Imaging and Analysis of Tissue Orientation and Growth Dynamics in the Developing Drosophila Epithelia During Pupal Stages
Summary
This protocol is designed for the imaging and analysis of the dynamics of cell orientation and tissue growth in the Drosophila abdominal epithelia as the fruit fly undergoes metamorphosis. The methodology described here can be applied to the study of different developmental stages, tissues, and subcellular structures in Drosophila or other model organisms.
Abstract
Within multicellular organisms, mature tissues and organs display high degrees of order in the spatial arrangements of their constituent cells. A remarkable example is given by sensory epithelia, where cells of the same or distinct identities are brought together via cell-cell adhesion showing highly organized planar patterns. Cells align to one another in the same direction and display equivalent polarity over large distances. This organization of the mature epithelia is established over the course of morphogenesis. To understand how the planar arrangement of the mature epithelia is achieved, it is crucial to track cell orientation and growth dynamics with high spatiotemporal fidelity during development in vivo. Robust analytical tools are also essential to identify and characterize local-to-global transitions. The Drosophila pupa is an ideal system to evaluate oriented cell shape changes underlying epithelial morphogenesis. The pupal developing epithelium constitutes the external surface of the immobile body, allowing long-term imaging of intact animals. The protocol described here is designed to image and analyze cell behaviors at both global and local levels in the pupal abdominal epidermis as it grows. The methodology described can be easily adapted to the imaging of cell behaviors at other developmental stages, tissues, subcellular structures, or model organisms.
Introduction
To achieve their roles, epithelial tissues fully rely on the spatial organization of their cellular components. In most epithelia, cells are not only packed against each other to create a precise cobblestone layer but they orient themselves relative to the body axes.
The functional importance of precise tissue organization is obvious in sensory epithelia, such as the vertebrate inner ear and retina. In the first case, hair and supporting cells align in a specific axial direction to efficiently sense mechanical inputs such as sound and motion1,2. Similarly, photoreceptor cell spatial organization is essential for achieving optimal optical properties by the retina3. Spatial control of cell position and orientation is thus of particular relevance for proper physiological function.
Drosophila is a holometabolous insect that undergoes a complete transformation of its larval body structures through metamorphosis, giving rise to its adult tissues. The Drosophila pupa is an excellent model for the noninvasive live imaging of a variety of dynamic events, including developmental cell migration4, cell division and growth dynamics5, muscle contraction6, cell death7, wound repair8, and cell orientation9. In the adult Drosophila, the external epithelium shows a high degree of order. This is easily observed on the arrangements of trichomes (i.e., cell protrusions originating from single epithelial cells) and sensory bristles all over the fly's body surface10. Indeed, trichomes are aligned in parallel rows guiding airflow11. The morphogenesis of the adult epithelia and the ordered arrangement of the individual cells starts during embryogenesis and culminates during pupal stages. While in embryos cell divisions, intercalations, and shape changes all decrease tissue order12,13, this is reverted at later stages of development, especially at pupal stages, when the fly approaches maturity9.
The immobile Drosophila pupa provides an ideal system to evaluate cell shape and orientation changes. The pupal abdominal epidermis presents special advantages. While the precursors of the adult head, thorax, genitalia, and appendages grow and get patterned from larval stages, the histoblasts, which are integrated into the larval epidermis, start growing and differentiating only at pupariation14. This feature allows the tracking of all spatiotemporal events involved in the establishment of tissue order in its entirety9.
Histoblasts are specified during embryonic development at contralateral positions in each presumptive abdominal segment. The dorsal abdominal epidermis of the adult derives from dorsolaterally located histoblast nests present at the anterior and posterior compartments15,16. As histoblasts expand, replacing the larval epithelial cells (LECs), the contralateral nests fuse at the dorsal midline forming a confluent sheet17,18,19,20.
This work describes 1) a methodology for dissection, mounting, and long-term live imaging of the Drosophila pupae, and 2) analytical methods to study the dynamics of cellular orientation and growth at high spatiotemporal resolution. A detailed protocol is provided here, covering all the steps required from the initial pupae preparation (i.e., staging and imaging) to the extraction and quantification of directionality and orientation features. We also describe how to infer local tissue properties from the analysis of cell clones. All the steps described are minimally invasive and allow long-term live analyses. The methods described here can be easily adapted and applied to other developmental stages, tissues, or model organisms.
Protocol
NOTE: This protocol is divided into five steps: (1) staging the pupae, (2) preparing the pupae for imaging, (3) live imaging of the growing abdominal epithelia, (4) generation of genetic mosaics, (5) data processing and analysis (including sections describing how to analyze cell orientation dynamics from cell junction outlines and growth dynamics from cell clones).
