The Optical Fractionator Technique to Estimate Cell Numbers in a Rat Model of Electroconvulsive Therapy
Summary
Here, we present a stereological method, the optical fractionator, used to quantify the formation of new neurons, and their survival, in the rat hippocampus following electroconvulsive stimulation. When correctly implemented, the sensitivity and efficiency of stereological methods ensures accurate estimates with a fixed and predetermined precision.
Abstract
Stereological methods are designed to describe quantitative parameters without making assumptions about size, shape, orientation and distribution of cells or structures. These methods have been revolutionary for quantitative analysis of the mammalian brain, in which volumetric cell populations are too high to count manually, and stereology is now the technique of choice whenever estimates of three-dimensional quantities need to be extracted from measurements on two-dimensional sections. All stereological methods are in principle unbiased; however, they rely on proper knowledge about the structure of interest and the characteristics of the tissue. Stereology is based on Systematic Uniformly Random Sampling (SURS), with adjustment of sampling to the most efficient level in respect to precision, providing reliable, quantitative information about the whole structure of interest. Here we present the optical fractionator in conjunction with BrdU immunohistochemistry to estimate the production and survival of newly-formed neurons in the granule cell layer (including the sub-granular zone) of the rat hippocampus following electroconvulsive stimulation, which is among the most potent stimulators of neurogenesis. The optical fractionator technique is designed to provide estimates of the total number of cells from thick sections sampled from the full structure. Thick sections provide the opportunity to observe cells in their full 3-D extent and thus, allow for easy and robust cell classification based on morphological criteria. When correctly implemented, the sensitivity and efficiency of the optical fractionator provides accurate estimates with a fixed and predetermined precision.
Introduction
Quantification of cells in a three-dimensional (3-D) structure may require obtaining estimates based on unbiased principles. Even today, studies frequently use two-dimensional (2-D) methods to report data of 3-D structures. However, these methods do not consider the heterogeneous nature of the structure in terms of shape and distribution of cells resulting in methodical limitations; they make assumptions about the 3-D structure of interest and the results do not refer to the complete structure. These limitations reduce the sensitivity and accuracy of the methods as well as increase the risk of errors1.
Bias in cell counting can be avoided by the application of design-based stereology, which is of general importance for obtaining quantitative information about the number of cells in a given tissue or structure. While stereology has formerly been a very time-consuming method, advances in procedures, software and imaging, has made it become much more efficient and widely applicable. Stereology entails statistical sampling principles and stochastic geometric theory to provide efficient tools for estimation of volume, surface area, length, and number of objects in a 3-D structure by sampling in 2-D sections2. Thus, stereology allows one to obtain unbiased quantitative data on structural changes in tissue sections, which give average estimations of the true numbers, without exhaustive analysis1. Stereology is defined as the statistical inference of geometric parameters from sampled information. This can be achieved by unbiased sampling, that is to say systematic random sampling, of sections as well as counting rules that ensure that all objects have equal probabilities of being counted3. While stereological results can have varying degrees of precision as indicated by the coefficient of error (CE), this can be adjusted to suit the inherent biological variance (coefficient of variance (CV)) by adjusting the amount of sampling as required4,5. A few previous stereological studies have examined the short-term effects of electroconvulsive stimulation (ECS) on hippocampal plasticity in rodents6,7. Data from these studies show an initial increase in the total number of neurons as well as elevated synaptic differentiation following repeated ECS. However, these studies do not estimate neurogenesis directly, as they do not use specific markers for cell proliferation.
