Co-Translational Insertion of Membrane Proteins into Preformed Nanodiscs
Summary
Co-translational insertion into pre-formed nanodiscs makes it possible to study cell-free synthesized membrane proteins in defined lipid environments without contact with detergents. This protocol describes the preparation of essential system components and the critical parameters for improving expression efficiency and sample quality.
Abstract
Cell-free expression systems allow the tailored design of reaction environments to support the functional folding of even complex proteins such as membrane proteins. The experimental procedures for the co-translational insertion and folding of membrane proteins into preformed and defined membranes supplied as nanodiscs are demonstrated. The protocol is completely detergent-free and can generate milligrams of purified samples within one day. The resulting membrane protein/nanodisc samples can be used for a variety of functional studies and structural applications such as crystallization, nuclear magnetic resonance, or electron microscopy. The preparation of basic key components such as cell-free lysates, nanodiscs with designed membranes, critical stock solutions as well as the assembly of two-compartment cell-free expression reactions is described. Since folding requirements of membrane proteins can be highly diverse, a major focus of this protocol is the modulation of parameters and reaction steps important for sample quality such as critical basic reaction compounds, membrane composition of nanodiscs, redox and chaperone environment, or DNA template design. The whole process is demonstrated with the synthesis of proteorhodopsin and a G-protein coupled receptor.
Introduction
Membrane proteins (MPs) are challenging targets in protein production studies due to their insolubility in aqueous environments. Conventional MP production platforms comprise cell-based systems such as E. coli, yeast, or eukaryotic cells. The synthesized recombinant MPs are either extracted from cell membranes or refolded from inclusion bodies1. After detergent solubilization, MPs can be transferred into suitable membrane environments by established in vitro reconstitution protocols. Besides vesicles and liposomes, MP reconstitution into planar membranes in the form of nanodiscs2 or salipro3 particles have become routine techniques in recent times. However, all these strategies imply detergent contact with MPs that can result in destabilization, dissociation of oligomers, and even loss of protein structure and activity4. Screening for optimal detergent solubilization and reconstitution conditions can therefore be tedious and time consuming5.
The open nature of cell-free (CF) systems allows the expression reaction to be directly supplied with preformed membranes with a defined lipid composition. In this lipid-based expression mode (L-CF), the synthesized MPs have the opportunity to co-translationally insert into the provided bilayers6,7 (Figure 1). Nanodiscs consisting of a membrane scaffold protein (MSP) surrounding a planar lipid bilayer disc8 appear to be particularly suitable for this strategy9,10. Nanodiscs can routinely be assembled in vitro with a variety of different lipids, they are very stable, and stocks can be concentrated up to 1 mM. However, MSP expression in E. coli and its purification is necessary. As an alternative strategy, MSP can be co-expressed together with the target MP in CF reactions supplied with liposomes11,12,13. DNA templates for both MSP and MP are added into the reaction and MP/nanodiscs can form upon expression. While MSP production is avoided, the co-expression strategy requires careful fine-tuning of the final DNA template concentrations and higher variations in the efficiency of sample production can be expected.
The co-translational insertion of MPs into membranes of preformed nanodiscs is a non-physiological and still largely unknown mechanism independent from translocon machineries and signal sequences13,14,15,16. Major determinants of the insertion efficiency are the type of membrane protein as well as the lipid composition of the provided membrane, with a frequent preference for negatively charged lipids15,17. As the nanodisc membranes are relatively confined in size, a substantial amount of lipids is released upon MP insertion18. Variation of nanodisc size enables insertion and tuning of higher oligomeric MP complexes15,18. Among others, the correct assembly of homooligomeric complexes was shown for the ion channel KcsA, for the ion pump proteorhodopsin (PR) and for the multidrug transporter EmrE15,18. MPs are likely to enter the symmetric nanodisc membrane from both sides at relatively equal frequency. It should therefore be considered that different monomers or oligomers inserted into one nanodisc may have opposite orientations. However, a bias in orientation could be caused by cooperative insertion mechanisms as reported for the formation of PR hexamers and KcsA tetramers18. A further bias in MP orientation might result from orientation switches of inserted MPs probably at the rim of the nanodisc membranes.
The production of CF lysates from E. coli strains is a reliable routine technique and can be performed in almost any biochemical laboratory19,20. It should be considered that besides disulfide bridge formation, most other post-translational modifications are absent if a MP is synthesized using E. coli lysates. While this might generate more homogenous samples for structural studies, it may be necessary to exclude potential effects on the function of individual MP targets. However, the efficient production of high quality samples of G-protein coupled receptors (GPCR)14,21,22, eukaryotic transporters23 or large heteromeric assemblies24 de E. coli CF lysates indicates their suitability for even complex targets. Preparative scale amounts (≈ 1 mg/mL) of a sample can be obtained with the two-compartment continuous exchange cell-free (CECF) configuration, composed of a reaction mixture (RM) and a feeding mixture (FM) compartment. The FM volume exceeds the RM volume 15 to 20-fold and provides a reservoir of low-molecular weight energy compounds and precursors19. The expression reaction is thus extended for several hours and the final yield of the MP target is increased. The RM and FM compartments are separated by a dialysis membrane with a 10-14 kDa cutoff. The two compartments require a special design of the CECF reaction container (Figure 1). Commercial dialysis cassettes as RM containers in combination with tailored plexiglass containers holding the FM are suitable examples. MP yields can further be manipulated by modifying the RM:FM ratios or by exchanging the FM after a certain period of incubation.
Yield and quality of a MP frequently require intense optimization of reaction parameters. An important advantage of CF expression is the possibility to modify and fine tune almost any compound according to the individual requirements of a MP. A straightforward strategy is to focus first on improving the yield of a MP by establishing a basic production protocol and then to optimize MP quality by fine tuning reaction and folding conditions. The absence of physiological processes in CF lysates and their reduced complexity result in high success rates for the efficient production of MPs25. Routine considerations for DNA template design and optimization of Mg2+ ion concentration are in most cases sufficient to obtain satisfactory yields26. Depending on expression mode, optimization of MP quality can become time consuming, as a larger variety of parameters need to be screened14,17,22.
To establish the described CF expression platform, protocols are necessary for the production of E. coli CF lysate (i), T7 RNA polymerase (ii), nanodiscs (iii), and the basic CECF reaction compounds (iv) (Figure 1). The E. coli K12 strain A1927 or BL21 derivatives are frequently used for the preparation of efficient S30 (centrifugation at 30,000 x g) lysates. Besides S30 lysates, corresponding lysates centrifuged at other g-forces (e.g. S12) may be used. The lysates differ in expression efficiency and in proteome complexity. The proteome of the S30 lysate prepared by the described protocol has been studied in detail28. The S30 proteome still contains some residual outer membrane proteins which could give background problems upon expression and analysis of ion channels. For such targets, the use of S80-S100 lysates is recommended. A valuable modification during lysate preparation is the induction of the SOS response by combined heat shock and ethanol supply at mid-log growth phase of the cells. The resulting HS30 lysates are enriched in chaperones and can be used in blends with S30 lysates for improved MP folding22. CF expression in E. coli lysates is operated as a coupled transcription/translation process with DNA templates containing promoters controlled by T7 RNA polymerase (T7RNAP). The enzyme can be efficiently produced in BL21(DE3) Star cells carrying the pAR1219 plasmid29.
Two copies of MSP1E3D1 assemble into one nanodisc with a diameter of 10-12 nm, but the protocol described below may also work for other MSP derivatives. However, nanodiscs larger than those formed with MSP1E3D1 tend to be less stable while smaller nanodiscs formed with MSP derivatives such as MSP1 may not accommodate larger MPs or MP complexes. MSP1E3D1 nanodiscs can be assembled in vitro with a large variety of different lipids or lipid mixtures. Preformed nanodiscs are usually stable for > 1 year at -80 °C, while stability may vary for different lipid components. For the screening of lipid effects on folding and stability of a MP, it is necessary to prepare a comprehensive set of nanodiscs assembled with a representative variety of lipids/lipid mixtures. The following lipids may give a good starting selection: 1,2-Dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DMPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2 dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (DOPG), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG) and 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC).
A protocol for the preparation of a 3 mL CECF reaction is described. Further up or down scaling in a 1:1 ratio is possible. For the two-compartment CECF configuration, a RM containing all compounds and a FM containing only the low-molecular weight compounds have to be prepared. Commercial 3 mL dialysis devices with 10-14 kDa MWCO can be used as RM containers, which are then placed into custom made plexiglass containers holding the FM (Figure 1D)30. The ratio of RM:FM is 1:20, so 60 mL of FM have to be prepared for 3 mL RM. Quality, concentration, or type of several components can be critical for the final yield and/or quality of the synthesized MP (Table 1). DNA templates should be prepared according to published guidelines and codon optimization of the reading frame of the target can further significantly improve product yield26. For preparative scale CECF reaction, an established protocol for the production of PR is described. To establish expression protocols for new MPs, it is usually necessary to perform optimization screens of certain compounds (Table 1) to improve yield and quality. For Mg2+ ions, a well-focused concentration optimum does exist that frequently shows significant variation for different DNA templates. Other CF reaction compounds such as new batches of T7RNAP or S30 lysates may further shift the optimal Mg2+ concentration. As an example, a typical Mg2+ screen within the range of 14-24 mM concentration and in steps of 2 mM is described. Each concentration is screened in duplicates and in analytical scale CECF reactions. As CECF reaction container, custom-made Mini-CECF Plexiglas containers30 holding the RM are used in combination with standard 24-well microplates holding the FM (Figure 1B). Alternatively, commercial dialysis cartridges in combination with 96-deep well microplates or other commercial dialyzer devices in appropriate setups may be used (Figure 1C).