1. Staging of Drosophila pupae before imaging
- Culture flies of the appropriate genotype on standard medium in plastic vials at 25 °C for 5 days (±12 h) after egg laying (AEL).
NOTE: Metamorphosis starts within the confinement of the third-instar larvae into the pupal case at 120 h AEL until 0 h after puparium formation (APF). This transition is easily identifiable, because larvae stop feeding and moving and the opercular region is formed (Figure 1A) at 0-12 h APF. The puparium is initially soft and white, but progressively hardens and tans. - Transfer white prepupae (0 h APF) to a fresh plastic vial using a moisten paintbrush. Animals can be kept at different temperatures depending on the designed experiment until the desired age.
NOTE: Pupa formation (i.e., pupation) occurs at 12 h APF, when the head and the appendages of the adult fly are totally everted (Figure 1A). By this time the pupal case is fully separated from the pupa, allowing its complete removal (Figure 1A).
2. Preparing pupae for live imaging
NOTE: After staging, the pupae are dissected and mounted as described below (see also Figure 1).
- Remove staged pupae from the wall of the vial with the help of the forceps.
- Glue the ventral side of each pupa on a glass slide covered with double-sided sticky tape. Gently tap on the head spiracles (i.e., opercular region) and the dorsal surface of the pupa with the tips of the forceps to assure the adhesion of the pupal case to the tape (Figure 1A,B).
NOTE: The dorsal surface should face up to facilitate the dissection of the case and the recovery of the pupa. - Begin dissection under a stereomicroscope by gently removing the operculum from the puparium with the forceps (Figure 1C).
- Insert one tip of the forceps in a shallow angle between the pupal case and the pupa surface through the opercular opening. Tear the case from head to tail laterally in one or more swings, avoiding pinching the pupa (Figure 1D). Fold back the cracked pupal case to the lateral sides as you keep proceeding to the posterior end (Figure 1E).
NOTE: The pupal case is quite rigid and cracks easily. In case of high humidity or in particular genotypic backgrounds, the pupal case becomes softer and tearing is more difficult. In these cases, the cracking of the pupal case can be helped by pricking its free edges with both tips of the forceps. - Remove the pupa from the opened-up pupal case by carefully inserting the forceps under the animal and gently pulling up (Figure 1F). The pupa will stick to the tip of the forceps (Figure 1G−H).
- Transfer the pupa with the help of the forceps to a glass-bottom dish and deposit it on top of a small drop of gas-permeable halocarbon oil (Figure 1I). Hold the pupa gently by the ventral side to avoid any possible tissue damage.
NOTE: The drop of halocarbon oil has to be small, with a diameter approximately less than half the length of the pupa. Such amount is sufficient to adhere the pupa to the glass by capillarity and to correct the optics for oil immersion objectives. - Roll a piece of wet tissue paper at the edges of the dish to maintain humidity. Cover the dish to avoid dehydration of the pupae during imaging.
NOTE: Both female and male pupae can be employed for imaging. We recommend employing the third abdominal segments (AIII) as a reference abdominal metamere since it is almost identical in both sexes in terms of size, shape, and patterning.
3. Live imaging of growing abdominal epithelia
NOTE: An inverted laser scanning confocal microscope equipped with a 40x/1.3 NA oil immersion objective was used to image pupae at different developmental stages.
- Orient the pupa over the oil drop on the glass-bottom dish according to the domain and the process to be evaluated (e.g. dorsolaterally for long-term live imaging of the early expansion of the dorsal nests, or dorsally to image their late expansion and tissue remodeling). See Figure 1J,K and Figure 4.
- Transfer the glass-bottom dish containing the mounted pupae to the microscope stage and focus on the surface of the abdominal area using the transmitted light.
NOTE: Even if this protocol is optimized for imaging on inverted microscopes, it is also possible to perform imaging on an upright microscope. In that case, the sample is placed on the microscope stage with the glass-bottom surface facing up. The halocarbon oil holds each pupa on a meniscus. - Set the acquisition parameters: 1) the number of Z-slices are usually between 20−40 to allow appropriate two-dimensional (2D) reconstruction of the abdominal epidermis; 2) step size between each slice (e.g., 1 micron); 3) time interval for recording (a 5 min interval is suitable for high fidelity analyses of cell orientation dynamics); and 4) frame resolution (e.g., 1024 x 1024).
- Turn on the appropriate lasers (i.e., 488 nm and 561 nm to visualize GFP and RFP fluorophores respectively) and adjust the laser power and gain/offset settings to visualize the marked cells. Use the lowest possible laser power (in the range 5%−20%) to minimize photobleaching and phototoxicity.
