Demonstration of Proteolytic Activation of the Epithelial Sodium Channel (ENaC) by Combining Current Measurements with Detection of Cleavage Fragments
Summary
Proteolytic activation of the epithelial sodium channel (ENaC) heterologously expressed in Xenopus laevis oocytes can be demonstrated by combining current measurements with a biotinylation approach to investigate the appearance of ion channel cleavage products at the cell surface. Functionally important cleavage sites can be identified by using site-directed mutagenesis.
Abstract
The described methods can be used to investigate the effect of proteases on ion channels, receptors, and other plasma membrane proteins heterologously expressed in Xenopus laevis oocytes. In combination with site-directed mutagenesis, this approach provides a powerful tool to identify functionally relevant cleavage sites. Proteolytic activation is a characteristic feature of the amiloride-sensitive epithelial sodium channel (ENaC). The final activating step involves cleavage of the channel’s γ-subunit in a critical region potentially targeted by several proteases including chymotrypsin and plasmin. To determine the stimulatory effect of these serine proteases on ENaC, the amiloride-sensitive whole-cell current (ΔIami) was measured twice in the same oocyte before and after exposure to the protease using the two-electrode voltage-clamp technique. In parallel to the electrophysiological experiments, a biotinylation approach was used to monitor the appearance of γENaC cleavage fragments at the cell surface. Using the methods described, it was demonstrated that the time course of proteolytic activation of ENaC-mediated whole-cell currents correlates with the appearance of a γENaC cleavage product at the cell surface. These results suggest a causal link between channel cleavage and channel activation. Moreover, they confirm the concept that a cleavage event in γENaC is required as a final step in proteolytic channel activation. The methods described here may well be applicable to address similar questions for other types of ion channels or membrane proteins.
Introduction
Proteases are enzymes that are involved in various physiological reactions ranging from the well-known proteolytic degradation of proteins, in the context of digestion, to highly sophisticated protease cascades involved in complex regulatory signaling pathways. Proteases are classified into seven groups according to their catalytic active site: aspartate, asparagine, cysteine, glutamic acid, metallo, serine, and threonine proteases. Different proteases target distinct cleavage sites which are not always easy to predict from the primary structure of a protein. The MEROPS database (http://merops.sanger.ac.uk/) provides detailed information on a wide range of proteases and their preferential cleavage sites. Functionally relevant cleavage sites can be identified using site-directed mutagenesis.
It is well established that proteolytic processing of ENaC is an important mechanism of activation of this particular ion channel1,2. Interestingly, there is evidence that the function of the related acid-sensing ion channel 1a (ASIC1a) may also be modified by proteases3-5. At present it remains an open question whether proteolytic channel cleavage plays a relevant physiological role in regulating the activity of other ion channels or transporters. However, it is well established that proteolytic cleavage activates a group of G protein-coupled receptors, the protease-activated receptors (PARs)6. Several serine proteases (e.g. channel-activating proteases (CAP1-3), chymotrypsin, trypsin, furin, plasmin, neutrophil elastase, and kallikrein) have been shown to proteolytically activate ENaC2. In addition to serine proteases, other groups of proteases may be involved in proteolytic ENaC activation. Indeed, recent data shows that the metalloproteinase meprin-β7 and the cysteine protease cathepsin-S8 can also activate ENaC. However, the (patho-)physiologically relevant proteases for ENaC activation remain to be determined and may differ from tissue to tissue.
Proteases are known to preferentially cleave at particular sites in the amino acid sequence. For instance, the serine protease chymotrypsin shows a specific cleavage pattern cleaving after the aromatic amino-acid residues phenylalanine and tyrosine. In contrast, the serine protease trypsin preferentially cleaves after the basic residues lysine or arginine. Using mutant human γENaC constructs generated by site-directed mutagenesis, functionally relevant cleavage sites in ENaC heterologously expressed in the oocyte expression system could be identified8-13.
By injecting cRNA for the three ENaC subunits (αβγ) into isolated oocytes, ENaC can be functionally expressed in these cells and the activity of channels present at the plasma membrane can be measured by using the two-electrode voltage-clamp technique. By using the diuretic amiloride, a specific ENaC inhibitor, the amiloride-sensitive ENaC-mediated whole-cell current component (ΔIami) can be separated from unspecific leak currents or from currents conducted by other ion channels. Thus, ΔIami values reflect overall ENaC activity and can be determined by subtracting whole-cell currents measured in the presence of amiloride from the corresponding whole-cell currents recorded in the absence of amiloride. To test whether a protease has a stimulatory effect on ENaC, ΔIami is measured twice in the same oocyte, i.e. before and after incubation of the oocyte in a protease containing solution. An increase of ΔIami from the first to the second measurement indicates proteolytic ENaC activation. Chymotrypsin or trypsin are known to maximally stimulate ENaC in the oocyte expression system2,14 and can be used to confirm that proteolytic ENaC activation is detectable in a given batch of oocytes.