Protocol
1. Preparation of S30 lysate
- Day 1: Streak out cells from glycerol stocks on an LB agar plate and incubate at 37 °C overnight.
- Day 2: Inoculate 200 mL of LB medium with the cells from the agar plate and incubate at 37 °C for 12-14 h.
- Day 3: Inoculate 10 L of sterile YPTG medium (10 g/L yeast extract, 16 g/L tryptone, 5 g/L NaCl, 19.8 g/L glucose, 4.4 mM KH2PO4, 8 mM K2HPO4) tempered to 37 °C in a 15 L stirred tank reactor with 100 mL of the pre-culture (1:100). Cultivate at 37 °C, 500 rpm and high aeration (~ 3 air volumes per minute). To prevent excessive foaming, add sterile antifoam.
- Measure optical density (OD) at 600 nm in regular time intervals. After approximately 2 h the culture will enter the log-phase.
- Modification for HS30 lysate: When cells are in mid log phase (OD600 ≈ 3.6-4.2) add 300 mL of ethanol to the culture medium and proceed cultivation at 42 °C, 500 rpm, and high aeration (approximately 3 air volumes per minute) for another 45 min. Then proceed with cooling and harvesting of the cell culture as described in step 1.6. For the production of standard S30 lysate, skip this step and proceed with step 1.6.
- When cells are in mid log phase (OD600 ≈ 3.6-4.2), cool fermenter below 20 °C within < 30 min. The final OD600 should be around 4.5-5.5. Harvest cells at 4,500 x g for 20 min at 4 °C and discard the supernatant. Keep cells at 4 °C throughout the following steps.
NOTE: During cooling, excessive foaming can occur. In this case, either add antifoam or reduce stirring speed and/or decrease the air stream in the bioreactor. - Suspend cells completely in 300 mL of S30-A buffer (10 mM Tris-HCl, pH 8.2, 14 mM Mg(OAc)2, 60 mM KCl, 6 mM 2-mercaptoethanol) using a spatula followed by pipetting the suspension up and down until homogeneity. Centrifuge at 8,000 x g for 10 min at 4 °C. Repeat this step twice.
CAUTION: 2-mercaptoethanol is toxic. Avoid contact with skin or respiratory tract. If possible, work under a hood. Weigh the empty centrifuge beaker before the last washing step. After centrifugation, weigh the wet cell pellet and either proceed with step 1.8 or store the pellet until further use.
NOTE: The weight of the wet cell pellet is 5-7 g per 1 L bioreactor culture. At this point the protocol may be paused and the pellet can be frozen in liquid nitrogen and stored at -80 °C for 4-8 weeks. - Suspend cells thoroughly in 1.1 volumes (1 g = 1 mL) of S30-B buffer (10 mM Tris-HCl, pH 8.2, 14 mM Mg(OAc)2, 60 mM KOAc, 1 mM DTT, 1 tablet of cOmplete protease inhibitor). Fill the suspension into a pre-cooled French press pressure cell and disrupt cells at 1,000 psig. Pass cells through the French press once.
- Centrifuge the lysate at 30,000 x g for 30 min at 4 °C. Transfer the supernatant into a fresh tube and repeat the centrifugation step.
NOTE: A loose and slimy layer may be present on top of the pellet after the first centrifugation, which should not be transferred with the supernatant. - Apply a high salt and heat step to remove unstable lysate components and to release mRNA from ribosomes. Adjust the lysate to 0.4 M NaCl and incubate at 42 °C for 45 minutes in a water bath.
NOTE: This step will cause significant precipitate formation. Flip or invert the tube during incubation from time to time. - Transfer the turbid suspension (usually 50-80 mL) into dialysis tubes with a 10-14 kDa cutoff and dialyze against 5 L S30-C buffer (10 mM Tris-HCl, pH 8.2, 14 mM Mg(OAc)2, 60 mM KOAc, 0.5 mM DTT). Exchange the S30-C dialysis buffer after 2-3 h and dialyze for another 12-14 h.
- Day 4: Transfer the dialyzed suspension into centrifuge tubes and centrifuge at 30,000 x g for 30 min at 4 °C. Transfer supernatant (S30 lysate) into fresh 50 mL tubes and gently mix. The protein concentration of the lysate should be approximately 30-40 mg/mL. Shock freeze aliquots (e.g. 0.5 mL, 1 mL) in liquid nitrogen and store at -80 °C. The aliquots are stable for > 1 year.
2. Expression and purification of T7 RNA polymerase
- Day 1: Inoculate 200 mL LB medium containing 100 µg/mL ampicillin with freshly plated BL21(DE3) Star x pAR1219 cells and incubate at 37 °C and 180 rpm for 12-16 h.
- Day 2: Inoculate 1 L terrific broth (12 g/L tryptone, 24 g/L yeast extract, 4 mL/L glycerol) in a 2 L baffled flask with 10 mL of the pre-culture and incubate at 37 °C and 180 rpm agitation. The starting OD600 should be around 0.1.
- When the OD600 reaches 0.6-0.9, add IPTG to a final concentration of 1 mM to induce T7RNAP expression and continue cultivation for another 5 h.
- Harvest cells by centrifugation at 4,500 x g for 20 min at 4 °C. Freeze cells in liquid nitrogen and store at -80 °C.
NOTE: The protocol may be paused here. - Day 3: Add 30 mL of ice-cold suspension buffer (30 mM Tris-HCl, pH 8.0 with one tablet of cOmplete protease inhibitor) to the frozen cell pellet and thaw the pellet in a water bath at room temperature. Then, suspend the pellet by pipetting up and down until homogeneity.
- Perform cell disruption using a French press as described in the previous section or use a sonication device according to manufacturer’s recommendations. Subsequently, centrifuge at 20,000 x g for 30 min at 4 °C and transfer supernatant into a fresh tube.
- To precipitate genomic DNA, add streptomycin sulfate slowly and under gentle agitation to the supernatant until a final concentration of 3% (w/v) is reached. Incubate at 4 °C for 5-10 min under gentle agitation. Centrifuge at 20,000 x g for 30 min at 4 °C.
- Filter the supernatant with a 0.45 µm filter and load it on a Q-Sepharose column with a column volume (CV) of 40 mL equilibrated with 2 CV equilibration buffer (30 mM Tris-HCl, pH 8.0, 10 mM EDTA, 50 mM NaCl, 5% glycerol and 10 mM 2-mercaptoethanol) using a peristaltic pump at a flow rate of 4 mL/min. Then, connect the column to a UV detector and continue elution with equilibration buffer until the UV signal at 280 nm reaches a stable baseline.
- Elute T7RNAP with a gradient from 50 mM to 500 mM NaCl per CV at a flow rate of 3 mL/min. Collect 1 mL fractions and prepare samples for SDS-PAGE analysis by mixing 10 µL of each fraction with 2 x SDS sample buffer. Run SDS-PAGE and stain the gel with Coomassie Blue R. T7RNAP should appear as a prominent band at approximately 90 kDa along with numerous additional bands of impurities.
- Pool fractions containing T7RNAP and dialyze using membranes with a 12-14 kDa MWCO for 12-16 h in dialysis buffer (10 mM K2HPO4/KH2PO4, pH 8.0, 10 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 5% (w/v) glycerol).
- Day 4: Collect T7RNAP solution and concentrate using filter units with 12-14 MWCO to a final concentration of 3-10 mg/mL. T7RNAP may start to precipitate during concentration. Stop concentration instantly as soon as turbidity in the sample occurs. Add glycerol to a final concentration of 50% (w/v) and mix gently. Shock-freeze 0.5-1 mL aliquots and store at -80 °C. Working T7RNAP aliquots can be stored at -20 °C.
NOTE: Approximately 20-40 mL of 5 mg/mL partially purified T7RNAP with 20,000-40,000 units should be obtained from a 1 L fermentation. To test the optimum amount of each T7RNAP batch, CF expression reactions of green fluorescent protein (GFP) containing 0.02 mg/mL-0.1 mg/mL of the prepared T7RNAP sample should be performed.