- Manually set the position and the appropriate Z-stack limits for multiple pupae using the attached motorized stage and the microscope multiposition acquisition software.
4. Generation of genetic mosaics to follow behaviors of cell clones
NOTE: We employ mitotic recombination to induce genetic mosaics in the abdominal epithelium via site-specific recombination (FLP/FRT system21,22) (Figure 2).
- Cross virgin females carrying a heat shock-inducible Flippase transgene (hs-FLP), a FLP recognition target (FRT) site at a specific genomic location (e.g., FRT site at position 40A at the L arm of chromosome 2), and a recognizable cellular marker (e.g., Ubi-RFP.nls or Ubi-GFP.nls) distal to the FRT site, to mutant males carrying an FRT site at the equivalent genomic location (Figure 2A−C).
NOTE: Autonomous and nonautonomous effects within or outside clones for any gene loss of function could be studied employing specific recessive alleles distal to the FRT site. - Generate FLP/FRT somatic clones in the histoblasts by heat shock treatment at the third instar larval stage of the progeny of the cross. This is performed by submerging the plastic vials containing the animals in a water bath at 37 °C for 45 min to 1 h at the wandering larvae (LIII) stage.
NOTE: The sensitive period for mitotic recombination is the G2 phase of the cell cycle. Histoblasts are arrested in G2 during the whole larval development. - Score twin clones for absence (i.e., mutant cells) or enhanced (i.e., wild type twin-spot cells) levels of the fluorescent protein marker from 16 h APF onward (Figure 2D).
NOTE: On average, 45 min to 1 h at 37 °C renders approximately only 2−3 twin clones per region of interest (e.g., the abdominal hemisegment). Pupae showing too a high clone density (e.g., more than four twin clones per hemisegment) should be discarded from further quantitative analyses. - Upon clone identification, image living pupae at the desired stage and for the desired length of time as described in the previous section.
5. Data processing and analyses
NOTE: Data are processed using ImageJ (imagej.nih.gov/ij/).
- Distinguish cell orientation dynamics from cell junction outlines.
- Project the Z-stack slices acquired by confocal microscopy in 2D using the Maximum Intensity Projection (MIP) function of ImageJ.
NOTE: The number of slices per stack should be kept to a minimum to avoid the out of focus noise generated by macrophages patrolling under the epidermis. - Set a planar coordinate system identifying reliable tissue landmarks (e.g., A/P compartment boundaries) for analysis of each dataset (Figure 3A).
NOTE: Employing the same planar references for each data set will allow multiple measurements to be compared. - Use the OrientationJ plugin (bigwww.epfl.ch/demo/orientation/) of ImageJ23,24 on local cell edges to obtain qualitative and quantitative orientation values. The plugin renders color-coded overlays on input images based on local orientations and provides numerical values when used in quantitative mode (Figure 3B−B'' and Figure C−C'''').
NOTE: The OrientationJ plugin is based on structure tensors, 2 x 2 matrix of eigenvalues derived from gradient and directional derivatives (See references23,24 for a detailed description). - Use the OrientationJ Distribution option to color-code cell edge orientations relative to the set planar coordinate system (i.e., cell edge orientation maps) (Figure 3B). The Distribution option is found under the plugin menu in ImageJ. The settings to employ are: Gaussian window sigma = 1 pixel; Cubic Spline = Gradient; Minimum Coherency = 20%; Minimum Energy = 1%. Cell edge orientations are displayed as a color-coded image employing the Color Survey option of the plugin (Hue = orientation; Saturation = coherency; and Brightness = input image).
NOTE: Areas that contain background fluorescence do not provide any directional information and must be manually excluded from the analysis. High threshold settings reduce the pixels considered in the processed images. - Use the OrientationJ Measure option to quantify cellular orientations and directional cell-cell alignment (i.e., coherency) (Figure 3C). The Measure option is found under the plugin menu in ImageJ. Generate small adjacent non-overlapping regions of interest (ROIs) of uniform weight (64 x 64 pixels, about 20 µm x 20 µm) within the area occupied by the histoblasts (Figure 3C).
- Calculate the dominant local orientation (i.e., the averaged orientation between neighboring cells-averaged cell edge orientation map) and coherency from the ROIs. The software computes the predominant orientation and local coherency within each ROI (Figure 3C).
NOTE: The largest and smallest eigenvalues of the structure tensor estimated by OrientationJ are employed to calculate coherency as the ratio between their difference and their sum. Coherency is bounded between values of 0−1. A value of 1 indicates full alignment uniformity, while a value of 0 indicates isotropic areas with no alignment. - Statistically analyze the calculated orientation and coherency values from multiple images using free software packages such as PAST25. Axial data such as orientation values (in degrees) are appropriately described by directional statistics.