Here we present an application of a stereological method, the optical fractionator technique8,9, for quantifying the total number of newly-formed neurons in the rat hippocampal granule cell layer (GCL), including the sub-granular zone (SGZ) as well as their long-term survival10,11. We subjected rats to a clinical relevant schedule of ECS (three times a week for 3 weeks), which is one of the most potent stimulators of neurogenesis12. Control animals were treated using the same procedure, but without passing of current. On each of the 21 days of experimentation, all rats were injected intraperitoneally with 5-bromo-2`-deoxyuridine (BrdU) that incorporates into DNA in place of thymidine during cell division13. Following experimentation, the rats were maintained for four different time periods (24 hours, 3 months, 6 months and 12 months). Using the fractionator principle, we obtained a known fraction of the hippocampus through uniform random sampling of sections, and applied optical disectors (3-D probes) to the sampled and immunostained thick tissue sections. It is especially noted that frozen sections usually collapse in the z-axis during processing, and that optical disectors based upon the number-weighted mean section thickness should be used in this particular case14. As the focal plane of the optical disectors was moved a known distance down through the section, BrdU-positive neurons were counted when their feature of interest was clearly recognized. In the field of stereology there is some debate about the total number of particles that should be counted15; typically 150-200 neurons are counted in the structure of interest to obtain adequate precision i.e. a coefficient of 7-8%8. The BrdU-positive neuronal counts were completed using stereological software with imaging by a standard microscope equipped with camera and the following objectives: 2X, 4X and 10X as well as a 100X oil-immersion objective (numerical aperture = 1.40), and a motorized stage. Movements in the z-direction were measured with a digital microcator. The final magnification was 3000X.
Protocol
All the animal procedures were performed in accordance with guidelines from the Danish Animal Experimentation Inspectorate and approved by the local ethical committee for Experimental Animals.
1. Tissue processing
NOTE: Following transcardial perfusion and post fixation in 4% paraformaldehyde (PFA) diluted in 0.2 M phosphate buffer overnight at 4 °C, brains are transferred to 30% sucrose in phosphate buffered saline (PBS) for three days at 4 °C. Thereafter, the brains are frozen on powdered dry ice and stored at -80 °C until sectioning.
- Separate the right and left hemispheres along the midline of the brain using a scalpel and then choose one or the other hemisphere randomly in the first brain, thereafter shift systematically between right and left hemisphere (Figure 1A). Mount the sampled hemisphere onto a specimen disc with the long axis perpendicular to the disc using a mounting medium.
- Place numbered multidish containers for pre-cooling in the cryostat.
- Mount the specimen disc into the cryostat and cut the entirety of the hippocampus (Bregma 1.80 to - 7.0416) into 80 µm thick sections in the coronal plane (Figure 1B).
- Place all sampled sections consecutively in the cooled multidish containers, to ensure that the sections are kept in cutting-order.
- Cover the sections completely with cryoprotectant and hold the containers at -20 °C until further use.
2. Immunostaining
NOTE: To keep track of the individual sections throughout the staining procedure, place the sampled sections in 25-well staining nets. Use every 5th section, giving a mean final number of 12 (8-16) sections per hippocampus for immunostaining and subsequent cell counting.
- Prior to subsampling every nth section with a random start between section one and section n (Figure 1C) transfer the containers from -20 °C to room temperature (RT).
- Use e.g. a paintbrush, to transfer the sections in numerical order to the 25-well staining nets placed in petri dishes filled with PBS.
NOTE: From this point, keep the sections in the staining nets which are placed in matching glass dishes placed on an orbital shaker (section 2.3 to 2.9). - Transfer the staining nets to the matching glass dishes and wash the sections twice for 10 min in PBS at RT before incubation in 3% hydrogen peroxide (H2O2) diluted in distilled water (dH2O) for 20 min.
- Transfer the sections to three different solutions of hydrochloric acid (HCl) (1 M at 0 °C for 10 min, 2 M at RT for 10 min, and 2 M at 37 °C for 20 min) before neutralization in 0.1 M sodium tetraborate at RT for 20 min.
- After 3 × 10 min incubations in 1% Triton X-100 diluted in PBS (washing buffer), transfer the sections to washing buffer containing 10% fetal bovine serum (blocking solution) for 1 h at RT.
- Incubate the sections for 48 h at 4 °C in mouse-anti-BrdU diluted 1:100 in blocking solution.
- Wash the sections 3 × 10 min in washing buffer, and incubate for 48 h in horse radish peroxidase diluted 1:10 in washing buffer.
- Transfer the sections to PBS for 5 × 10 min, and then for 7 min in 0.01% diaminobenzidine (DAB) diluted in PBS before they are transferred to a similar DAB solution containing 0.02% H2O2 for 10 min at RT.
- Complete a series of washes (2 × 10 min in PBS and 2 × 10 min in phosphate buffer without added sodium chloride) and mount the sections in numerical order on microscope slides.
- Dry the sections for approximately 30 min before counterstaining with cresyl violet.