3. Expression and purification of MSP1E3D1
- Day 1: Transform E. coli BL21 (DE3) Star cells with the pET28(a)-MSP1E3D1 vector31 using standard heat shock transformation or electroporation protocols. Streak out or plate transformed cells on LB agar containing 30 µg/mL kanamycin and incubate at 37 °C for 12-16 h.
- Day 2: Inoculate 200 mL of LB medium supplemented with 0.5% (w/v) glucose and 30 µg/mL kanamycin with a single colony picked from the agar plate and incubate at 37 °C at 180 rpm for 12-16 h.
- Day 3: Inoculate 10 L LB medium supplemented with 0.5% (w/v) glucose and 30 µg/mL kanamycin with 100 mL of the pre-culture in a stirred-tank bioreactor. Conduct fermentation at 37 °C, 500 rpm and aeration of approximately 3 bioreactor volumes per minute. In the case of excessive foaming, add antifoam.
NOTE: Baffled shaking flasks may be used instead of a fermenter. - At OD600 of 6.5-7.5, add IPTG to a final concentration of 1 mM and continue incubation at 37 °C for 1 h. Harvest cells by centrifugation at 4,500 x g for 20 min at 4 °C. Wash cell pellets with 200 mL of 0.9% (w/v) NaCl and centrifuge again at 8,000 x g for 10 min at 4 °C. Discard supernatant and transfer cells into 50 mL tubes using a spatula. The expected wet pellet weight is 8-12 g per L of bioreactor culture. Shock-freeze cell pellets in liquid nitrogen and store at -80 °C until further use.
NOTE: The protocol can be paused here. - Day 4: For purification, thaw cell pellets in a water bath at RT. Suspend the cells in MSP-A buffer (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1% (w/v) Triton X-100, 1 tablet of c0mplete protease inhibitor cocktail per 50 mL cell suspension) to yield a 30-40% (w/v) cell suspension.
- Disrupt cells by sonication or using a French press and centrifuge at 30,000 x g for 30 min at 4 °C. Transfer the supernatant into a fresh tube and filter through a 0.45 µm filter. Apply the filtrate on a Ni2+ loaded nitrilotriacetic acid column (CV > 15 mL) equilibrated with 5 CV of MSP-B buffer (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1% (w/v) Triton X-100) by using a peristaltic pump.
NOTE: To facilitate filtration, the supernatant can be sonicated for another minute to break down large DNA fragments. - After sample loading, wash the column with 5 CV MSP-B, 5 CV MSP-C (40 mM Tris-HCl, pH 8.9, 300 mM NaCl, 50 mM cholic acid), 5 CV MSP-D (40 mM Tris-HCl, pH 8.9, 300 mM NaCl), 5 CV MSP-E (40 mM Tris-HCl, pH 8.0, 300 mM NaCl) and 5 CV MSP-F (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 50 mM imidazole).
NOTE: Cholic acid in the MSP-C buffer is important to remove lipids attached to MSP. Cholic acid is not completely soluble at low pH values. The pH value must therefore be adjusted to 8.9 in MSP-C and MSP-D. It is important to wash the column with MSP-D buffer, as a lower pH might cause residual cholic acid to precipitate and to block or damage the column. - Elute MSP with MSP-G (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 300 mM imidazole). Collect fractions of 1 mL and monitor UV absorption at 280 nm. Collect and pool the fractions of the elution peak.
- Transfer pooled MSP fractions into 12-14 kDa MWCO dialysis membrane tubes and dialyze against 5 L of MSP-H (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% (w/v) glycerol) for 2-3 h at 4 °C. Change to fresh 5 L MSP-H buffer and dialyze for another 12-16 h at 4 °C.
- Day 5: Transfer MSP solution into centrifuge tubes and centrifuge at 18,000 x g for 30 min at 4 °C to remove precipitate. Transfer supernatant into a fresh tube and concentrate to 200-800 µM using ultrafiltration devices with 12-14 kDa MWCO at 4 °C. Measure concentration using a UV/VIS measuring device. Use extinction coefficient of 27.31 M-1 cm-1 and the molecular mass of 31.96 kDa for calculation of concentration.
NOTE: Expression in a bioreactor usually yields 15-30 mg of MSP1E3D1 per L culture. - Shock-freeze aliquots of the concentrated MSP solution in liquid nitrogen and store at -80 °C.
4. Assembly of MSP1E3D1 nanodiscs
- Day1: Prepare 1-2 mL of 50 mM lipid stocks (DMPG, DOPG, POPG DMPC, DOPC or POPC) in 100 mM sodium cholate. Store at -20 °C if not used on the same day.
NOTE: The lipid solution must be clear. If the solution is not clear, sodium cholate concentration may be increased to 150 mM. - Mix the MSP1E3D1 solution with the lipid solution. Add dodecyl phosphocholine (DPC) at a final concentration of 0.1% (w/v). Assembly reactions may be adjusted to final volumes of 3 mL (Table 2) or 12 mL corresponding to typical volumes of dialysis cassettes (10 kDa MWCO). Incubate assembly reactions for 1 h at 21 °C under gentle agitation.
NOTE: For each lipid, a specific MSP1E3D1:lipid ratio has to be used to ensure homogeneous nanodisc preparation (Table 2). For new lipids or lipid mixtures, the optimal ratio should be determined with a pilot experiment and size exclusion chromatography analysis14. - Pre-wet the membrane of a dialysis cassette with disc formation (DF) buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl). Transfer the assembly mixture into the dialysis cassette with a syringe. Potential air bubbles can be removed by aspiration using the syringe.
- Days 1-4: Dialyze against 3 x 5 L DF buffer for 10-20 h at RT under agitation using a stirring bar. Then transfer the assembly mixture from the dialysis cassette into centrifugal filter units with 10 kDa MWCO equilibrated with DF buffer. Concentrate at 4,000 x g at 4 °C.
NOTE: A turbid dialysis mix could indicate a wrong stoichiometric ratio of MSP:lipid. Precipitate must be removed at 18,000 x g for 20 min at 4 °C before concentration of sample. To avoid precipitation during sample concentration, use ultrafiltration units with a large deadstop. - Concentrate the assembled nanodiscs to a concentration of at least 300 µM. Measure concentration using an UV/VIS reader. Consider that one nanodisc is formed by two MSP1E3D1 molecules and thus, the concentration of MSP must be divided by 2 to determine the correct amount of nanodiscs.
NOTE: The described setup (Table 2) will yield 0.6-1 mL of a 300 µM nanodisc solution. - Centrifuge the concentrated nanodisc preparation at 18,000 x g for 20 min at 4 °C to remove precipitate. Then, aspirate the supernatant and shock-freeze 50 µL aliquots in liquid nitrogen and store at -80 °C.
NOTE: It is advisable to evaluate the success of nanodisc formation using size exclusion chromatography. The sample should be monodisperse and should contain only a low amount of aggregates. Aggregates indicate that the amount of lipid in the setup is too high. As a reference, purified MSP1E3D1 can be used. If the nanodisc preparation shows a peak at the height of the reference MSP1E3D1 peak, the chosen lipid to MSP1E3D1 ratio was too low.
5. Preparative scale 3 mL CECF reaction setup
- Day 1: Prepare all necessary stock solutions as listed (Table 3). Stocks are stored at -20 °C and thawed at room temperature. Make sure to thoroughly mix all stocks after thawing and before pipetting. Calculate the required volumes for all stocks and make a pipetting scheme (Table 4). High-molecular weight compounds will only be added into the RM. For the low-molecular weight compounds, a combined 3 x mastermix for RM and FM can be prepared.
NOTE: Final volumes of compound mixtures might be less than calculated due to volume loss during mixing. This can be avoided by adding an excess volume of 2-5% to compensate for volume loss. - Equilibrate the membrane of a 3 mL dialysis cassette in 100 mM Tris-acetate, pH 8.0. Make sure to remove excess buffer.
- Using a syringe to transfer the RM into the dialysis cassette. Remove excessive air in the RM compartment by aspiration with the syringe. Place the dialysis cassette into the dialysis chamber.
- Fill up the dialysis chamber with 60 mL of FM. Place the lid on the chamber and tighten the screws to fix it. Incubate the chamber for 12-16 h at 30 °C at 200 rpm.
NOTE: Make sure that the chamber remains in the upright position during agitation, otherwise exchange of low molecular compounds will be hampered. - Day 2: Using a syringe, aspirate the RM from the dialysis chamber. Transfer the RM to fresh tubes and centrifuge at 16,000 x g at 4 °C for 10 min to remove precipitate. Transfer the supernatant into fresh tubes. The protein in the supernatant can now be further analyzed.
NOTE: In some cases, a pellet will be present after centrifugation. This can provide important information on the suitability of the analyzed nanodiscs or the reaction compounds to keep the synthesized MP soluble. It is therefore advisable to analyze the amount of target potentially present in the precipitate by using SDS-PAGE, Western Blot, or by visual evaluation of the pellet size.
6. Analytical scale 60 µL CECF reaction setup for Mg2+ ion screening
- Day 1: Mix all components of the respective 3x master mix and distribute 60 µL to the RM and 825 µL to the FM. Fill up FM with H2O and mix FM by vortexing. Transfer FM into 3 wells of the 24 well microplate (Table 5).