- Through the software, calculate the mean direction and circular variance for each set of orientation distributions. The statistical significance of the difference between the distribution of the orientations between different genotypes or conditions is determined using the nonparametric Mardia-Watson-Wheeler test (W-test) for equal distributions.
- Calculate the statistical significance of the difference in coherency applying the non-parametric Kolmogorov-Smirnov test (K-S test). Display data graphically as desired (polar plots, bar charts, box plots, etc.).
- Project the Z-stack slices acquired by confocal microscopy in 2D using the Maximum Intensity Projection (MIP) function of ImageJ.
- Growth dynamics from cell clones
NOTE: The following steps allow retrieval of geometrical and shape parameters for cells from 2D MIP images containing the clone(s) of interest. For comparisons between multiple clones, images must be acquired with the same settings.- Segment clone areas by drawing their contours with the ImageJ Freehand Selection tool.
- Calculate geometrical and shape parameters by using the Set Measurements tool of ImageJ under the Analyze menu. Activate the Area, Perimeter, Fit Ellipse, and Shape Descriptor options.
NOTE: This will allow retrieval of diverse geometrical parameters including the area (sum of the pixels within the clone), the perimeter (sum of the pixel of the clone border), the aspect ratio (AR, the ratio between the major and minor axes of the best-fit Legendre ellipse inscribed into the clone border), and the angle of orientation (i.e, the angle of the major axis of the clone relative to the anteroposterior boundary). - Calculating non-dimensional ratios from these measurements retrieves shape parameters. These include roundness (4 x [area]/π x [major axis]2), roughness (solidity – area/convex area), and circularity (4π x [area]/[perimeter]2).
NOTE: Each of the shape parameters represents the degree of deviation of the clones from ideal shapes such as a circle or a bounded convex hull, and they are all bounded between values of 0 and 1. Values equal to 1 indicate maximum symmetry (i.e., minimal complexity). - Statistically analyze geometrical and shape parameters between different genotypes or conditions using Microsoft Excel and/or PAST. Statistical significance of the difference is determined using an unpaired two-tailed Student's t-test for equal mean or the non-parametric Kolmogorov-Smirnov K-S-test for equal distributions between conditions. Data can be graphically displayed as desired (e.g., bar charts, box plots, etc.). See Figure 5.
Representative Results
The protocol described above covers the preparation of Drosophila pupae for long-term live imaging and the procedures for the analysis of cell orientation and growth dynamics of the abdominal epidermis. By applying this methodology it is possible to generate high-resolution movies of the developing pupae for periods of up to 48 h without significant photobleaching or phototoxicity. Snapshots depicting the abdominal epidermis (e.g., histoblasts and LECs) at different time points and from pupae oriented at different angles are shown in Figure 4. The subsequent analysis of these movies allows identification and quantification of the dynamics of local and global changes in the main geometrical and shape parameters modulated during the expansion of histoblasts and the replacement of LECs. These analyses can be performed in different scenarios and specific mutant backgrounds. They can also be employed for clones in which gene expression is altered, leading to autonomous loss or gain of function conditions. This would allow the exploration of nonautonomous responses in surrounding cells, facilitating the identification of cross talk or cell communication mechanisms (Figure 5). This approach has been recently employed in the identification of the Fat/Dachsous/Four jointed pathway as a key element directing and orienting cells alignment during the deploying of the planar polar pattern of the abdominal epidermis of the adult9.

Figure 1: Dissecting and mounting pupae for live imaging. (A) From left to right: dorsal view of a prepupa at 0 h APF (left) and of a pupa at 14 h APF before (middle) and after (right) the removal of the opaque pupal case. The opercular region is indicated (white arrowheads). (B) The essential toolkit for dissection is shown. From left to right: glass slide, double-sided sticky tape, a pair of forceps, and a glass-bottom dish. (C) Staged pupae are immobilized on double-sided sticky tape on glass slides dorsal side up. The pupal case of each pupa is opened from the opercular region with surgical forceps. (D−E) The peeling of the pupal case is gradual. The case is torn and folded to the pupal sides from the head to the abdomen. (F−H) The pupa is gently lifted from the ventral side with the tips of the forceps (G) and transferred to a glass-bottom dish over a minimal drop of halocarbon oil. (I) The pupa is then appropriately oriented (dorsolaterally, top; dorsally, bottom). Multiple pupae can be mounted simultaneously by repeating the steps shown in panels C−I. (J) Image showing the outline of the cells of the dorsal histoblast nest of the AIII segment expressing the junctional marker Atpα::GFP. The pupa was oriented dorsolaterally and imaged at 14 h APF. Scale bar = 22 µm. (K) Image equivalent to (J) but from a dorsal point of view. The dynamics of tissue development can be visualized for several hours. See also Figure 4. Please click here to view a larger version of this figure.