- Place the microscope slides in a slide rack.
- Rehydrate the sections for 10 min in slide staining dishes containing dH2O, and then transfer the slides to 0.02% cresyl violet in dH2O for 15 min.
- Repeat step 2.12 before the sections are dehydrated three times in ethanol (96% for 5 min and 99% for 2 × 2 min).
- Place the sections in xylene for 2 × 15 min and cover-slip the slides using a rapid drying medium for mounting.
- Finally, leave the cover-slipped slides for approximately 24 h before they can be used for microscopy.
3. Estimation of the total number of BrdU-labelled neurons using the optical fractionator
NOTE: Conduct a pilot study including a few animals to determine the optimal sampling parameters, such as the number of sections to be analyzed and the number of optical disectors within the sampled sections. This pilot also provides a preliminary CV (standard deviation/mean) and the possibility to adjust the CE (see section 3.9.3) to obtain a satisfactory precision of the estimates determined by the investigator (for more details see below). Likewise perform a z-distribution analysis to address the following points: shrinkage of the tissue, quality of the staining throughout the section and distribution of the cells in the z-axis14.
- Check the thickness of the tissue in the sections to confirm that they are suitable for the use of the optical fractionator, e.g. is thick enough for the chosen disector height including guard zones (see below). NOTE: The tissue thickness is measures in several places within the region of interest.
- Place the slides on the motorized stage of the microscope and turn on the preferred stereological software.
- Delineate the area of interest using a low-magnification objective (2X or 4X) before changing to a 100X oil-immersion objective (see Figure 2A).
- Using a specific point of a counting frame (e.g. at or adjacent to a corner), locate the top of the section by moving along the focal plane until some feature of the section appears in focus. Register this z-position as 0 (see Figure 2B).
- Move the focal plane down through the tissue until the same specific point of the counting frame is at the last z-level of tissue in focus and mark this position. The local tissue thickness is defined from 0 to this endpoint and can be read on the z-axis. Register the tissue thickness (see Figure 2B).
- Document full penetration of the stain and distribution of the cells in the full thickness of the section.
NOTE: The distribution of BrdU-labelled neurons is used as a guide for determining the required guard zones. Although a uniform distribution of cells is critical, it is acceptable to observe fewer cells near the top and bottom of the section, indicating the effect of lost caps (for further details see Dorph-Petersen et al., 2009).- Sample BrdU-labelled neurons and register their z-position together with the local section thickness measured in the selected corner of each counting frame.
- Plot the number of BrdU-positive neurons as a function of their z-position.
- Fix the height of the disector and guard zones based on the mean section thickness and the distribution of cells (see 3.1 and 3.2).
NOTE: The height of the disector should be less than the section thickness to avoid artifacts close to the surfaces. This risk is circumvented by including guard zones to the top and bottom of the section. - Determine the step length between the disectors.
- Delineate the region of interest on all the sections to be analyzed and register the areas.
- Sum these areas and then divide by the number of disectors to be placed within this area.
NOTE: Based on previous experience a good starting point entails 75 probes. This will provide an approximation of the area and the corresponding sampling positions (Astep) (for further details see West, 2012). - Take the square root of Astep, which will provide the x-y-step size.
- Determine the size of the counting frame.
- Set the size of the counting frame to an arbitrary unit and perform a pilot sampling of BrdU-positive neurons using the parameters obtained in section 3.3 and 3.4.
NOTE: The size has to be determined empirically by trial and error, but it is recommended that it is adjusted to enable sampling of 2-3 cells per disector8.
- Set the size of the counting frame to an arbitrary unit and perform a pilot sampling of BrdU-positive neurons using the parameters obtained in section 3.3 and 3.4.
- Following this final step of the pilot study begin cell sampling.
NOTE: The obtained sampling parameters should result in counts of approximately 150-200 cells in each animal which is sufficient to obtain an efficient stereological design8. - Count the BrdU-positive neurons using a 100× oil-immersion objective and a final magnification of 2000-3000×. In each optical disector, identify any BrdU-labelled neurons that are clearly recognized by the feature of interest (in the present study, the criterion is when the leading edge of the BrdU-labelled neuron comes into focus for the first time), are located inside the counting frame or touch the inclusion lines (Figure 2B). Do not count the BrdU-positive neurons that, when clearly recognized by the feature of interest inside the disector height, touch the exclusion lines. It is critically important to follow scrupulously these counting rules throughout the entire stereological quantification.