NOTE: Since the customized Mini-CECF container might not be accessible, the volumes in Table 5 are calculated for a reaction carried out in dialysis cartridges (10-14 kDa MWCO, RM: 100 µL) with a FM volume of 1.7 mL in 96 deep-well (2 mL) plates (Figure 1). - Cut regenerated cellulose dialysis tubes with a 10-14 kDa MWCO into approximately 25 x 20 mm sized pieces and store in H2O with 0.05% (w/v) sodium azide.
CAUTION: Sodium azide is very toxic. Avoid any contact with skin or mucous membrane. Do not use metal containers as sodium azide can react with metals to form explosive metal azides. - Before Mini-CECF container assembly, remove excessive H2O from the dialysis membranes with a tissue. Place one membrane piece on each container and fix the membranes with a polytetrafluorethylene ring.
- Transfer the Mini-CECF container into the wells of a 24-well plate with 825 µL of FM. Avoid air bubbles below the dialysis membrane of the Mini-CECF container.
- Add high molecular components to the RM as described (Table 5) and mix by pipetting up and down. Add 60 µL to the reaction container. Avoid air bubbles during transfer of the viscous solution.
- Cut 2 8 x 10 cm sheets of a sealing thermoplastic film and wrap them around the 24-well plate with the reaction containers inside. This will reduce evaporation from the reaction mixture. Now place the lid of the cell culture plate on the plate and fix it with tape. Incubate the plate at 30 °C and 200 rpm agitation for 12-16 h.
- Day 2: Harvest the reaction mix by piercing through the dialysis membrane with the pipet tip at the Mini-CECF container and aspirate the RM. Transfer RM into fresh tubes and centrifuge at 16,000 x g at 4 °C for 10 min to remove precipitates. Transfer the supernatant into fresh tubes. The protein in the supernatant can now be further analyzed.
NOTE: In some cases, after centrifugation a pellet will be present. This can provide important information on the suitability of the analyzed nanodiscs or other reaction compounds to keep the synthesized MP soluble. It is therefore advisable to analyze the amount of target potentially present in the precipitate by SDS-PAGE, Western Blot, or visual evaluation of the pellet size.
Representative Results
The impact of fine-tuning reaction compounds on the final yield or quality of synthesized MPs is exemplified. A frequent routine approach is to adjust the optimal Mg2+ concentration in CF reactions by expression of green fluorescent protein (GFP) as a convenient monitor of system efficiency. As an example, GFP was synthesized from a pET-21a(+) vector at Mg2+ concentrations between 14 and 24 mM (Figure 2). SDS-PAGE analysis identified the optimal Mg2+ concentration at 20 mM (Figure 2A), which is in good accordance with complementary fluorescence measurements (excitation at 485 nm, emission measurement at 535 nm) of the CF reaction supernatant (Figure 2B). For the CF expression of the bacterial PR expressed from pIVEX 2.3 vector and of the turkey β1 adrenergic receptor (Tβ1AR) expressed from pET-21a(+) vector, the optimal Mg2+ concentration was identified at 20-22 mM.
As a further example, quality refinement of synthesized MPs with the described strategy is shown. Tunable parameters for the co-translational insertion of MPs into preformed nanodiscs are (i) the lipid composition of the nanodisc membrane and (ii) the final nanodisc concentration in the reaction. It is well known that the hydrophobic environment is an important parameter for correct folding, oligomeric assembly, and stability of MPs. Since lipid composition in nanodiscs can be modulated, the described approach enables a straightforward systematic screening of lipid effects on structure and function of a MP. In initial experiments, it is recommended to screen lipids with phosphatidylcholine (PC) and phosphatidylglycerol (PG) headgroups and to test different chain lengths and saturations. The impact of membrane composition and nanodisc concentration on the solubilization efficiency and quality of two different MPs is shown. PR is a light activated proton pump and a very stable and highly synthesized MP reaching final concentrations of > 100 µM in the RM. Thus, it is recommended to use it as a positive control to ensure correct reaction setup as PR concentration can be easily assessed by measurement of absorption at 530 nm in the RM supernatant. In contrast, Tβ1AR is an example of the large family of eukaryotic G-protein coupled receptors (GPCRs) and is a complex and unstable MP. For convenient monitoring, a Tβ1AR-GFP fusion construct was synthesized and the total concentration of nanodisc solubilized receptor in the CF reactions was determined via GFP fluorescence (Figure 3A). The fraction of functionally folded and ligand-binding active receptor was further determined using a filter binding assay and the labeled ligand [3H]-dihydroalprenolol (Figure 3B). The filter binding assay was performed as described previously14. Briefly, Tβ1AR-GFP concentration in the RM was determined via its fluorescence. The GPCR was incubated with the radiolabelled ligand for 1 hour at 20 °C, applied to a filter, and unbound ligand was washed off. For determination of unspecific [3H]-dihydroalprenolol binding, the receptor was saturated with unlabeled alprenolol in a control reaction. The counts of the radioactive ligand in the filters were measured in a scintillation counter and the amount of bound ligand was determined via its specific label activity. Percent GPCR binding activity was then calculated from the amount of bound ligand, the Tβ1AR-GFP concentration, and the assay volume. The overall synthesis and nanodisc solubilization of Tβ1AR-GFP is similar with all analyzed membrane compositions and within the range of 8-13 µM (Figure 3A). In contrast, a much higher variation is detectable in the quality of the synthesized GPCR. Lowest activity with less than 10% active fraction is obtained with the lipids DMPC and POPC. With DOPG and POPG, the active fraction of Tβ1AR-GFP could be increased to approximately 30% (Figure 3B). The results indicate that charge of the lipid headgroup as well as flexibility of the fatty acid chain are important modulators for folding and activity of this GPCR.
Besides nanodisc composition, their final concentration in the CF reaction can be an important factor for MP quality. Once a suitable membrane composition of a nanodisc has been identified, its concentration during MP synthesis should be screened. It is obvious that the nanodisc concentration needs to be adjusted according to the expression efficiency of a MP. CF synthesis of Tβ1AR-GFP receptor and PR result in final concentrations of approximately 10 µM and 100 µM in the RM, respectively. If the nanodisc concentration is screened within a range of 3.75-60 µM, a complete solubilization of the GPCR is obtained at approximately 30 µM nanodiscs, giving a ratio of Tβ1AR:nanodisc of 1:3 (Figure 4A). In contrast, complete solubilization of PR is already achieved with approximately 10 µM nanodiscs and gives a PR:nanodisc ratio of 10:1 (Figure 4B). An excess of nanodiscs is therefore necessary to achieve almost complete solubilization of Tβ1AR-GFP, while an inverted ratio in the case of PR indicates the insertion of multiple PR copies into one nanodisc. Subsequent studies of purified PR/nanodisc complexes with native mass spectrometry confirmed this observation and found a prevalence of higher oligomeric forms of PR if synthesized at lower nanodisc concentrations15,18. The titration of CF synthesized MPs with nanodiscs can therefore be used as a tool to trigger oligomeric assemblies and to study their effects on MP function.
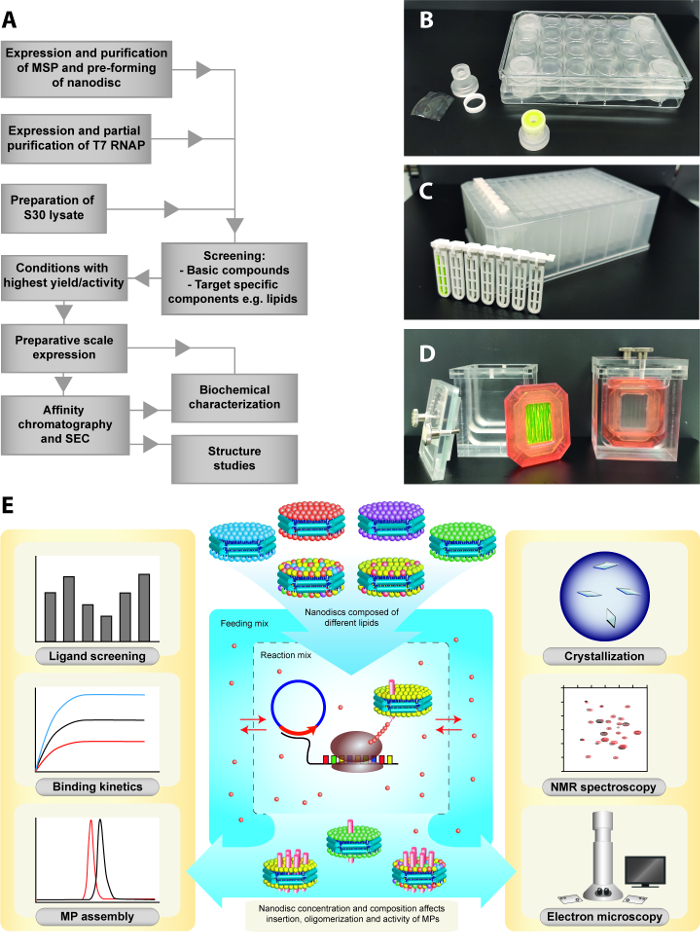
Figure 1: CECF expression strategy for the insertion of membrane proteins into nanodiscs. (A) Basic workflow illustrating the major steps of the process. (B) Customized analytical scale reaction vessels for CECF expression. (C) Commercially available dialysis cartridges for analytical scale CECF setup. (D) Preparative scale setup (3 mL RM) including a 3 mL dialysis cassette and a customized plexiglass FM container. (E) Co-translational insertion of MPs into preformed nanodiscs and lipid screening in the CECF configuration. The RM contains the required transcription/translation machinery and nanodiscs while low molecular weight compounds are present in both compartments. Biochemical and structural applications of the produced MP/nanodisc samples are further illustrated. Please click here to view a larger version of this figure.