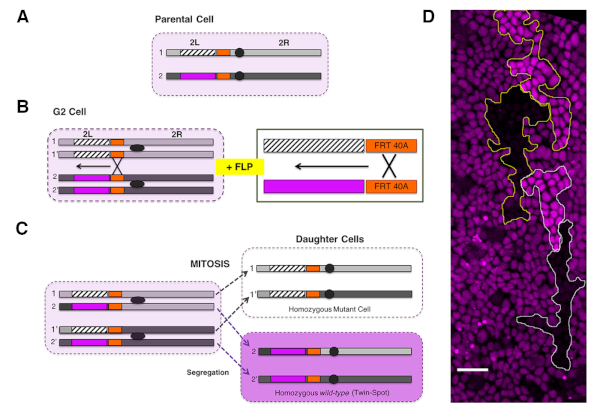
Figure 2: Generation of genetic mosaics via FLP/FRT system. (A) A parental heterozygous cell (pale magenta) carries a recessive mutant allele (dashed rectangle) on one chromosome arm and a gene encoding for a fluorescent marker (magenta) in the other chromosome arm [the left arm of the chromosome 2 (2L) in this example]. FRT sites (orange rectangles) are engineered in both arms at the same chromosomal positions proximal to the centromere (grey ovals). (B) The activation of FLP recombinase by heat shock in cells paused in G2 leads to the recombination between the FRTs of sister chromatids 1-1' and 2-2'. As a result, the region distal to the FRT sites (FRT40A on 2L) is exchanged. A magnified view is shown on the right panel. (C) Upon the polar segregation of the rearranged regions (1-1' and 2-2') during mitosis, two genetically different sister cells are generated. One daughter is homozygous for the recessive allele and for the entire chromosome arm distal to the site of recombination. This cell lacks the gene that encodes for the fluorescent marker, negatively marked in white. The other daughter cell will be homozygous for the wild type arm, giving rise to a twin-spot and expressing two copies of the genes encoding for the fluorescent marker (dark magenta). For simplicity, segregations of rearranged regions to opposite poles of the mitotic cell (i.e., 1−2 and 1'−2') are not shown. These give rise to cells phenotypically indistinguishable from the parental cell (heterozygous background, pale magenta). (D) Image showing clones in the A compartment generated via FLP/FRT somatic recombination at the third instar larvae stage and visualized at 26 h APF. Clones of cells are marked by the absence of RFP.nls (black) and homozygous expression of RFP.nls (bright magenta, twin spots) in an otherwise heterozygous RFP.nls background (dim magenta). Equivalent events can be achieved for FRT sites located in other chromosomal locations (2R, 3L, 3R, and X). See also Figure 5. Please click here to view a larger version of this figure.
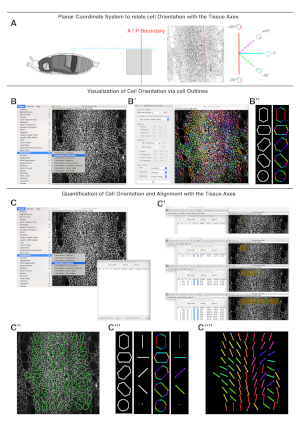
Figure 3: Illustration of cell orientation measurements. To extract information on cell orientations, first a planar coordinate system to relate cell orientation with tissue axis is set (A). Second, cell orientations are extracted from cell outlines (B). Last, cell orientations and alignments with the tissue axis are quantified (C). (A) On the left, a diagram of a lateral view of a pupa oriented according to the planar coordinate system. The red solid and the cyan dashed lines define the Cartesian plane representing the anteroposterior (A/P) boundary and the dorsal midline respectively. In the middle, an inverted image showing a dorsolateral view of the expanding histoblasts at 26 h APF outlined by the junctional marker Atpα::GFP (input image). The position of the A/P boundary is highlighted with a red line. On the right, an axial compass showing the color code applied to describe cell edge orientations (polygons) or averaged cell orientations (bars). (B) Display of the Plugins/OrientationJ menu and the OrientationJ Distribution window. (B') Display of the OrientationJ Distribution window showing the settings of the parameters employed to obtain the cell edge orientation map on the right. (B'') Illustration of the color-coded cell outlines on idealized cells. A circle does not show any preferred color. (C) Display of the OrientationJ Measure window. (C') Sequential screenshots of the OrientationJ Measure window showing how to measure local orientation and coherency in consecutive ROIs of uniform weight. The local orientation angle and coherency for each region are displayed as ellipsoids. (C'') Representation of the final results of the orientation measurement. Ellipsoids visually display the orientation (i.e., angle of the ellipsoid longest axis with respect to the A/P boundary) and coherency (i.e., ratio between the longest and the shorter axis of the ellipsoid). The numerical values of both parameters are saved in a spreadsheet for further analyses. (C''') Illustration of the color-coded cell outlines and averaged cell orientation (bars) on idealized cells. A circle does not show any preferred orientation. (C'''') Representation of the preferred local orientation of each ROI (locally averaged orientation map). The colors highlight the orientation of each region. Anterior is to the left. Scale bar = 22 μm. Please click here to view a larger version of this figure.