NOTE: As BrdU incorporates into all dividing cells, morphology is used to distinguish between neurons and other types of cells. Neuron characteristics are a clearly defined nucleus with a neutral cytoplasm and a dark nucleolus. BrdU-positive cells are recognized as newly formed neurons based on their relatively larger size compared to glial cells. It should be mentioned that in studies of short-term proliferation (e.g. < 24 h) double-staining using immature neuronal markers may be a prerequisite17. - Estimate the total number of BrdU-positive neurons in each brain by multiplying the total number of cells counted (ΣQ–) with the reciprocal sampling fractions:
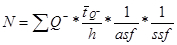
for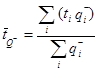
where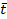 Q- is the number-weighted mean section thickness, h is the height of the disector, asf is the area sampling fraction, ssf is the section sampling fraction and ti is the section thickness in the ith counting frame with a cell count of
Q- is the number-weighted mean section thickness, h is the height of the disector, asf is the area sampling fraction, ssf is the section sampling fraction and ti is the section thickness in the ith counting frame with a cell count of 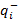 in the disector14,18.
in the disector14,18. - Calculate the coefficient of error (CE)8 which is the sampling variance of BrdU-positive neurons in systematic uniformly random sampling (SURS).
- First, calculate the noise, which is equal to the total number of BrdU-positive neurons counted:
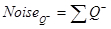
- Next, calculate the variance in SURS:
NOTE: We have judged the area and cell counts in the hippocampus to change smoothly from section to section and consequently used "smoothness class" m = 1 (1/240)8,19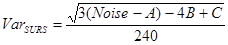
where
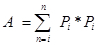
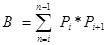
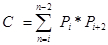
Pi is the number of points counted for the object in the ith section and n is the number of sections4,20. - Finally, calculate the CE of the estimate:
NOTE: In general, optimal precision is typically achieved when the CE value is less than half of the observed CV, as OCV2 = ICV2 + CE2, where OCV is the observed CV and ICV is the inherent CV4.
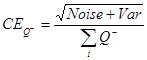
- First, calculate the noise, which is equal to the total number of BrdU-positive neurons counted:
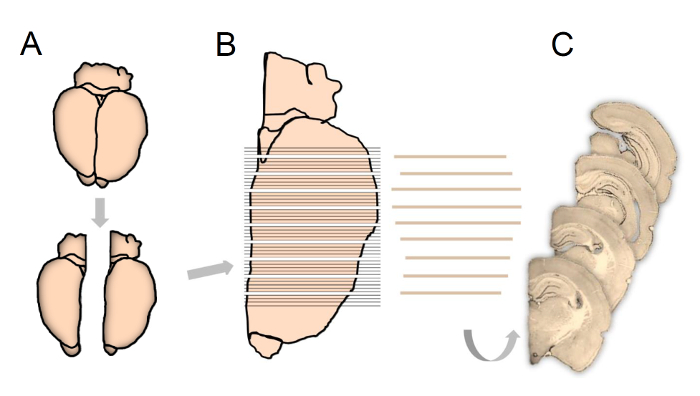
Figure 1: Schematic illustration showing tissue processing according to the fractionator principles used in the present study.
Following experimentation, the hemispheres are separated and right or left hemisphere chosen systematically and randomly (A). The complete brain extending over the entire hippocampus is cut into 80-µm-thick coronal sections (B), whereupon every 5th section is sub-sampled, starting randomly between section 1 to 5, for final immunostaining and stereological quantification (C). Please click here to view a larger version of this figure.