In parallel to whole-cell current measurements, a biotinylation approach9 was used to investigate whether the increase in ΔIami detected upon exposure of the oocytes to proteases correlates with the appearance of ENaC cleavage fragments at the cell surface. Proteins at the cell surface are labeled with biotin and can be separated from intracellular proteins by binding the biotinylated proteins to Neutravidin-labeled agarose beads. The biotinylated proteins can be analyzed by western blot. γENaC cleavage fragments at the cell surface can be detected using a specific antibody directed against an epitope in the C-terminus of the γENaC. To identify functionally relevant cleavage site(s), predicted cleavage sites can be mutated using site-directed mutagenesis. Wildtype and mutant channels are compared in parallel experiments using oocytes from the same batch.
With this methodological approach it was demonstrated for the first time that proteolytic activation of ENaC-mediated whole-cell currents correlates with the time-dependent appearance of ENaC cleavage fragments at the cell surface. These results suggest a causal link between channel cleavage and channel activation. Moreover, using site-directed mutagenesis of putative cleavage sites in combination with the two-electrode voltage-clamp technique, functionally relevant cleavage sites for plasmin, chymotrypsin13 and cathepsin-S8 were identified.
Protocol
1. Isolation of Xenopus Oocytes and Microinjection of cRNA
- Obtain oocytes from adult female Xenopus laevis. Anaesthetize animals in 0.2% MS222, and resect ovarian lobes through a small abdominal incision.
- Isolate oocytes from the ovarian lobes by enzymatic digestion at 19 °C for 3-4 hr with 600-700 U/ml of type 2 collagenase from Clostridium histolyticum dissolved in calcium free OR2 solution (recipe in Table 1).
- For selection, place the defolliculated oocytes in a Petri dish under a binocular microscope in a high sodium containing solution (ND96: recipe in Table 1).
- Select stage V-VI oocytes and place them in another Petri dish with a Pasteur pipette. NOTE: Blunt Pasteur pipette by flaming to prevent oocyte injury.
- Inject oocytes with cRNA (e.g. 0.2 ng per αβγENaC subunit). Dissolve cRNAs in RNase-free water. NOTE: Total volume injected into each oocyte is 46 nl.
- Store injected oocytes at 19 °C in a low sodium solution to prevent sodium loading of the oocytes (ND9: recipe in Table 1). Supplement the solution with 100 U/ml sodium penicillin and 100 μg/ml streptomycin sulphate to prevent bacterial growth. Carefully handle the oocytes to limit the amount of damaged or dead oocytes and maintain them in individual small groups in a 12-well-plate wells filled with bath solution during the two days after cRNA injection.
| OR2 | 82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, adjusted to pH 7.4 with NaOH |
| ND96 | 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, adjusted to pH 7.4 with Tris |
| ND9 | 9 mM NaCl, 87 mM NMDG-Cl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, adjusted to pH 7.4 with Tris |
| biotinylation buffer | 10 mM triethanolamine, 150 mM NaCl, 2 mM CaCl2, adjusted to pH 9.5 with HCl |
| quench buffer | 192 mM glycin, 25 mM Tris-Cl, adjusted to pH 7.5 with HCl |
| lysis buffer | 500 mM NaCl, 5 mM EDTA, 50 mM Tris-Cl, adjusted to pH 7.4 with HCl |
Table 1. Solutions
2. Performing Two-electrode Voltage-clamp Experiments
- Measure the oocytes two days after injection.
- Fill one syringe of a gravity fed perfusion system with ND96 solution and another syringe with ND96 solution containing amiloride (2 µM). Mount syringes 50 cm above the oocyte bath chamber. NOTE: The concentration of the ENaC inhibitor amiloride was chosen to be 20 times higher than its IC50 (100 nM).
- Turn on a 150 W halogen cold light source and adjust it to 10 cm above the oocyte bath chamber allowing good visualization with the binocular microscope. Then turn on suction and adjust the suction tube at the end of the oocyte bath chamber. Locate suction tube opposite to superfusion tubes’ adapter entering the oocyte bath. NOTE: Suction power must be sufficient to support continuous flow of the solution superfusing the oocyte.
- Adjust superfusion speed of each solution to 3-5 ml/min by using the i.v. gravity flow control device. Connect the superfusion tubes with an adapter to the oocyte bath chamber.
- Pull glass capillaries with a micropipette puller to obtain tip diameters of <1 µm. Then fill capillaries to ~1/4 with 3 M KCl. NOTE: Make sure that the chlorinated part of the silver wire of the electrode holder is immersed in KCl solution. Check for air bubbles in the tip of the capillary. Air bubbles impair the measurement by increasing resistance stray capacitance.
- Insert the capillaries into the electrode holders of the current and the voltage electrode and place them into ND96 containing amiloride (2 µM) solution using the micro-manipulators.
3. Measurement of Amiloride-sensitive Whole-cell Currents
- Zero the electrode potential of the voltage electrode (Vm) and the current electrode (Ve) by adjusting the Vm and Ve offsetbuttons. NOTE: The resistance should be 1-2 MΩ for the electrode to measure Vm and 0.5-1 MΩ for the current injection electrode.