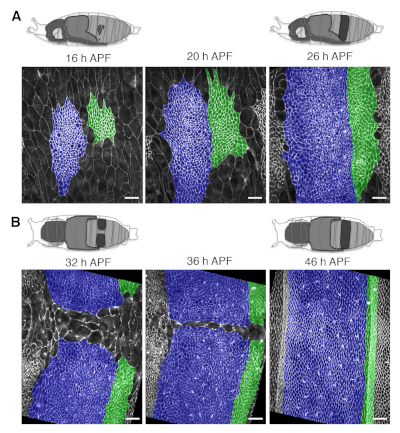
Figure 4: Long-term live imaging of growing abdominal epithelia. (A) Representative snapshots from long-term imaging movies from early to late phases of histoblast expansion. Top: Schematic views of a pupa oriented for dorsolateral imaging at 16 h and 26 h APF. The territory occupied by the nests visible from the dorsolateral side of the pupa is highlighted in dark grey at 16 and 26 h APF. Bottom: Images showing cell outlines from both histoblasts and LECs labeled by the ubiquitous expression of the junctional marker Atpα::GFP. The A/P boundary lies in between the two highlighted compartments (Fake colored cells in blue = anterior compartment; green = posterior compartments). (B) Representative snapshots from long-term imaging movies from early to late phases of nests confluence. Top: Schematic view of a pupa oriented for dorsal imaging at 32 h and 48 h APF. Bottom: Cell outlines (labeled and colored as in A). Note that the Atpα::GFP marker allows delineation of the shape of individual epithelial cells over time with high resolution. Scale bar = 22 μm. Please click here to view a larger version of this figure.
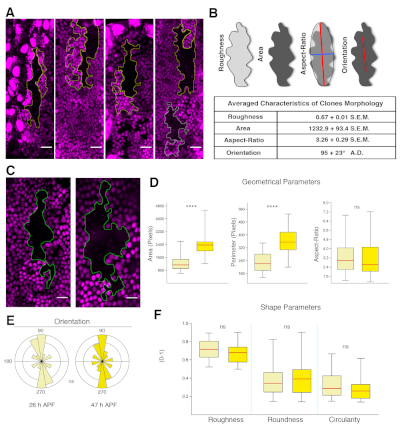
Figure 5: Tissue properties extracted from clonal analysis. (A) Examples of wild type clones in the A compartment marked by the absence of RFP.nls (black) and their twin spots (bright magenta) at 26 h APF. The clones elongate along the segmental boundaries. Twin clones arrange in parallel or in tandem. Scale bar = 22 μm. (B) Top: Images illustrating the parameters quantified from the clone outlines. Bottom: Summary table reporting the average values for the indicated parameters for wild type animals (n = 29). (C) Morphology of a wild type clone at 26 h (left) and 47 h (right) APF. The clone shows complex border morphology at both stages. (D) Box and whisker plots for geometrical parameters at 26 h (light yellow) and 47 h APF (dark yellow). The averaged area and perimeter increase significantly in this time window. (E) Polar plots representing the clones orientation (bin size 18°, abundance proportional to the area). Orientation is sustained during expansion and remodeling. (F) Box and whisker plots for shape parameters at 26 h (light yellow) and 47 h APF (dark yellow). Roughness (solidity), roundness, and circularity barely change. The median values are shown with a red horizontal line and whiskers extend to the minimum and maximum values of the distribution. Statistics was performed with K-SM test or W-tests (p < 0.0001****, p > 0.05 not significant). Anterior is to the left, dorsal is up. Scale bar = 16 μm. Genotype is hsflp1.22;FRT40A/FRT40A Ubi.RFP.nls. Please click here to view a larger version of this figure.