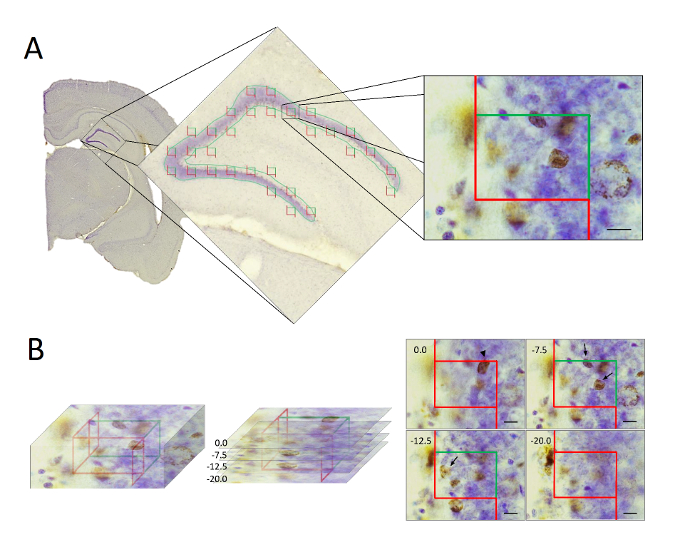
Figure 2: Illustration showing the stereological sampling procedure using optical disectors.
(A) Following histological processing the hippocampal GCL/SGZ was delineate and optical disectors applied uniformly and randomly over the region. (B) In the optical disectors (left), the BrdU-positive neurons were counted only if their feature of interest were clearly recognized within the disector height, and located inside the counting frame or touching the inclusion line (right). Neurons touching the exclusion line were not counted. Scale bars = 10 µm. Please click here to view a larger version of this figure.
Representative Results
Sampling parameters
By analyzing the z-distribution in a pilot study (Figure 3), we found a uniform distribution of BrdU-positive neurons in the disector height. Shrinkage of the sections was measured; final mean thickness was 26.4 μm (19.0-32.0 μm) in sections originally cut at 80 μm. Based on the tissue thickness and distribution of BrdU labelled neurons the disector height was set to 10 μm, and the guard zones at the top and bottom of the section was set to 5-μm and 4-17 μm respectively. Depending of the density of the cells, the area of the counting frames was 1414 or 5210 μm2. The step length was 220 μm in both the x and y directions and cell counting was performed in an average of 12 (range 8-16) sections per animal. These sampling parameters resulted in a mean of 133 (range 33-372) BrdU-positive cells counted in 173 (107-204) disectors in each brain hemisphere.
As an example, we obtained the following CV and CE values for BrdU-positive neuronal estimations in ECS treated rats: 24 hours: CV = 0.59, CE = 0.11; 3 months: CV = 0.34, CE = 0.12; 6 months: CV = 0.43, CE = 0.09; 12 months: CV = 0.22, CE = 0.09.
We quantified the total number of newly-formed neurons and their long-term survival in the rat hippocampus following ECS using the optical fractionator in conjunction with a BrdU staining technique10,11. The final results showed a basal neurogenesis in the four groups of control animals, which did not significantly differ as a function of survival time (Figure 3). Furthermore, ECS induced a significant increase in the total number of BrdU-positive neurons at 24 hours (259%, p<0.0001), 3 (229%, p<0.001), 6 (152%, p<0.05) and 12 months (162%, p<0.05) following experimentation, as compared to controls animals.
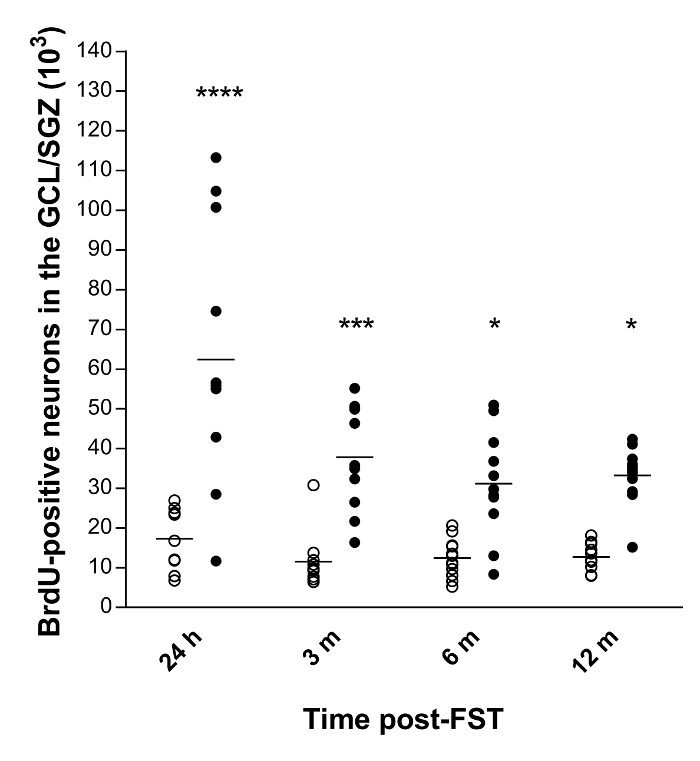
Figure 3: Quantitative data of the total number of BrdU-positive neurons in the GCL/SGZ of the rat hippocampus10,11.