- Place the oocyte into the bath chamber in close proximity to the voltage sensing electrode. NOTE: Do not damage the oocyte during any of these transfer steps. Use a Pasteur pipette to transfer the oocyte. To avoid damaging the oocyte the edges of the pipette should be blunted by flaming.
- Impale oocytes gently with both microelectrodes.
- Set the holding potential at the amplifier to -60 mV and turn on the chart recorder. Turn on the amiloride (2 µM) containing solution. NOTE: The current should be about 0 ± 0.5 µA. Larger leak currents indicate a leaky impalement. Therefore, these oocytes should be rejected. Moreover, the leak currents measured in the presence of amiloride (2 µM) should be similar in αβγ-WT expressing oocytes to those measured in αβγ-mutant ENaC expressing oocytes. This indicates that the mutations do not affect the amiloride-sensitivity of the channel.
- Start recording. If necessary adjust the gain.
- After the measured current reaches a stable plateau, change to amiloride-free solution. NOTE: Downward current deflections in the current traces correspond to inward currents, i.e. movement of positive charge (Na+) from the extracellular side into the cell.
- After a current plateau is reached (after ~60 sec), switch the superfusion back to the amiloride containing solution. After the current of the oocyte reaches the initial baseline current, turn off the voltage clamp and gently withdraw the electrodes.
- To allow resealing of the plasma membrane at the sites of impalement, place the oocyte into one well of a 96-well plate containing 100-150 µl of protease free ND96 solution.
- After 5 min, transfer oocyte to a protease containing solution or to a control solution without protease for a 30 min incubation time. NOTE: The incubation time depends on the protease and the studied channel.
- After the incubation step repeat the current measurement (see 3.2 and following). NOTE: It is possible to measure >90% of the oocytes after incubation in protease solution.
4. Biotinylation Assay
- Select and discard defective oocytes under the binocular microscope. NOTE: Use injected oocytes from the same batch for the current measurements and for the biotinylation experiments.
- Keep biotin at RT for at least 20 min before its use in the experiment.
- Prepare the solutions: ND96 and ND96 containing the appropriate protease. Prepare Pasteur pipettes by labeling them and by briefly flaming their tips to avoid injury of the oocytes. NOTE: Here, the protease chymotrypsin 2 µg/ml in ND96 is used. Treat each group with a separate pipette to avoid cross contamination of solutions.
- Fill each well of a 6-well plate with 2.5 ml control ND96 or ND96 containing a protease at RT. Then deposit 30 oocytes per well and incubate them for 30 min at RT. NOTE: For the subsequent procedures it is important to keep samples on ice at all times. All centrifugation steps are at performed at 4 °C.
- Fill each well of a new 6-well plate with 2.5 ml ND96 (each group needs 3 wells for the washing steps) and weigh the biotin. NOTE: 2.5 mg biotin per well (1 mg/ml) is required. Dissolve the biotin in the biotinylation buffer (i.e. 25 mg biotin (for 10 groups) in 25 ml biotinylation buffer (recipe in Table 1).
- Transfer each group of oocytes to a well filled with 2.5 ml ND96. Transfer oocytes sequentially into two additional wells with ND96 to wash off any remaining protease. Incubate the oocytes for 5 min in ND96.
- Transfer the oocytes into a well containing 2.5 ml biotin solution and incubate them with gentle agitation (‘shaker’) for 15 min. NOTE: Minimize the transferred ND96 with the pipette to avoid dilution of the biotin solution.
- Transfer each group of oocytes into a well containing 2.5 ml quench buffer (recipe in Table 1) to stop the biotinylation reaction. Then, transfer each group of oocytes into a second well also containing 2.5 ml quench buffer and incubate for 5 min with gentle agitation.
- Remove damaged or dead oocytes. NOTE: Choose the same number of oocytes per group for the following procedure.
- Transfer each group of oocytes to a 1.5 ml plastic microcentrifuge tube. NOTE: Minimize the quantity of quench buffer that is transferred.
- Subsequently, lyse the oocytes by passing them through a 27 G needle in 1 ml lysis buffer (recipe see Table 1) supplemented with protease inhibitors.
- Centrifuge the lysates for 10 min at 1,500 x g.
- Aspirate supernatant and transfer it to a 1.5 ml microcentrifuge tube containing 0.5% Triton-X-100 and 0.5 % NP40. Discard the remaining pellet. NOTE: The supernatant contains biotinylated plasma membrane proteins and non-biotinylated intracellular proteins.
- Incubate the microcentrifuge tubes for 20 min on ice. Repeatedly vortex the tubes during this period to completely dissolve the proteins in NP40 and Triton-X-100.
- Centrifuge 100 µl of agarose beads per oocyte group for 3 min at 1,500 x g. After the centrifugation remove supernatant from the beads solution and wash three times with lysis buffer to equilibrate the beads with buffer.
- Pipette 100 µl of the washed beads into each microcentrifuge tube containing the protein-detergent-solution prepared in 4.13 to allow binding of the biotinylated proteins to the beads.
- Incubate microcentrifuge tubes with overhead rotation O/N at 4 °C.
- Centrifuge the microcentrifuge tubes for 3 min at 1,500 x g. Then transfer the supernatant into a new tube. NOTE: Intracellular proteins are not labeled with biotin. Supernatant can be stored at -20 °C. Do not aspirate the beads.