Discussion
Long-range order is an essential characteristic of most functional physiological units. During morphogenesis, order is achieved through the integration of complex instructions implemented with high temporal and spatial precision. Multiple and multilevel constrains are integrated into stereotyped tissue arrangements.
Polarity and directionality are critical to ordered spatial arrangement during development. Polarity implies symmetry breaking during development. The achievement of asymmetry is necessary for the determination of the embryonic anteroposterior (A/P) and dorsoventral (D/V) axes and adult organization26. Beyond this early role, local asymmetries are essential for morphological diversity at all levels. Directionality is an essential complement of asymmetry and polarity during morphogenesis. Global order is implemented by the ability of cells to sense and transmit signals locally on a cell-to-cell base with a precise sense or orientation. Single cells asymmetries harmonize cooperatively over time by orienting positions or movements within particular directions in space. Cell communication implicates secreted factors, cell-cell contacts, and mechanical inputs. Signals act on fields of cells that first locally and then globally modify their behaviors27.
This work presents a simple way to analyze the coordinated behaviors of individual cells, including their planar orientation and growth parameters during the development of tissue order. The morphogenesis of the adult abdomen of Drosophila during pupation presents a series of technical advantages over other equivalent models such as germ band extension/ retraction28 or dorsal closure29 during fly embryogenesis, Drosophila wing morphogenesis ex vivo30, ventral enclosure in Caenorhabditis elegans31, or palatal fusion in mice32, amongst others.
First, abdomen morphogenesis constitutes a process that can be followed in its entirety in vivo. Continuous live imaging of the replacement of the LECs by the histoblasts can be performed from 12 h APF, when the pupa is formed, up to the completion of adult epithelial morphogenesis. Second, it allows the analyses of a full set of cell behaviors such as cell migration, division, shape changes, delamination, and intercalation. Last, it is amenable to genetic interference and clonal analysis. Autonomous and nonautonomous effects of loss and gain of function activities can be monitored live by employing appropriate markers. However, the pupa as a model system presents some minor limitations. Due to its ovoid shape, it is not possible to perform simultaneous live imaging of lateral and dorsal events with high resolution. This issue can be overcome by performing sequential imaging in dorsolaterally and dorsally oriented pupae or better, by employing multiple angles light sheet selective plane illumination microscopy (SPIM) microscopy. Another drawback is the presence of two different cell populations of different sizes in the analyses (i.e., polyploid LECs and diploid histoblasts). This can make image segmentation complex and the two cell populations must be analyzed separately.
In the future, the long-term imaging of Drosophila pupae can be easily adapted to study a full range of morphogenetic phenomena including the coordination of epidermal, muscular, and neural development during metamorphosis in wild type and mutant conditions. Moreover, the algorithms and plugins in use may be employed to study the organization, patterning, and dynamics of many other tissues.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank members of the Martín-Blanco laboratory for helpful discussions. We also thank Nic Tapon (The Crick Institute, London, UK), the Bloomington Stock Center (University of Indiana, USA) and FlyBase (for Drosophila gene annotation). Federica Mangione was supported by a JAE-CSIC predoctoral fellowship. The Martín-Blanco laboratory was funded from the Programa Estatal de Fomento de la Investigación Científica y Técnica de Excelencia (BFU2014-57019-P and BFU2017-82876-P) and from the Fundación Ramón Areces.
Materials
| Analysis Software | – | ImageJ | Analyzing data |
| Drosophila | Atpa::GFP | – | Strains employed for data collection |
| Drosophila | hsflp1.22;FRT40A/FRT40A Ubi.RFP.nls | – | Strains employed for data collection |
| Dumont 5 Forceps | FST | 11251-20 | 1.5 mm diameter for dissection |
| Glass Bottom Plates | Mat Tek | P35G-0.170-14-C | Mounting pupae for data collection |
| Halocarbon Oil 27 | Sigma-Aldrich | 9002-83-9 | mounting pupae |
| Inverted Confocal microscope | Zeiss | LSM700 | Data collection |
| Stereomicroscope | Leica | DFC365FX | Visualization of the pupae during dissection |
References
- Gillespie, P. G., Muller, U. Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell. 139, 33-44 (2009).
- Deans, M. R. A balance of form and function: planar polarity and development of the vestibular maculae. Seminars in Cellular and Developmental Biology. 24, 490-498 (2013).
- Stell, W. K. The structure and morphologic relations of rods and cones in the retina of the spiny dogfish, Squalus. Comparative Biochemistry and Physiology – Part A: Comparative Physiology. 42, 141-151 (1972).