Horizontal bars represent the mean values. Abbreviations: GCL, granule cell layer; SGZ, sub-granular zone; h, hours; m, months. ****p<0.0001, ***p<0.001 and *p<0.05 versus control (n=11-12 per group). Please click here to view a larger version of this figure.
Discussion
Using ECS as a methodology to induce neurogenesis, we find an immediate increase of 260% in the formation of new BrdU-positive neurons in the hippocampus. In this pool of acutely generated neurons we found 40% attrition from day 1 to three months, with nearly 50% of the newly formed neurons surviving at least 12 months following treatment10,11. The counting of BrdU-labelled neurons followed a strict sampling scheme, whereby the entire hippocampus was cut into 80-µm-thick sections followed by sub-sampling of every 5th section with a random sampling start between section one and section five. Providing these sections are chosen in a systematic random manner, this is demonstrably an excellent method to reduce the variance of the end result, without exhaustive counting1. This sampling scheme allowed us to count BrdU-positive neurons in an average of 12 (8-16) hippocampal sections in each rat brain, with a final precision of 9-11%.
When using the fractionator method, it is essential to know the fraction of the section height in which counting is performed, since tissue shrinkage and deformation frequently occurs during histological processing14. Indeed, we saw substantial shrinkage of thickness in this study. Furthermore, it should be noted that differential tissue shrinkage may occur, as presented in Dorph-Petersen et al. 2001. However, as long as the complete structure of interest is available for analysis, these limitations do not result in a bias of the total numbers of particles, i.e. BrdU-positive neurons. In studies where tissue shrinkage is a particular issue, it should always be stated that the results are obtained in deformed tissue. In this study, we counted in a disector height of 10 µm. The sections of the present study had a final mean thickness of 26 µm, a 5-µm guard zone at the top and a 11-µm (mean) guard zone at the bottom of the section. These parameters are acceptable since guard zones should be approximately the diameter of the sampled particles.
Following delineation of the GCL/SGZ, disectors were uniformly randomly placed within the delineated area. The disectors should be positioned with a fixed step length that optimizes sampling and counting to efficiently obtain estimates with a precision determined by the investigator. Optimal precision is typically achieved by counting up to 150-200 cells in each structure of interest5. In the present study, we counted a mean of 133 (range 33-372) BrdU-positive neurons in each rat hippocampus. Due to a low cell numbers in some of the control animals, our counts of BrdU-positive neurons fell below the generally acceptable number of cells required to obtain a suitable precision, which resulted in relatively high CE values for those cases. However, as we obtained CE-values of less than half of the CV-values (see representative result section) additional sampling and counting was not required. The high CV-values show that the greatest contributor to the observed variation originates from the biological variation. Indeed, we might assert that the average number of BrdU-positive neurons counted in the present study was higher than necessary. For example, in one group of rats we achieved a CE-value of 9%, and observed a CV-value of 43%. In this particular case, we could have aimed for a precision of approximately 20%. In summary, the precision of the total number of BrdU-positive neurons in the present study is sufficient in that it captures real treatment effects on the true numbers of particles. Due to the rather large biological variation in certain groups of animals the same estimates could have been obtained with an acceptable precision, despite less expenditure of effort. High biological variances within groups can only be compensated by increasing the number of animals.
One might argue that the gold standard for obtaining exact cell numbers entails exhaustive counting of all the objects of interest. However, in most studies of the brain this is not a possibility due to the vast numbers of cells. Although much more efficient than exhaustive cell counting, the optional fractionator is relatively time consuming in comparison to simple screening of differences in cell numbers which is preferred if the accurate number of cells is not essential. It is our experience that differences of more than 20-30% can be detected purely by screening procedures.