5. Detection of ENaC Cleavage Fragments at the Cell Surface by Western Blot Analysis
- Wash the beads three times with lysis buffer and add 100 µl of 2x SDS-PAGE sample buffer. NOTE: Samples can be stored at -20 °C or immediately prepared for western blot analysis.
- Boil samples for 5 min at 95 °C and then place tubes on ice.
- Centrifuge samples for 3 min at 20,000 x g and pipette the supernatant into a new microcentrifuge tube. NOTE: This supernatant contains the biotinylated plasma membrane proteins from the cell surface of the oocyte.
- Analyze 30 µl of this supernatant by western blot to investigate cleavage fragments at the cell surface.
- Separate the biotinylated proteins by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) using an appropriate gel (8%, 10%, 12% depending on the molecular weight of the cleavage fragments investigated).
- Transfer the proteins to polyvinylidene difluoride (PVDF) membranes by semi-dry blotting.
- Probe the membrane with a specific antibody against human γENaC directed against an epitope in the C-terminus (see Figure 3 and 13).
- Use horseradish peroxidase-labeled goat anti-rabbit antibody as secondary antibody.
- Detect chemiluminescent signals.
Representative Results
To investigate whether the serine protease plasmin can activate ENaC-mediated currents, ΔIami of individual ENaC-expressing oocytes was determined before and after 30 min incubation of the oocytes in protease-free (control) (Figure 2A) or plasmin containing solution (Figure 2B) using the two-electrode voltage-clamp technique (see Figure 1). Exposure to plasmin increased ΔIami in every oocyte measured. In contrast, in control experiments, 30 min incubation of ENaC-expressing oocytes in protease-free solution had a negligible effect (Figure 2 C,D). Thus, by using this method a stimulation of ENaC-mediated current by plasmin can be detected.
To study the effects of mutating putative cleavage sites upon the activation of ENaC-mediated currents, as well as upon channel cleavage, the effect of chymotrypsin on WT-ENaC was compared with that on a mutant ENaC with mutated prostasin and plasmin cleavage sites (γRKRK178AAAA;K189A). The time course of channel activation by chymotrypsin as well as the appearance of ENaC cleavage products at the cell surface was investigated by using different protease incubation times (Figure 4A). It was demonstrated that the mutant channel delays and reduces the activation of ENaC-mediated current by chymotrypsin. This is paralleled by a delayed appearance of a lower molecular weight γENaC cleavage fragment of 67 kDa corresponding to the fully cleaved subunit. Cleavage fragments were detected using a γENaC antibody directed against an epitope in the C-terminus (Figure 3). This methodological approach demonstrates that the time course of proteolytic activation of ENaC-mediated currents correlates with the appearance of a 67 kDa γENaC cleavage product at the cell surface (Figure 4 B,C). This supports the concept of a causal link between proteolytic channel cleavage and channel activation13. Moreover, by combining current measurements and the detection of γENaC fragments at the cell surface it was demonstrated that the mutated cleavage sites are functionally relevant for proteolytic channel activation.
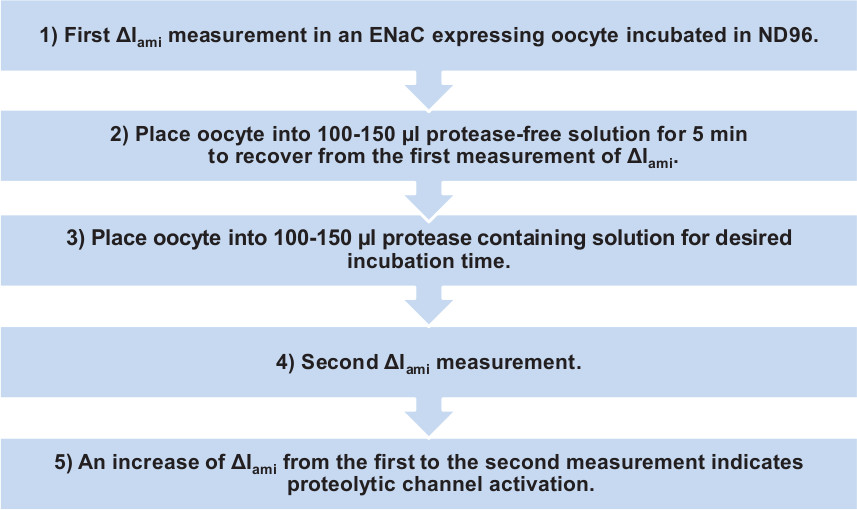
Figure 1. Procedure of determining the stimulatory effect of a protease on ENaC heterologously expressed in Xenopus laevis oocytes. ENaC activity is estimated by measuring the amiloride-sensitive whole-cell current component ΔIami.