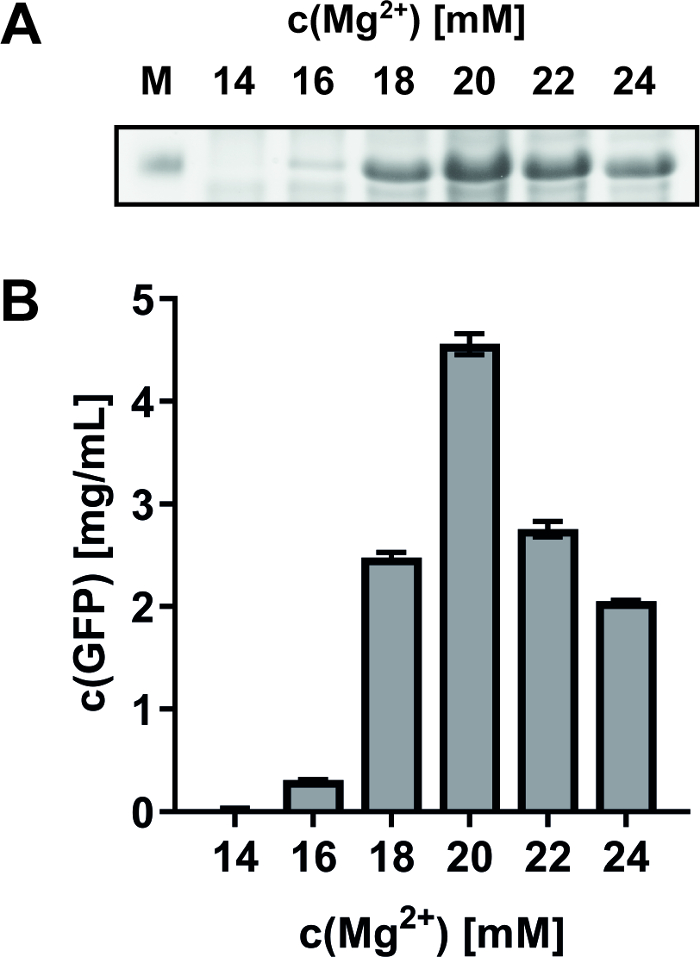
Figure 2: Effect of different Mg2+ concentrations on CF GFP synthesis. GFP was synthesized in CF reactions with Mg2+ concentrations within a range of 14-24 mM. (A) SDS-PAGE analysis shows the strongest band of GFP at 20 mM Mg2+. M: Marker. (B) GFP fluorescence after CF expression with different Mg2+ concentrations. The maximal GFP fluorescence at 20 mM Mg2+ corresponds to 4.6 mg/mL. Error bars represent the standard deviation of a duplicate measurement. Please click here to view a larger version of this figure.
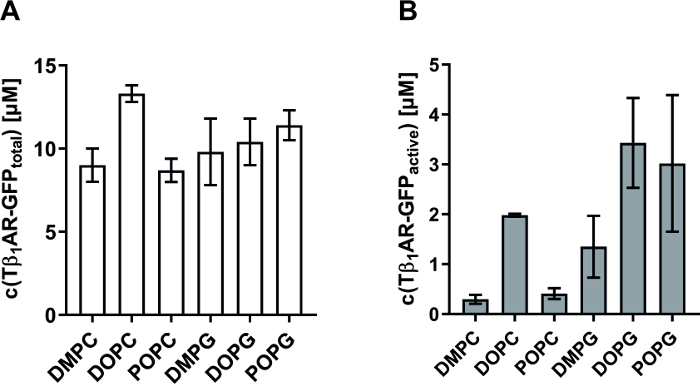
Figure 3: Effect of nanodisc membrane composition on solubilization and quality of a CF synthesized GPCR. Yield and activity of Tβ1AR-GFP synthesized in the presence of 30 µM nanodiscs containing different membrane compositions. The total synthesized protein (A) and fraction of ligand-binding active receptor (B) are given in µM. Total concentration was determined via fluorescence measurement of the GFP fusion. Activity was measured via filter binding assay with the radiolabeled ligand [3H]-dihydroalprenolol. Error bars represent standard deviations of independent triplicates. Data taken from previously published manuscript14. Please click here to view a larger version of this figure.
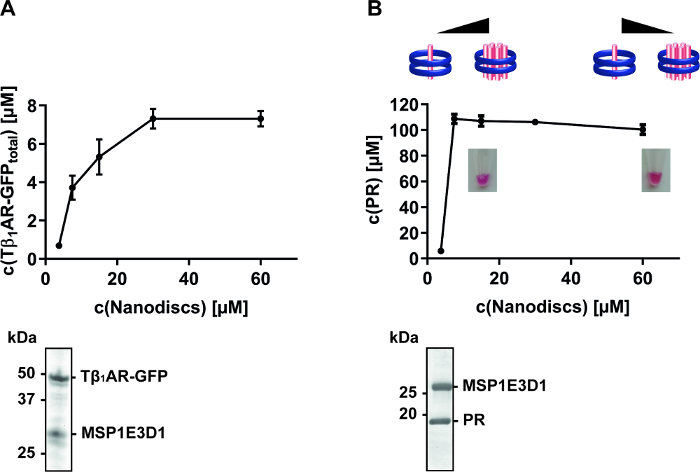
Figure 4: Solubilization screen of CF synthesized MPs with increasing nanodisc concentrations. The MPs were synthesized in the presence of supplied nanodiscs within a range of 3.75-60 µM. (A) Expression of Tβ1AR-GFP with nanodiscs (DMPC) in the CF reaction. Total concentration was determined via fluorescence measurement of the GFP fusion. Data taken from previously published manuscript14. Affinity-tag purified samples were analyzed by SDS-PAGE and Coomassie-Blue staining shows two prominent bands corresponding to Tβ1AR-GFP at approximately 50 kDa and MSP1E3D1 above 25 kDa (B) Expression of PR with nanodiscs (DOPG) in the CF reaction. Total concentration of PR was determined via absorption measurement at 530 nm. Data taken from previously published manuscript10,18. The photos show the red color of the final reactions due to the presence of folded PR. The pictograms illustrate the modulated PR assembly towards lower oligomeric conformations upon increased nanodisc concentrations, as revealed by subsequent native mass spectrometry18. Affinity-tag purified samples were analyzed by SDS-PAGE and Coomassie-Blue staining shows two prominent bands corresponding to PR at approximately 20 kDa and MSP1E3D1 above 25 kDa. Please click here to view a larger version of this figure.
| Compound | Final concentration range | Effect on MP sample |
| Mg2+ | 10 – 30 mM | yield |
| DTT | 1 – 20 mM | disulfide bridge formation, folding |
| 20 Amino acid mixture1 | 0.2 – 2 mM | yield, |
| GSSG : GSH2 | (1-10 µM) : (1-10 µM) | disulfide bridge formation, folding |
| Nanodiscs | 10 – 100 µM | solubilization, oligomeric assembly, folding |
| Lipid type | solubilization, oligomeric assembly, folding | |
| S30 : HS30 lysate3 | (50-100 %) : (50-100 %) | folding |
| S12 – S100 | 20 – 50 % | yield, MP background in channel assays |
| DNA template | 0.5 – 30 ng/µL RM | yield, oligomeric assembly, folding |
| DNA template design | yield | |
| 1, mixture may be varied according to the individual composition of a MP to e. g. improve yield or optimize NMR labelling schemes. 2, GSH should always be prepared freshly. 3, total CF lysate concentration in the RM should be at least 35 % with a S30 content of at least 50 % in order to ensure high expression levels. |
||
Table 1: Critical screening components of CF reactions.