If performed correctly, sampling using design-based stereology provides unbiased and precise estimates in an efficient manner1. The unbiased property of stereology produces estimates that, when replicated, approximate the true population mean, and enhance the reproducibility of the estimate21. Initially, a representative sample of the entire structure of interest (in this study the hippocampal GCL/SGZ) must be obtained, allowing for estimation in a statistically valid subset of sections4. To obtain an appropriate number of sections and counting probes we apply SURS, which reduces the variance compared to random sampling8. Furthermore, SURS ensures that the particles of interest within the structure are sampled with the same probabilities, independent of their size, shape, orientation and distribution in the structure. As such stereology is to be highly recommended when obtaining precise and unbiased estimates is a central aspect of the project.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Susanne Sørensen for the skillful technical assistance. This work was supported by grants from The Velux Foundation; The Jascha Foundation; Aase and Ejnar Danielsens Foundation; Hartmann Brothers Foundation; Director Jacob Madsen and wife Olga Madsens Foundation; Doctor Sofus Carl Emil Friis and wife Doris Friis' Trust; The Foundation of 1870; Torben and Alice Frimodts Foundation and The Foundation for Neurological Research. Manuscript editing was performed by Inglewood Biomedical Editing.
Materials
| Materials | |||
| 5'-bromo-2'deoxyuridine – BrdU | Sigma-Aldrich | 19-160 | |
| Cresyl Violet | Sigma-Aldrich | C5042 | Contains the following materials for 1 L: 0.2 g cresyl violet; 2.0 g sodiumacetate, 3 H2O; 3 ml glacial acetic acid; 1000 ml destilled water |
| Cryoprotectant | Capital Region Pharmacy, DK | 861334 | Contains the following materials: Sucrose; polyvinylpyrrolidon PVP 40; sodiumphosphatebuffer (0.2 M); ethylenglycol; demineralized water. |
| dH2O | Capital Region Pharmacy, DK | 817205 | |
| Diaminobenzidine – DAB | Sigma-Aldrich | D5637 | |
| Envision-HRP | DAKO | K4001 | Peroxidase labeled polymer conjugated to goat anti-mouse immunoglobulins |
| Ethanol – 70 % | Plum A/S | 201098 | |
| Ethanol – 96 % | Plum A/S | 201104 | |
| Ethanol – 99.9 % | Plum A/S | 201111 | |
| Fetal Bovine Serum | In Vitro | BI-04-003-1A | |
| H2O2 30% | Capital Region Pharmacy, DK | 896456 | |
| HCl | J. T. Baker | 6081 | |
| Mounting medium | Sakura | 4583 | Tissue Tek |
| Mouse-anti-BrdU | Becton-Dickinson | 347580 | |
| PBS buffer | Capital Region Pharmacy, DK | 853436 | pH = 7.4 |
| PFA | VWR | 28,794,295 | pH = 7.4 |
| Phosphate buffer | Capital Region Pharmacy, DK | 866533 | 0.1 mol/l, pH 7.4 |
| Rapid drying mounting medium | Histolab Products AB | 801 | Pertex |
| Sodium Tetraborate | Merck A/S | 1,063,080,500 | |
| Sucrose | Merck A/S | 1,076,515,000 | |
| Triton X-100 | Merck A/S | 1,086,031,000 | |
| Xylene | VWR | 28,973,363 | |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| Cover slips | Menzel-Gläser | 24×50 mm #0 | |
| Cryostat | Leica | CM1860 | |
| Digital Microcator | Heidenhain | VRZ 401 | |
| Glass dishes | Brain Resaerch Laboratories | 1256 | |
| Microscope | Nikon | Eclipse 80i | |
| Microscope camera | Olympus | DP72 | |
| Microscope slides | VWR | 48311-703 | SuperFrost+ |
| Motorized stage | Prior | H101A | |
| Multidish Containers | Sigma-Aldrich | D6315-1CS | Nuclon 12-wells |
| New-Cast software | Visiopharm | ver. 4.5.6.440 | |
| Objective 2x | Nikon | CFI Plan UW 2X | |
| Objective 4x | Nikon | CFI Plan Fluor 4X | |
| Objective 10x | Nikon | CFI Plan Fluor 10X | |
| Objective 100x oil immersion | Nikon | CFI Plan Apo Lambda DM 100X Oil | Numerical aperture 1.40 |
| Orbital shaker | Gemini BV | CM-9 | Sarstedt |
| Slide rack | Sakura | 4465 | |
| Slide staining dishes | Sakura | 4457 | Tissue-Tek |
| Specimen disc | Leica | 14037008637 | |
| Staining nets | Brain Research Laboratories | 2512 | 25-wells |
References
- Boyce, R. W., Dorph-Petersen, K. A., Lyck, L., Gundersen, H. J. Design-based stereology: Introduction to basic concepts and practical approaches for estimation of cell number. Toxicol. Pathol. 38 (7), 1011-1025 (2010).