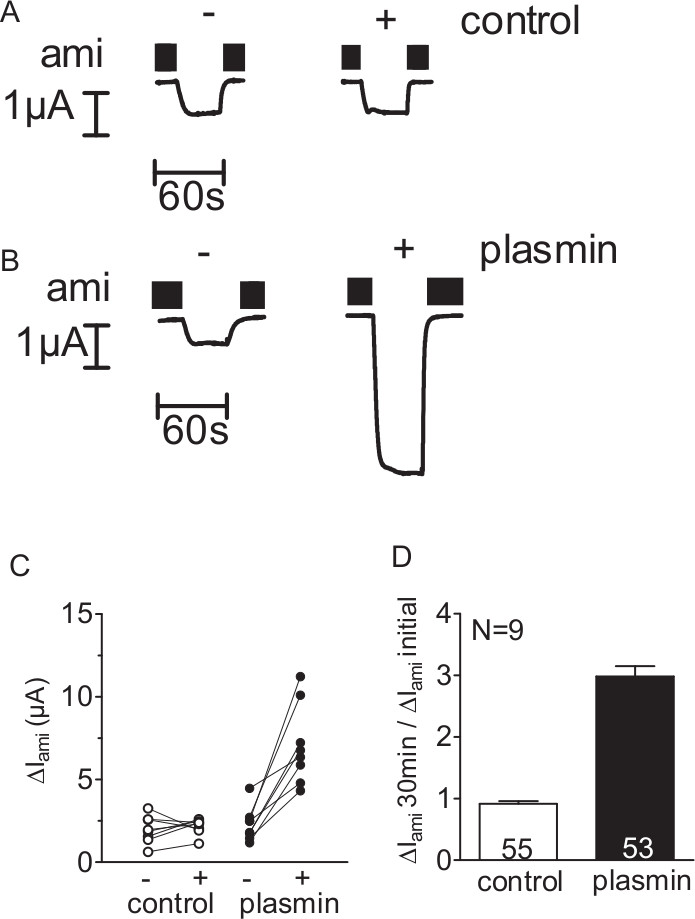
Figure 2. Plasmin stimulates ENaC-mediated currents in oocytes expressing ENaC. (A-D) Oocytes expressing human ENaC were incubated for 30 min in protease-free solution (control) or in solution containing plasmin (10 μg/ml). To determine ΔIami before (-) and after (+) incubation, oocytes were clamped at a holding potential of -60 mV. (A,B) Four representative whole-cell current traces from one batch of oocytes. Amiloride (ami) was present in the bath solution to specifically inhibit ENaC as indicated by black bars. (C) Data points obtained from an individual oocyte are connected by a line. (D) Summary of similar experiments as shown in C. Columns represent relative stimulatory effect on ΔIami calculated as the ratio of ΔIami measured after a 30 min incubation (ΔIami 30 min) to the initial ΔIami (ΔIami initial) measured before incubation. Numbers inside the columns indicate the number of individual oocytes measured. N indicates the number of different batches of oocytes. (This figure has been modified from [Haerteis et al. 2012 J Gen Physiol 140, 375-389, doi:10.1085/jgp.201110763])
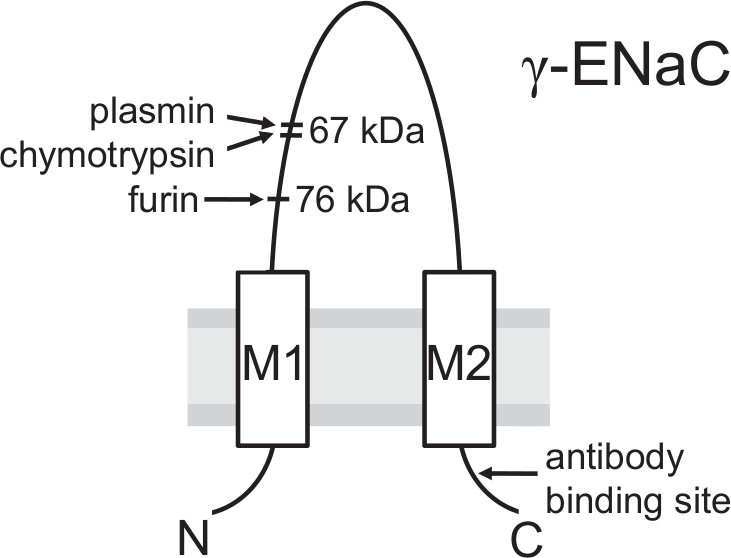
Figure 3. Model of the γENaC subunit showing cleavage sites for proteolytic activation and the binding site of the antibody used. Proteolytic cleavage by the Golgi-associated convertase furin is important for ENaC maturation in the biosynthetic pathway before the channel reaches the plasma membrane. After cleavage by furin a 76 kDa fragment can be detected at the cell surface using a biotinylation approach and an antibody against an epitope in the C-terminus of the γ-subunit. The pivotal final step in proteolytic ENaC activation probably takes place at the plasma membrane where γENaC is cleaved by extracellular proteases (e.g. plasmin or chymotrypsin) in a region distal to the furin site resulting in a 67 kDa cleavage fragment. (This figure has been modified from [Haerteis et al. 2012 J Gen Physiol 140, 375-389, doi:10.1085/jgp.201110763])
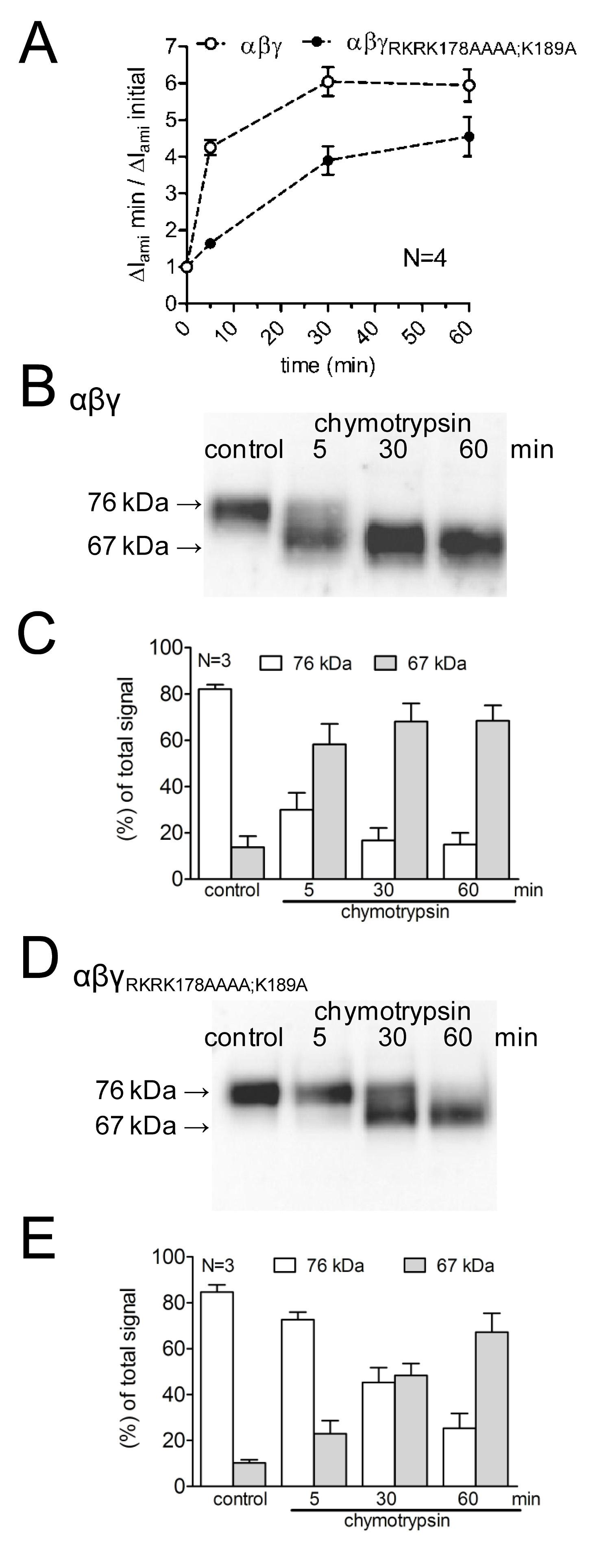