| MSP1E3D1 | Lipid | DPC | ||||||
| Ratio | (410 µM stock) | (50 000 µM stocks) | (10 % (w/v) stock) | |||||
| Lipid | Lipid : MSP | V(MSP) | c(MSPfinal) | V(lipid) | c(lipidfinal) | V(DPC) | c(DPCfinal) | DF buffer |
| [µL] | [µM] | [µL] | [µM] | [µL] | [%] | [µL] | ||
| DMPC | 115 : 1 | 1500.0 | 205.0 | 1415.0 | 23 575 | 30.0 | 0.1 | 55.5 |
| DOPC | 80 :1 | 1500.0 | 205.0 | 984.0 | 16 400 | 30.0 | 0.1 | 486.0 |
| POPC | 85 : 1 | 1500.0 | 205.0 | 1046.0 | 17 425 | 30.0 | 0.1 | 425.0 |
| DMPG | 110 : 1 | 1500.0 | 205.0 | 1353.0 | 22 550 | 30.0 | 0.1 | 117.0 |
| DOPG | 80 : 1 | 1500.0 | 205.0 | 984.0 | 16 400 | 30.0 | 0.1 | 486.0 |
| POPG | 90 : 1 | 1500.0 | 205.0 | 1107.0 | 18 450 | 30.0 | 0.1 | 363.0 |
Table 2: Lipid to MSP1E3D1 ratios for 3 mL in vitro assembly setups.
| Compound | Concentration | Preparation |
| DNA plasmid template1 | > 400 µg/mL in 10 mM Tris-HCl pH 8.0 | Midi/Maxi kit (e.g. Qiagen) preparation2 |
| Mixture of 20 amino acids3 | 25 mM each in H2O | precipitate remains4 |
| Mixture of 4 NTPs (75 x) | c(CTP) 240 mM, c(ATP) 360 mM, c(UTP) 240 mM, c(GTP) 240 mM in H2O. Adjust pH to 7-8 with KOH |
A little precipitate remains4 |
| Acetyl phosphate | 1 M in H2O, adjust pH 7-8 with KOH | precipitate remains4 |
| Phospho(enol)pyruvic acid (K+) | 1 M in H2O, adjust pH 7-8 with KOH | |
| Folinic acid | 10 mg/mL in H2O | precipitate remains4 |
| DTT | 0.5 M in H2O | |
| c0mplete protease inhibitor cocktail | 1 tablet per 1 mL in H2O | |
| Tris-acetate, pH 8.0 | 2.4 M in H2O | |
| Mg(OAc)2 | 1 M in H2O | |
| KOAc | 10 M in H2O | |
| Ribolock RNase-Inhibitor | 40 U/mL | Thermo Fisher Scientific |
| tRNA (E. coli) | 40 mg/mL in H2O | Roche (Germany) |
| T7RNAP | 3-7 mg/mL5 | see protocol section 2 |
| Pyruvate kinase | 10 mg/mL | Roche (Germany) |
| Nanodiscs (DMPG) | 0.2-1.0 mM6 in 10 mM Tris-Cl, pH 8.0, 100 mM NaCl | see protocol sections 3 and 4 |
| 1, PCR template could be used at similar concentrations. 2, the quality of “Mini”-kit prepared DNA is not satisfactory. 3, cysteine tends to be unstable and may be added separately. 4, solution is oversaturated, thorough mixing instantly before removing aliquots is necessary. 5, for each new T7RNAP batch, an initial screen is recommended to identify the best final concentration. 6, solubility of nanodiscs can depend on their lipid composition. |
||
Table 3: Preparation of CF stock solutions.
| Compound | ||
| For Mastermix (3.0 x) | c(final) | V [µL] |
| Mixture of 20 amino acids | 1 mM | 2520.0 |
| Mixture of 4 NTPs (75 x) | 1x | 840.0 |
| Acetyl phosphate | 20 mM | 1260.0 |
| Phospho(enol) pyruvic acid |
20 mM | 1260.0 |
| Folinic acid | 0.1 mg/mL | 630.0 |
| Tris-acetate, pH 8.0 | 100 mM | 2625.0 |
| c0mplete 50 x | 1x | 1260.0 |
| Mg(OAc)2 | 19.8 (= 20)1 mM | 1260.0 |
| KOAc | 180 (= 270)2 mM | 1140.0 |
| DTT | 2 mM | 252.0 |
| H2O | 7953.0 | |
| Total | 21 000 | |
| For RM | c(final) | V [µL] |
| 3x Mastermix | 1 x | 1000.0 |
| RNase inhibitor | 0.3 U/µL | 22.5 |
| tRNA (E. coli) | 0.5 mg/mL | 37.5 |
| Nanodiscs (DMPG)3 | 10 µM | 75.0 |
| DNA template4 | 0.015 ng/µL | 112.5 |
| Pyruvate kinase | 0.04 mg/mL | 12.0 |
| T7RNAP5 | 0.03 mg/mL | 15.0 |
| S30 lysate | 0.35 % | 1050.0 |
| All-trans retinal6 | 0.6 mM | 9.0 |
| H2O | – | 666.5 |
| Total | 3000.0 | |
| For FM | c(final) | V [µL] |
| Mastermix (3 x) | 1 x | 20 000 |
| H2O | – | 40 000 |
| Total | 60 000 | |
| 1, 0.2 mM Mg2+ are contributed from the S30 lysate. 2, 90 mM K+ are contributed from acetyl phosphate and phospho(enol) pyruvic acid. 3, calculated stock solution is 400 µM. 4, calculated stock solution is 400 µg/mL. 5, calculated stock solution is 6 mg/mL. 6, specific cofactor for PR, stock solution is 200 mM in DMSO. |
||
Table 4: Pipetting scheme for a CECF reaction with 3 mL of RM and 60 mL of FM
| Compound | ||||||||
| For Mastermix (3.0 x) | c(final) | V [µL] | ||||||
| Mixture of 20 amino acids | 1 mM | 864.0 | ||||||
| Mixture of 4 NTPs | 1x | 288.0 | ||||||
| Acetyl phosphate | 20 mM | 432.0 | ||||||
| Phospho(enol)pyruvic acid | 20 mM | 432.0 | ||||||
| Folinic acid | 0.1 mg/mL | 216.0 | ||||||
| Tris-acetate, pH 8.0 | 100 mM | 900.0 | ||||||
| c0mplete | 1x | 432.0 | ||||||
| Mg(OAc)2 | 13,8 (= 14) mM1 | 298.0 | ||||||
| KOAc | 180 (= 270) mM2 | 389.0 | ||||||
| DTT | 2 mM | 86.4 | ||||||
| H2O | – | 2862.6 | ||||||
| Total [µL]1 | – | 7200.0 | ||||||
| Mg2+ concentration | ||||||||
| 14 mM | 16 mM | 18 mM | 20 mM | 22 mM | 24 mM | |||
| For RM | c(final) | V [µL] | V [µL] | V [µL] | V [µL] | V [µL] | V [µL] | |
| 3x Mastermix | 1 x | 67.0 | 67.0 | 67.0 | 67.0 | 67.0 | 67.0 | |
| 0.1 M Mg(OAc)2 | 14-24 mM | 0.0 | 4.0 | 8.0 | 12.0 | 16.0 | 20.0 | |
| RNase inhibitor | 0.3 U/µL | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | |
| tRNA (E. coli) | 0.5 mg/mL | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | |
| DNA template3 | 0.015 ng/µL | 7.5 | 7.5 | 7.5 | 7.5 | 7.5 | 7.5 | |
| Pyruvate kinase | 0.04 mg/mL | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | |
| T7RNAP4 | 0.03 mg/mL | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| S30 lysate | 35 % | 70.0 | 70.0 | 70.0 | 70.0 | 70.0 | 70.0 | |
| H2O | – | 49.7 | 45.7 | 41.7 | 37.7 | 33.7 | 29.7 | |
| Total [µL]5 | – | 200.0 | 200.0 | 200.0 | 200.0 | 200.0 | 200.0 | |
| Mg2+ concentration | ||||||||
| 14 mM | 16 mM | 18 mM | 20 mM | 22 mM | 24 mM | |||
| For RM | c(final) | V [µL] | V [µL] | V [µL] | V [µL] | V [µL] | V [µL] | |
| 3x Mastermix | 1 x | 1133.0 | 1133.0 | 1133.0 | 1133.0 | 1133.0 | 1133.0 | |
| 0.1 M Mg(OAc)2 | 14-24 mM | 0.0 | 68.0 | 136.0 | 204.0 | 272.0 | 340.0 | |
| H2O | – | 2267.0 | 2199.0 | 2131.0 | 2064.0 | 1995.0 | 1927.0 | |
| Total [µL]5 | – | 3400.0 | 3400.0 | 3400.0 | 3400.0 | 3400.0 | 3400.0 | |
| 1, 0.2 mM Mg2+ are contributed from the S30 lysate. The mastermix is adjusted to the lowest screening concentration. 2, 90 mM K+ are contributed from acetly phosphate and phospho(enol) pyruvic acid. 3, calculated stock solution is 400 µg/mL. 4, calculated stock solution is 6 mg/mL. 5, reactions are performed in duplicates. |
||||||||
Table 5: Pipetting scheme for Mg2+ concentration screen with 100 µL of RM and 1.7 mL of FM.
