilization. Next, the nuclei should be centrifuged at minimum speed (200 × g), the supernatant carefully discarded, and the samples pooled together in 450 µL of 1x restriction buffer before proceeding with digestion. Second, the estimation of digestion efficiency is important to provide enough DNA fragments for fill-in and ligation. If upon qualitative assessment, the digestion is found to be inefficient, a second round of digestion should be performed with the restriction enzyme for a period ranging from 4 h to overnight.
Third, the estimation of ligation efficiency is important. If upon qualitative assessment, the ligation is found to be inefficient (i.e., instead of the high molecular weight band, a smear similar to that observed for the digested sample is observed), the ligation should be repeated by centrifuging the nuclei at 200 × g and resuspending them in ligation mix using fresh 10x ligation buffer and ligase. Fourth, the percentage of Hi-C valid products should be estimated by digesting a PCR amplicon of an expected interaction with Cla I (for Mbo I original digestion). The efficient amplification and digestion of the amplicon of the expected interaction confirms successful ligation and formation of Hi-C junctions. If amplicon digestion is not efficient, the majority of the molecules will be 3C instead of Hi-C products, and this should be taken into consideration if the library will be sequenced. This can also be confirmed by performing the final library Cla I digestion control, as described in the representative results section. Finally, selection of the lower number of PCR cycles is important to avoid PCR duplicates. If upon sequencing, the percentage of read pair duplicates is found to be high, the number of PCR cycles should be decreased further.
This in-nucleus Hi-C technique has some limitations. First, the protocol described here represents the Hi-C experiment performed for a cell population. Therefore, the signal of the frequency of genomic contacts represents millions of genomes with variable individual conformations. To obtain the set of genomic contacts from a single genome, a single-cell Hi-C experiment22 is recommended. Second, Hi-C is based on the ligation of proximal DNA fragments. Thus, if genomic regions are part of a large protein-chromatin complex, the distance between fragments could impede ligation. For example, it has been shown that trans contacts are poorly represented in Hi-C38. Moreover, Hi-C finishes with paired-end sequencing, thus retrieving pairs of genomic contacts. However, several DNA fragments can simultaneously interact in the same chromatin complex. To obtain the identity of multiple DNA fragments in a chromatin complex, alternative sequencing methods can be applied to Hi-C39, or different experimental strategies can be employed in which ligation is not performed38,40,41. Finally, although Hi-C measures genomic contacts, it does not reveal the identity of the proteins mediating the interactions. Alternative methods have to be applied to identify the genomic interactions mediated by a particular protein of interest42 or the identity of the ensemble of proteins at specific genomic elements43.
In conclusion, with a high quality Hi-C experiment as the one described here for the Drosophila genome (Table 2), matrices can be built at a wide range of resolutions (from 1, 5 kb, 50 kb or lower; see Figure 4). Additionally, if a particular region of the genome has to be evaluated at the restriction fragment level, the data can be used to build a virtual 4C landscape of the desired viewpoint (e.g., the Notch gene 5' UTR in Figure 4E). The Hi-C other-ends tool in SeqMonk is a very user-friendly option that enables the visualization of this landscape. Applying the 4C quantification tool, also a part of SeqMonk, to this landscape can yield statistically significant contacts.
Applying the in-nucleus Hi-C experiment described here to a collection of mutant cell lines with altered AP DNA-binding sites at the TAD boundaries (Figure 4) revealed that genetic elements are needed at the boundaries to structure the Drosophila genome in domains and sustain gene expression regulation as fully discussed by Arzate-Mejía et al.25. Thus, genetic editing of regulatory elements with the CRISPR/Cas9 system, combined with high-resolution profiling of genomic interactions using the in-nucleus Hi-C protocol described here, can be a powerful strategy to test the structural function of genetic elements.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by UNAM Technology Innovation and Research Support Program (PAPIIT) grant number IN207319 and the Science and Technology National Council (CONACyT-FORDECyT) grant number 303068. A.E.-L. is a master's student supported by the Science and Technology National Council (CONACyT) CVU number 968128.
Materials
| 16% (vol/vol) paraformaldehyde solution | Agar Scientific | R1026 | |
| Biotin-14-dATP | Invitrogen | CA1524-016 | |
| ClaI enzyme | NEB | R0197S | |
| COVARIS Ultrasonicator | Covaris | LE220-M220 | |
| Cut Smart | NEB | B72002S | |
| Dulbecco's Modified Eagle Medium (DMEM) 1x | Life Technologies | 41965-039 | |
| Dynabeads MyOne Streptabidin C1 | Invitrogen | 65002 | |
| Fetal bovine serum (FBS) sterile filtered | Sigma | F9665 | |
| Klenow Dna PolI large fragment | NEB | M0210L | |
| Klenow exo(-) | NEB | M0210S | |
| Ligation Buffer | NEB | B020S | |
| MboI enzyme | NEB | R0147M | |
| NP40-Igepal | SIGMA | CA-420 | Non-ionic surfactant for addition in lysis buffer |
| PE adapter 1.0 | Illumina | 5'-P-GATCGGAAGAGCGGTTCAGCAG GAATGCCGAG-3' |
|
| PE adapter 2.0 | Illumina | 5'-ACACTCTTTCCCTACACGACGCT CTTCCGATCT-3' |
|
| PE PCR primer 1.0 | Illumina | 5'-AATGATACGGCGACCACCGAGAT CTACACTCTTTCCCTACACGACG CTCTTCCGATCT-3' |
|
| PE PCR primer 2.0 | Illumina | 5'-CAAGCAGAAGACGGCATACGAG ATCGGTCTCGGCATTCCTGCTGA ACCGCTCTTCCGATCT-3' |
|
| Phenol: Chloroform:Isoamyl Alcohol 25:24:1 | SIGMA | P2069 | |
| Primer 1 (known interaction, Figure 2A) | Sigma | 5'-TCGCGGTAATTTTGCGTTTGA-3' | |
| Primer 2 (known interactions, Figure 2A) | Sigma | 5'-CCTCCCTGCCAAAACGTTTT-3' | |
| Protease inhibitor cocktail tablet | Roche | 4693132001 | |
| Proteinase K | Roche | 3115879001 | |
| Qubit | ThermoFisher | Q33327 | |
| RNAse | Roche | 10109142001 | |
| SPRI Beads | Beckman | B23318 | |
| T4 DNA ligase | Invitrogen | 15224-025 | |
| T4 DNA polymerase | NEB | M0203S | |
| T4 polynucleotide kinase (PNK) | NEB | M0201L | |
| TaqPhusion | NEB | M0530S | DNA polymerase |
| Triton X-100 | Non-ionic surfactant for quenching of SDS |
References
- Cremer, T., Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Review Genetics. 2, 292-301 (2001).