Figure 4: Mutating both the plasmin (K189) and the prostasin cleavage site (RKRK178) delays the activation of ENaC-mediated currents and the appearance of a 67 kDa cleavage product of the channel’s γ-subunit. Oocytes expressing WT (open symbols) and γRKRK178AAAA;K189AENaC mutant channel (closed symbols) were incubated for 30 min in protease-free solution (control) or for 5, 30, or 60 min in a solution containing chymotrypsin (2 μg/ml). (A) To determine ΔIami before and after incubation, oocytes were clamped at a holding potential of -60 mV. Circles represent the ratio of ΔIami measured after 5, 30, or 60 min incubation (ΔIami min) to the initial ΔIami (ΔIami initial) measured before incubation. Each data point represents the mean ΔIami measured in 22-24 individual oocytes of four different batches. (B-D) In parallel to the detection of ΔIami, expression of biotinylated γENaC at the cell surface was analyzed by SDS-PAGE. γENaC was detected with an antibody against an epitope in the C-terminus of human γENaC. Representative western blots from one batch of oocytes are shown. (C-E) Densitometric analysis of three western blots similar to those shown in B or D. For each lane, the signals detected in the regions of 76 kD (open columns) and 67 kD (gray columns) were determined and normalized to the sum of the total signal detected. N indicates the number of different batches of oocytes. Click here to view larger image.
Discussion
In this manuscript a methodological approach which was successfully applied to study the mechanisms underlying the activation of ENaC by proteases is described8,13. The well established Xenopus laevis oocyte expression system was used to functionally express ENaC. ENaC function was assessed with the conventional two-electrode voltage-clamp technique. Site-directed mutagenesis was employed to identify functionally relevant protease cleavage sites. Biotinylation experiments performed in parallel with the electrophysiological measurements made it possible to correlate the appearance of ENaC cleavage products at the cell surface with proteolytic current activation. A correlation between the time course of current activation and the appearance of proteolytic cleavage fragments at the cell surface supports the concept of proteolytic channel activation.