Discussion
The setup and strategies to optimize the CF expression and co-translational insertion of functional MPs into nanodiscs are described. The required equipment comprises a bioreactor, a French press device or similar, an UV/VIS and fluorescence reader, CF reaction vessels suitable for a two-compartment configuration setup, and a temperature-controlled incubator. Further standard equipment are centrifuges for harvesting E. coli cells as well as tabletop centrifuges reaching at least 30,000 x g for preparation of S30 lysates. If S80-S100 lysates should be prepared, ultracentrifuges are necessary. The listed equipment is regularly available in biochemical labs and no larger initial investments are required for getting started with CF expression. Furthermore, fermentation and handling of E. coli cells for CF lysate and T7RNAP preparations are common and robust techniques. The most expensive compounds are CF lysate, T7RNAP, and nanodiscs. They can be prepared by reliable and efficient protocols and aliquots are stable for years at -80 °C. The protocols require three days each for CF lysate and T7RNAP preparation and approximately one week for expression and purification of MSP and preforming of nanodiscs. Target template DNA can be supplied using pET-21, pIVEX, or similar vector systems. For the setup and optimization of CF expression systems, the production of GFP variants such as shifted GFP (GFP+) or superfolderGFP can be used as monitor20,32. For MP production conditions, the expression of PR is a good model system or positive control as it folds under many different conditions and can easily be monitored by absorbance at 530 nm10,15,18. To establish an efficient CF expression protocol for a new MP, codon optimization of the target reading frame and using fusions with C-terminally attached GFP is recommended25,26. Further required materials include chemicals and enzymes that are all commercially available. These components need to be obtained in high quality and an overall cost calculation for the described CF reaction with 3 mL of RM and 60 mL of FM would amount to approximately 150-200 €. A prime application for CF expression systems is therefore the production of proteins difficult to obtain in classical cell-based expression systems such as many MPs or toxins. CF systems are furthermore core platforms in synthetic biology and continuously growing fields of applications make them increasingly indispensable as a tool in molecular research. Among others, routine applications are the labelling of proteins, high-throughput processes or synthetic cell design.
The demonstrated strategy enables detergent-free production of pure MPs already inserted into desired lipid environments of highly soluble nanodiscs. Once the CF expression protocol for a MP is established and optimized, the production is very fast and pure samples can be obtained in less than 24 h even in preparative scale. The MP/nanodisc complexes are purified directly from the CF reaction via streptavidin II or polyhistidine tags attached to the MP. The process allows parallel functional and structural analysis of identical MP samples by an array of different techniques33. Speediness and efficiency of the strategy is therefore competitive to conventional approaches employing cell-based expression systems and detergent extraction of MPs from cell membranes followed by routine in vitro reconstitution5,34. The open accessibility of CF reactions further facilitates numerous unique mechanistic studies of MP folding and membrane insertion15,16,18, MP complex assembly15,18,24 or functional regulation23.
An important difference of CF expression over cell-based expression is the absence of quality control systems for the synthesized protein product. Any translated polypeptide independent of its functional folding will end up in the final product sample. Furthermore, the insertion process into nanodisc membranes is artificial and translocon independent and may result either in incomplete integration or may not work at all with a number of MP targets. The finally solubilized product fraction in a CF reaction could thus be quite heterogeneous containing fully inserted but also only partially integrated or membrane associated MPs. Consequently, only a small fraction of a purified sample of MP/nanodisc complexes might be functionally folded. Modifying membrane composition and the careful fine-tuning of MP to nanodisc ratios by modulating the concentrations of nanodiscs and DNA templates are suitable tools for optimization. Nevertheless, downstream quality control approaches such as size exclusion profiling and target specific quantitative functional assays are always necessary for optimization of CF expression protocols. Availability of such assays can therefore limit applications, particularly in projects including MPs that require liposome environments for function such as transporters, channels, or pumps. Furthermore, the size limitations of nanodiscs could restrict the insertion of large MP assemblies.
In the documented examples, the variation in the functionally folded fraction of the GPCR Tβ1AR-GFP is within the range of a few percent, up to approximately 30%. Functional folding requires careful adjustment of a number of parameters14 and a similar individual and tedious optimization process might be necessary for other GPCRs as well22. However, the finally achieved yield of functionally folded GPCR is competitive with other production systems and allows the setup of > 1,000 radioassays for ligand binding studies from a single 60 µL reaction in an analytical scale Mini-CECF reactor. It is worth mentioning that ligand binding studies of GPCR-GFP fusion constructs do not require any purification steps. If necessary, purification can be achieved by taking advantage of affinity-tags attached to the synthesized MP, such as His-tags or Strep-tag II. The RM is then diluted in the desired buffer and loaded on the corresponding affinity chromatography column that has been pre-equilibrated accordingly. The structural evaluation of PR and other CF synthesized MPs by NMR spectroscopy or crystallization have already helped to establish CF expression systems as core platforms in MP research. The described strategy for production of MP/nanodisc complexes could become particularly interesting for future structural studies by electron microscopy.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank the Deutsche Forschungsgemeinschaft (DFG) grant BE1911/8-1, the LOEWE project GLUE, and the collaborative research center Transport and Communication across Membranes (SFB807) for financial support.
Materials
| 1,2-dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (DMPG) | Avanti Polar Lipids (USA) | 840445P | |
| 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) | Avanti Polar Lipids (USA) | 850345C | |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (sodium salt) (DOPC) | Avanti Polar Lipids (USA) | 850375C | |
| 1,2 dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (sodium salt) (DOPG) | Avanti Polar Lipids (USA) | 840475C | |
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids (USA) | 850457C | |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (POPG) | Avanti Polar Lipids (USA) | 840034C | |
| 2-Amino-2-(hydroxymethyl)-propan-1,3-diol (Tris) | Carl Roth (Germany) | 4855 | |
| 2-Mercaptoethanol | Carl Roth (Germany) | 4227 | |
| 2-Propanol | Carl Roth (Germany) | 9781 | |
| [3H]-dihydroalprenolol Hydrochloride | American Radiolabeled Chemicals (USA) | ART0134 | |
| Acetyl phosphate lithium potassium salt (ACP) | Merck (Germany) | 1409 | |
| Adenosine 5’-triphosphate (ATP) | Sigma Aldrich (Germany) | A9251 | |
| Alprenolol hydrochloride | Merck (Germany) | A0360000 | |
| Anion exchange chromatography column material: Q-sepharose® | Sigma-Aldrich (Germany) | Q1126 | |
| Autoclave Type GE 446EC-1 | Gettinge (Germany) | ||
| Bioreactor Type 884 124/1 | B.Braun (Germany) | ||
| Centrifuge | Sorvall RC12BP+; Thermo Scientific (Germany); Sorvall RC-5C; Thermo Scientific (Germany); Mikro 22 R; Hettich (Germany) | ||
| Cholic acid | Carl Roth (Germany) | 8137 | |
| Coomassie Brilliant Blue R250 | Carl Roth (Germany) | 3862 | |
| Culture flasks 500 ml baffled flasks, 2 l baffled flasks | Schott Duran (Germany) | ||
| Cytidine 5'-triphosphate disodium salt | Sigma-Aldrich (Germany) | C1506 | |
| D-glucose monohydrate | Carl Roth (Germany) | 6780 | |
| Di-potassiumhydrogen phosphate trihydrate | Carl Roth (Germany) | 6878 | |
| Dialysis tubing SpectrumTM Labs Spectra/PorTM 12-14 kD MWCO Standard RC tubing | Fisher Scientific (Germany) | 8700152 | |
| Dithiothreit | Carl Roth (Germany) | 6908 | |
| Ethanol | Carl Roth (Germany) | K928 | |