- West, M. J. . Introduction to stereology. 2012 (8), 843-851 (2012).
- West, M. J. Stereological methods for estimating the total number of neurons and synapses: issues of precision and bias. Trends Neurosci. 22 (2), 51-61 (1999).
- Gundersen, H. J., Jensen, E. B. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 147 (Pt 3), 229-263 (1987).
- West, M. J., Slomianka, L., Gundersen, H. J. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat. Rec. 231 (4), 482-497 (1991).
- Kaae, S. S., Chen, F., Wegener, G., Madsen, T. M., Nyengaard, J. R. Quantitative hippocampal structural changes following electroconvulsive seizure treatment in a rat model of depression. Synapse. 66 (8), 667-676 (2012).
- Chen, F., Madsen, T. M., Wegener, G., Nyengaard, J. R. Repeated electroconvulsive seizures increase the total number of synapses in adult male rat hippocampus. Eur Neuropsychopharmacol. 19 (5), 329-338 (2009).
- Gundersen, H. J., Jensen, E. B. V., Kiëu, K., Nielsen, J. The efficiency of systematic sampling in stereology – reconsidered. J.Microsc. 193 (3), 199-211 (1999).
- West, M. J. Design based stereological methods for estimating the total number of objects in histological material. Folia Morphol.(Warsz). 60 (1), 11-19 (2001).
- Olesen, M. V., Wörtwein, G., Pakkenberg, B. Electroconvulsive stimulation, but not chronic restraint stress, causes structural alterations in adult rat hippocampus – a stereological study. Hippocampus. 25 (1), 72-80 (2015).
- Olesen, M. V., Wortwein, G., Folke, J., Pakkenberg, B. Electroconvulsive stimulation results in long-term survival of newly generated hippocampal neurons in rats. Hippocampus. 27 (1), 52-60 (2017).
- Madsen, T. M., et al. Increased neurogenesis in a model of electroconvulsive therapy. Biol Psychiat. 47 (12), 1043-1049 (2000).
- Cameron, H. A., Mckay, R. D. G. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J.Comp. Neurol. 435 (4), 406-417 (2001).
- Dorph-Petersen, K. A., Nyengaard, J. R., Gundersen, H. J. G. Tissue shrinkage and unbiased stereological estimation of particle number and size. J.Microsc. 204 (3), 232-246 (2001).
- Schmitz, C., Hof, P. R. Recommendations for straightforward and rigorous methods of counting neurons based on a computer simulation approach. J Chem. Neuroanat. 20 (1), 93-114 (2000).
- Paxinos, G., Watson, C. . The rat brain in stereotaxic coordinates. , (1986).
- Kempermann, G., Gast, D., Kronenberg, G., Yamaguchi, M., Gage, F. H. Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development. 130 (2), 391-399 (2003).
- Fabricius, K., Wortwein, G., Pakkenberg, B. The impact of maternal separation on adult mouse behaviour and on the total neuron number in the mouse hippocampus. Brain Struct. Funct. 212 (5), 403-416 (2008).
- Dorph-Petersen, K. A., et al. Volume and neuron number of the lateral geniculate nucleus in schizophrenia and mood disorders. Acta neuropathologica. 117 (4), 369-384 (2009).
- Eriksen, N., et al. Application of stereological estimates in patients with severe head injuries using CT and MR scanning images. Br. J. Radiol. 83 (988), 307-317 (2010).
- Gundersen, H. J. Stereology: the fast lane between neuroanatomy and brain function–or still only a tightrope?. Acta Neurol. Scand. Suppl. 137, 8-13 (1992).