Two-electrode voltage-clamp recordings require the impalement of an oocyte with two microelectrodes. This procedure is usually performed only once in an individual oocyte. However, it was feasible to remove the microelectrodes after an initial whole-cell current recording without apparent damage to the oocyte. Indeed, the plasma membrane at the sites of impalements appears to reseal within a few minutes. Thus, after completing a first two-electrode voltage-clamp measurement, it is possible to transfer the oocyte from the experimental flow chamber of the two-electrode voltage-clamp setup to a microfuge tube or a well of a 96-well plate filled with a small volume of test or control solution. Afterwards, the same oocyte can be transferred back to the flow chamber and can be impaled again to perform a second two-electrode voltage-clamp measurement. Remarkably, ENaC-mediated currents did not vary much between the first and second measurement when the oocyte was maintained in control solution. In contrast, incubation of the oocyte in a protease containing solution after the first measurement resulted in increased ENaC-mediated current in the second measurement (Figure 2). This finding indicates proteolytic channel activation.
Performing two separate current measurements in a single oocyte offers the advantage that the oocyte can be exposed to proteases or other pharmacological agents between the two measurements for a variable length of time in a small volume of test solution. This is important when using agents which are expensive and/or unavailable in large quantities, e.g. purified protease preparations. The limited availability of agents may make it impossible (or unaffordable) to use them in continuous two-electrode voltage-clamp recordings because of the large volumes of test solution required for continuously superfusing the oocytes with flow rates of several milliliters per minute. Moreover, continuous two-electrode voltage-clamp measurements are limited by the well-known phenomenon of spontaneous channel rundown also described for ENaC 15. In contrast, exposing oocytes to test solutions between two separate measurements for up to an hour or more does not generally pose a problem (see Figure 4A). Finally, two sequential measurements performed in the same oocyte allow paired observations of drug effects. This has an advantage over unpaired measurements from two separate groups of oocytes (protease-treated and vehicle-treated), because it reduces the problem of high variability between oocytes, usually observed in ion channel expression. With paired observations and the possibility to normalize the data to the first measurement, fewer oocytes are needed per experimental group to demonstrate a significant effect of a pharmacological agent. Normalization of the data also makes it easy to summarize data from different batches of oocytes with different ion channel expression levels and hence different baseline currents (Figure 2D). Obviously, control experiments are necessary for this approach to demonstrate that the ion channel activity of interest remains stable in vehicle-treated control oocytes from the first to the second measurement (see Figure 2).
To demonstrate that proteolytic current activation correlates with the appearance of ENaC cleavage products at the cell surface, a biotinylation approach originally described by Harris et al.9 can be used. This procedure (as detailed in the protocol section and shown in Figure 4) was adapted to demonstrate that exposure of channels to proteases and subsequent activation of ENaC-mediated currents is paralleled by the time-dependent appearance of cleavage fragments. The biotinylation method also allows the analysis of an overall increase or decrease of membrane proteins at the cell surface. Thus, this method is suitable to investigate the effect of proteases and other pharmacological agents upon channel insertion into the plasma membrane or upon channel retrieval. Moreover, western blot analysis of the biotinylated plasma membrane proteins allows detection of protein fragments (e.g. proteolytic ENaC fragments) or changes in the glycosylation pattern which may be functionally relevant.
In conclusion, the combination of methods used to investigate the stimulatory effect of proteases on ENaC-mediated whole-cell currents and to demonstrate a correlation with the occurrence of ENaC cleavage products at the cell surface may be useful for a broad range of applications. In particular, these methods may be suitable to address similar questions regarding the regulation of other ion channels, transporters or transmembrane receptors (e.g. protease-activated receptors PARs).
Divulgations
The authors have nothing to disclose.
Acknowledgements
The expert technical assistance of Céline Grüninger, Christina Lang, Sonja Mayer, and Ralf Rinke is gratefully acknowledged. We thank Dr. Morag K. Mansley for carefully reading the manuscript. This project was supported by a grant of the Deutsche Forschungsgemeinschaft (Grant SFB 423: Kidney Injury: Pathogenesis and Regenerative Mechanisms, to C. Korbmacher), grants of the Interdisziplinäres Zentrum für Klinische Forschung (to S. Haerteis and M. Krappitz), the ELAN program (to S. Haerteis) of the Friedrich-Alexander-Universität Erlangen-Nürnberg, and the University Library of Erlangen-Nürnberg.