- Ninov, N., Chiarelli, D. A., Martin-Blanco, E. Extrinsic and intrinsic mechanisms directing epithelial cell sheet replacement during Drosophila metamorphosis. Development. 134, 367-379 (2007).
- Bosveld, F., et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science. 336, 724-727 (2012).
- Puah, W. C., Wasser, M. Live imaging of muscles in Drosophila metamorphosis: Towards high-throughput gene identification and function analysis. Methods. 96, 103-117 (2016).
- Teng, X., Qin, L., Le Borgne, R., Toyama, Y. Remodeling of adhesion and modulation of mechanical tensile forces during apoptosis in Drosophila epithelium. Development. 144, 95-105 (2017).
- Weavers, H., et al. Systems Analysis of the Dynamic Inflammatory Response to Tissue Damage Reveals Spatiotemporal Properties of the Wound Attractant Gradient. Current Biology. 26, 1975-1989 (2016).
- Mangione, F., Martin-Blanco, E. The Dachsous/Fat/Four-Jointed Pathway Directs the Uniform Axial Orientation of Epithelial Cells in the Drosophila Abdomen. Cell Reports. 25, 2836-2850 (2018).
- Casal, J., Struhl, G., Lawrence, P. A. Developmental compartments and planar polarity in Drosophila. Current Biology. 12, 1189-1198 (2002).
- Wootton, R. How flies fly. Nature. 400, 112-113 (1999).
- Zallen, J. A., Wieschaus, E. Patterned gene expression directs bipolar planar polarity in Drosophila. Developmental Cell. 6, 343-355 (2004).
- Gibson, M. C., Patel, A. B., Nagpal, R., Perrimon, N. The emergence of geometric order in proliferating metazoan epithelia. Nature. 442, 1038-1041 (2006).
- Robertson, C. W. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. Journal of Morphology. 59, 351-399 (1936).
- Mandaravally Madhavan, M., Schneiderman, H. A. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux’s archives of Developmental Biology. 183, 269-305 (1977).
- Kornberg, T. Compartments in the abdomen of Drosophila and the role of the engrailed locus. Biologie du développement. 86, 363-372 (1981).
- Garcia-Bellido, A., Merriam, J. R. Clonal parameters of tergite development in Drosophila. Biologie du développement. 26, 264-276 (1971).
- Roseland, C. R., Schneiderman, H. A. Regulation and metamorphosis of the abdominal histoblasts of Drosophila melanogaster. Wilhelm Roux’s archives of Developmental Biology. 186, 235-265 (1979).
- Madhavan, M. M., Madhavan, K. Morphogenesis of the epidermis of adult abdomen of Drosophila. Journal of Embryology and Experimental Morphology. 60, 1-31 (1980).
- Bischoff, M., Cseresnyes, Z. Cell rearrangements, cell divisions and cell death in a migrating epithelial sheet in the abdomen of Drosophila. Development. 136, 2403-2411 (2009).
- Golic, K. G., Lindquist, S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 59, 499-509 (1989).
- Xu, T., Rubin, G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117, 1223-1237 (1993).
- Fonck, E., et al. Effect of aging on elastin functionality in human cerebral arteries. Stroke. 40, 2552-2556 (2009).
- Rezakhaniha, R., Fonck, E., Genoud, C., Stergiopulos, N. Role of elastin anisotropy in structural strain energy functions of arterial tissue. Biomechanics and Modeling in Mechanobiology. 10, 599-611 (2011).
- Hammer, &. #. 2. 1. 6. ;., Harper, D. A., Ryan, P. D. PAST: paleontological statistics software package for education and data analysis. Palaeontologia electronica. 4, 1-9 (2001).
- Gray, R. S., Roszko, I., Solnica-Krezel, L. Planar cell polarity: coordinating morphogenetic cell behaviors with embryonic polarity. Developmental Cell. 21, 120-133 (2011).
- Vogg, M. C., Wenger, Y., Galliot, B. How Somatic Adult Tissues Develop Organizer Activity. Current Topics in Developmental Biology. 116, 391-414 (2016).
- Collinet, C., Rauzi, M., Lenne, P. F., Lecuit, T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nature Cell Biology. 17, 1247-1258 (2015).
- Martin-Blanco, E., et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes and Development. 12, 557-570 (1998).
- Dye, N. A., et al. Cell dynamics underlying oriented growth of the Drosophila wing imaginal disc. Development. 144, 4406-4421 (2017).
- Williams-Masson, E. M., Malik, A. N., Hardin, J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 124, 2889-2901 (1997).
- Ferguson, M. W. Palate development. Development. 103, 41-60 (1988).