| Folinic acid calcium salt hydrate | Sigma-Aldrich (Germany) | 47612 | |
| French pressure cell disruptor | SLM; Amico Instruments (USA) | ||
| Glycerol | Carl Roth (Germany) | 3783 | |
| Guanosine 5'-triphosphate di-sodium salt (GTP) | Sigma-Aldrich (Germany) | G8877 | |
| Hydrochloric Acid | Carl Roth (Germany) | K025 | |
| IMAC column: HiTrap IMAC HP 5 mL | GE Life Sciences (Germany) | GE17-5248 | |
| Imidazole | Carl Roth (Germany) | 3899 | |
| Isopropyl-β-D-thiogalactopyranosid (IPTG) | Carl Roth (Germany) | 2316 | |
| Kanamycin | Carl Roth (Germany) | T832 | |
| L-Alanine | Carl Roth (Germany) | 3076.1 | |
| L-Arginine | Carl Roth (Germany) | 6908 | |
| L-Asparagine | Carl Roth (Germany) | HN23 | |
| L-Aspartic Acid | Carl Roth (Germany) | T202 | |
| L-Cysteine | Carl Roth (Germany) | T203 | |
| L-Glutamic Acid | Carl Roth (Germany) | 3774 | |
| L-Glutamine | Carl Roth (Germany) | 3772 | |
| L-Glycine | Carl Roth (Germany) | 3187 | |
| L-Histidine | Carl Roth (Germany) | 3852 | |
| L-Isoleucine | Carl Roth (Germany) | 3922 | |
| L-Leucine | Carl Roth (Germany) | 1699 | |
| L-Lysine | Carl Roth (Germany) | 4207 | |
| L-Methionine | Carl Roth (Germany) | 9359 | |
| L-Proline | Carl Roth (Germany) | 1713 | |
| L-Phenylalanine | Carl Roth (Germany) | 1709 | |
| L-Serine | Carl Roth (Germany) | 4682 | |
| L-Threonine | Carl Roth (Germany) | 1738 | |
| L-Tryptophane | Carl Roth (Germany) | 7700 | |
| L-Tyrosine | Carl Roth (Germany) | T207 | |
| MD100 dialysis units | Scienova (Germany) | 40077 | |
| N-2-Hydroxyethylpiperazine-N'-2-ethansulfonic acid (HEPES) | Carl Roth (Germany) | 6763 | |
| n-dodecylphosphocholine | Antrace (USA) | F308S | |
| PAGE chamber: Mini-Protean Tetra Cell | Biorad (Germany) | ||
| PAGE gel casting system: Mini Protean Handcast systems | Biorad (Germany) | ||
| PAGE gel power supply: Power Pac 3000 | Biorad (Germany) | ||
| Tryptone/peptone from caseine | Carl Roth (Germany) | 6681 | |
| Peristaltic pump: ip-12 | Ismatec (Germany) | ||
| Phosphoenol pyruvate monopotassium salt | Sigma Aldrich (Germany) | 860077 | |
| Potassium dihydrogen phosphate | Carl Roth (Germany) | P018 | |
| Potassium acetate | Carl Roth (Germany) | 4986 | |
| Potassium chloride | Carl Roth (Germany) | 6781 | |
| Pyruvate Kinase | Roche (Germany) | 10109045001 | |
| Scintillation counter: Hidex 300 SL | Hidex (Finland) | ||
| SDS pellets | Carl Roth (Germany) | 8029 | |
| Sodium azide | Sigma-Aldrich (Germany) | K305 | |
| Sodium chloride | Carl Roth (Germany) | P029 | |
| Spectrophotometer Nanodrop | Peqlab (Germany) | ||
| Spectrophotometer/fluorescence reader Spark® | Tecan (Switzerland) | ||
| tRNA (E. coli) | Roche (Germany) | 10109550001 | |
| Ultra sonificator | Labsonic U, B. Braun (Germany) | ||
| Uridine 5’-triphosphate tri-sodium salt (UTP) | Sigma-Aldrich (Germany) | U6625 | |
| Y-30 antifoam | Sigma-Aldrich (Germany) | A6457 | |
| Yeast extract | Carl Roth (Germany) | 2904 |
References
- Pandey, A., Shin, K., Patterson, R. E., Liu, X. Q., Rainey, J. K. Current strategies for protein production and purification enabling membrane protein structural biology. Biochemistry and Cell Biology. 94 (6), 507-527 (2016).
- Ritchie, T. K., et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods in Enzymology. 464 (09), 211-231 (2009).
- Frauenfeld, J., et al. A novel lipoprotein nanoparticle system for membrane proteins. Nature Methods. 13 (4), 345-351 (2016).
- Yang, Z., et al. Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains. Protein Science. 23 (6), 769-789 (2014).
- Seddon, A. M., Curnow, P., Booth, P. J. Membrane proteins, lipids and detergents: not just a soap opera. Biochimica et Biophysica Acta. 1666 (1-2), 105-117 (2004).
- Junge, F., et al. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. New Biotechnology. 28 (3), (2011).
- Smolskaya, S., Logashina, Y. A., Andreev, Y. A. Escherichia coli extract-based cell-free expression system as an alternative for difficult-to-obtain protein biosynthesis. International Journal of Molecular Sciences. 21 (928), 1-21 (2020).
- Bayburt, T. H., Grinkova, Y. V., Sligar, S. G. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters. 2 (8), 853-856 (2002).
- Katzen, F., Peterson, T. C., Kudlicki, W. Membrane protein expression: no cells required. Trends in Biotechnology. 27 (8), 455-460 (2009).
- Roos, C., et al. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase. Biochimica et Biophysica Acta-Biomembranes. 1818 (12), 3098-3106 (2012).
- Cappuccio, J. A., et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Molecular and Cellular Proteomics. 7 (11), 2246-2253 (2008).
- Patriarchi, T., et al. Nanodelivery of a functional membrane receptor to manipulate cellular phenotype. Scientific Reports. 8 (1), 1-11 (2018).
- Shelby, M. L., He, W., Dang, A. T., Kuhl, T. L., Coleman, M. A. Cell-free co-translational approaches for producing mammalian receptors: expanding the cell-free expression toolbox using nanolipoproteins. Frontiers in Pharmacology. 10 (744), 1-12 (2019).
- Rues, R. B., Dötsch, V., Bernhard, F. Co-translational formation and pharmacological characterization of beta1-adrenergic receptor/nanodisc complexes with different lipid environments. Biochimica et Biophysica Acta-Biomembranes. 1858 (6), 1306-1316 (2016).
- Henrich, E., et al. Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife. 6, 1-19 (2017).
- Harris, N. J., Pellowe, G. A., Booth, P. J. Cell-free expression tools to study co-translational folding of alpha helical membrane transporters. Scientific Reports. 10 (1), 1-13 (2020).
- Henrich, E., et al. Lipid requirements for the enzymatic activity of MraY translocases and in vitro reconstitution of the lipid II synthesis pathway. Journal of Biological Chemistry. 291 (5), 2535-2546 (2016).
- Peetz, O., et al. Insights into co-translational membrane protein insertion by combined LILBID-mass spectrometry and NMR spectroscopy. Analytical Chemistry. 89 (22), 12314-12318 (2017).
- Kigawa, T., et al. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. Journal of Structural and Functional Genomics. 5 (1-2), 63-68 (2004).
- Schwarz, D., et al. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nature Protocols. 2 (11), 2945-2957 (2007).
- Yang, J. P., Cirico, T., Katzen, F., Peterson, T. C., Kudlicki, W. Cell-free synthesis of a functional G protein-coupled receptor complexed with nanometer scale bilayer discs. BMC Biotechnology. 11 (57), (2011).
- Rues, R. B., Dong, F., Dötsch, V., Bernhard, F. Systematic optimization of cell-free synthesized human endothelin B receptor folding. Methods. 147 (1), 73-83 (2018).
- Keller, T., Gorboulev, V., Mueller, T. D., Dötsch, V., Bernhard, F., Koepsell, H. Rat organic cation transporter 1 contains three binding sites for substrate 1-methyl-4-phenylpyridinium per monomer. Molecular Pharmacology. 95 (2), 169-182 (2019).
- Matthies, D., et al. Cell-free expression and assembly of ATP synthase. Journal of Molecular Biology. 413 (3), 593-603 (2011).
- Schwarz, D., Daley, D., Beckhaus, T., Dötsch, V., Bernhard, F. Cell-free expression profiling of E. coli inner membrane proteins. Proteomics. 10 (9), 1762-1779 (2010).
- Haberstock, S., et al. A systematic approach to increase the efficiency of membrane protein production in cell-free expression systems. Protein Expression and Purification. 82, 308-316 (2012).
- Falkinham, J. O., Clark, A. J. Genetic analysis of a double male strain of Escherichia coli K12. Génétique. 78 (2), 633-644 (1974).
- Foshag, D., et al. The E. coli S30 lysate proteome: A prototype for cell-free protein production. New Biotechnology. 40, 245-260 (2018).
- Davanloo, P., Rosenberg, A. H., Dunn, J. J., Studier, F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 81 (7), 2035-2039 (1984).
- Reckel, S., et al. Strategies for the cell-free expression of membrane proteins. Methods in Molecular Biology. 607, 187-212 (2010).
- Denisov, I. G., Baas, B. J., Grinkova, Y. V., Sligar, S. G. Cooperativity in cytochrome P450 3A4: Linkages in substrate binding, spin state, uncoupling, and product formation. Journal of Biological Chemistry. 282 (10), 7066-7076 (2007).
- Scholz, O., Thiel, A., Hillen, W., Niederweis, M. Quantitative analysis of gene expression with an improved green fluorescent protein. European Journal of Biochemistry. 267 (6), 1565-1570 (2000).
- Henrich, E., et al. From gene to function cell-free electrophysiological and optical analysis of ion pumps in nanodiscs. Biophysical Journal. 113 (6), 1331-1341 (2017).
- Shen, H. H., Lithgow, T., Martin, L. L. Reconstitution of membrane proteins into model membranes: Seeking better ways to retain protein activities. International Journal of Molecular Sciences. 14 (1), 1589-1607 (2013).