- Lieberman-Aiden, E., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326, 289-293 (2009).
- Dixon, J. R., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485, 376-380 (2012).
- Sexton, T., et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 148 (3), 458-472 (2012).
- Dixon, J. R., Gorkin, D. U., Ren, B. Chromatin domains: the unit of chromosome organization. Molecular Cell. 62, 668-680 (2016).
- Bonev, B., Cavalli, G. Organization and function of the 3D genome. Nature Reviews Genetics. 17, 661-678 (2016).
- Lupiáñez, D. G., Spielmann, M., Mundlos, S. Breaking TADs: how alterations of chromatin domains result in disease. Trends in Genetics. 32, 225-237 (2016).
- Phillips, J. E., Corces, V. G. CTCF: master weaver of the genome. Cell. 137 (7), 1194-1211 (2009).
- Hong, S., Kim, D. Computational characterization of chromatin domain boundary-associated genomic elements. Nucleic Acids Research. 45, 10403-10414 (2017).
- Zuin, J., et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences of the United States of America. 111 (3), 996-1001 (2014).
- Guo, Y., et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 162, 900-910 (2015).
- Lupiáñez, D. G., et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 161, 1012-1025 (2015).
- Van-Steensel, B., Furlong, E. E. M. The role of transcription in shaping the spatial organization of the genome. Nature Reviews Molecular Cell Biology. 20, 327-337 (2019).
- Merkenschlager, M., Nora, E. P. CTCF and cohesin in genome folding and transcriptional gene regulation. Annual Review of Genomics and Human Genetics. 17 (1), 17-43 (2016).
- Hansen, A. S., Pustova, I., Cattoglio, C., Tjian, R., Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife. 6, 1-10 (2017).
- Van Bortle, K., et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biology. 15, 82 (2014).
- Ramírez, F., et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nature Communications. 9, 189 (2018).
- Ulianov, S. V., et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Research. 26, 70-84 (2016).
- Rowley, M. J., et al. Evolutionarily conserved principles predict 3D chromatin organization. Molecular Cell. 67, 837-852 (2017).
- Gavrilov, A. A., Golov, A. K., Razin, S. V. Actual ligation frequencies in the chromosome conformation capture procedure. PLoS One. 8, 60403 (2013).
- Gavrilov, A. A., et al. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Research. 41, 3563-3575 (2013).
- Nagano, T., et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 502, 59-64 (2013).
- Rao, S. S. P., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159 (7), 1665-1680 (2014).
- Nagano, T., et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biology. 16, 175 (2015).
- Arzate-Mejía, R., et al. In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila. Nature Communications. 11, 894 (2020).
- Schoenfelder, S., et al. Promoter capture Hi-C: high-resolution, genome-wide profiling of promoter interactions. Journal of Visualized Experiments. (136), e57320 (2018).
- Servant, N., et al. HiC-Pro: an optimized and flexible pipeline for Hi-C processing. Genome Biology. 16, 259 (2015).
- Philip, E., Måns, M., Sverker, L., Max, K. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 10, 1093 (2016).
- Wingett, S., et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000 Research. 4, 1310 (2015).
- Imakaev, M., et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature Methods. 9 (10), 999-1003 (2012).
- Akdemir, K. C., Chin, L. HiCPlotter integrates genomic data with interaction matrices. Genome Biolog. 16, 198 (2015).
- Ramirez, F., et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nature Communications. 9, 189 (2018).
- Ando-Kuri, M., et al. The global and promoter-centric 3D genome organization temporally resolved during a circadian cycle. bioRxiv. , (2020).
- Cuellar-Partida, G., et al. Epigenetic priors for identifying active transcription factor binding sites. Bioinformatics. 28 (1), 56-62 (2012).
- Zhang, Y., et al. Model-based analysis of ChIP-Seq (MACS). Genome Biology. 9, 137 (2008).
- Ong, C. -. T., et al. Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell. 155 (1), 148-159 (2013).
- Fresán, U., et al. The insulator protein CTCF regulates Drosophila steroidogenesis. Biology Open. 4 (7), 852-857 (2015).
- Quinodoz, S., et al. RNA promotes the formation of spatial compartments in the nucleus. Cell. 174, 744-757 (2018).
- Olivares-Chauvet, P., et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature. 540 (7632), 296-300 (2016).
- Beagrie, R., et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 543, 519-524 (2017).
- Redolfi, J., et al. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nature Structural and Molecular Biology. 26, 471-480 (2019).
- Maxwell, R., et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nature Methods. 13, 919-922 (2016).
- Gao, X., et al. C-BERST: Defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nature Methods. 15 (6), 433-436 (2018).