Materials
| Bath Clamp Headstage for OC-725C-V | Warner Instrument Corporation | – | |
| Cold light source – Schott KL 1500 LCD | Schott | #SCOC150200EU | brightness 4; mechanical aperture: D; color temperature: 3000 K |
| E Series Electrode Holder (Str, Vent, Ag Wire, 1.2 mm) | ADinstruments | #ESW-F10v | |
| left micromanipulator; MM-33L | Warner Instrument Corporation | #64-0055 | |
| LIH 1600 – computer interface | HEKA | – | |
| magnetic valve system (ALA BPS-8) in combination with a TIB14 interface (HEKA) | ALA Scientific Instruments, HEKA | – | |
| OC-725C amplifier for two-electrode voltage-clamp recordings | Warner Instrument Corporation | – | |
| P-97 FLAMING/BROWN Micropipette Puller | Sutter Instruments | – | heat=550; velocity=22; time=200 |
| right micromanipulator; MM-33R | Warner Instrument Corporation | #64-0056 | |
| Series Electrode Holder (45°, Vent, Handle, Ag Wire, 1.2 mm) | ADinstruments | #E45w-f10vh | |
| STAT 2 IV Gravity Flow Controller | Conmed | #P-S2V-60 | |
| vacuum generator ejector SEG – for suction to remove bath solution | Schmalz | – | |
| Material | |||
| INFUJECT 60ml pump syringes for solutions | Braun | #22050 | |
| Injekt-F for lysing the oocytes | Braun | #9166033V | |
| standard wall borosilicate tubing with filament | Sutter Instruments | #BF150-86-10 | outside diameter: 1.50 mm; inside diameter: 0.86 mm; length: 10 cm |
| Reagent | |||
| complete, Mini, EDTA-free protease inhibitor cocktail tablets | Roche Applied Science | #11836170001 | |
| EZ-Link Sulfo-NHS-Biotin | Thermo Scientific | #21217 | |
| Horseradish peroxidase-labeled secondary goat anti-rabbit antibody | Santa Cruz Biotechnology | #sc-2004 | |
| NeutrAvidin Agarose | Thermo Scientific | #29200 | Neutravidin-labeled agarose beads |
| NP40 (Nonidet P-40) | Sigma-Aldrich | #I8896 | |
| Roti-Load 1 (2× SDS-PAGE sample buffer) | Carl Roth | #K929.2 | |
| SuperSignal West Femto Chemiluminescent Substrate for detection of chemiluminescent signals | Thermo Scientific | #34095 | |
| Triton-X-100 | Sigma-Aldrich | #T8787 | |
References
- Kleyman, T. R., Carattino, M. D., Hughey, R. P. ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. The Journal of Biological Chemistry. 284, 20447-20451 (2009).
- Rossier, B. C., Stutts, M. J. Activation of the epithelial sodium channel (ENaC) by serine proteases. Annu Rev Physiol. 71, 361-379 (2009).
- Poirot, O., Vukicevic, M., Boesch, A., Kellenberger, S. Selective regulation of acid-sensing ion channel 1 by serine proteases. The Journal of Biological Chemistry. 279, 38448-38457 (2004).
- Vukicevic, M., Weder, G., Boillat, A., Boesch, A., Kellenberger, S. Trypsin cleaves acid-sensing ion channel 1a in a domain that is critical for channel gating. The Journal of Biological Chemistry. 281, 714-722 (2006).
- Clark, E. B., Jovov, B., Rooj, A. K., Fuller, C. M., Benos, D. J. Proteolytic cleavage of human acid-sensing ion channel 1 by the serine protease matriptase. The Journal of Biological Chemistry. 285, 27130-27143 (2010).
- Ossovskaya, V. S., Bunnett, N. W. Protease-activated receptors: contribution to physiology and disease. Physiological reviews. 84, 579-621 (2004).
- Garcia-Caballero, A., et al. Activation of the epithelial sodium channel by the metalloprotease meprin β-subunit. Channels (Austin. 5, 14-22 (2011).
- Haerteis, S., et al. Proteolytic activation of the epithelial sodium channel (ENaC) by the cysteine protease cathepsin-S. Pflügers Archiv: European Journal of Physiology. 464, 353-365 (2012).
- Harris, M., Firsov, D., Vuagniaux, G., Stutts, M. J., Rossier, B. C. A novel neutrophil elastase inhibitor prevents elastase activation and surface cleavage of the epithelial sodium channel expressed in Xenopus laevis oocytes. The Journal of Biological Chemistry. 282, 58-64 (2007).
- Passero, C. J., Mueller, G. M., Rondon-Berrios, H., Tofovic, S. P., Hughey, R. P., Kleyman, T. R. Plasmin activates epithelial Na+ channels by cleaving the γ-subunit. The Journal of Biological Chemistry. 283, 36586-36591 (2008).
- Svenningsen, P., et al. Plasmin in nephrotic urine activates the epithelial sodium channel. Journal of the American Society of Nephrology : JASN. 20, 299-310 (2009).
- Patel, A. B., Chao, J., Palmer, L. G. Tissue kallikrein activation of the epithelial Na channel. American Journal of Physiology. Renal Physiology. 303, (2012).
- Haerteis, S., Krappitz, M., Diakov, A., Krappitz, A., Rauh, R., Korbmacher, C. Plasmin and chymotrypsin have distinct preferences for channel activating cleavage sites in the γ-subunit of the human epithelial sodium channel. The Journal of General Physiology. 140, 375-389 (2012).
- Chraibi, A., Vallet, V., Firsov, D., Hess, S. K., Horisberger, J. D. Protease modulation of the activity of the epithelial sodium channel expressed in Xenopus oocytes. The Journal of General Physiology. 111, 127-138 (1998).
- Volk, T., Konstas, A. A., Bassalay, P., Ehmke, H., Korbmacher, C. Extracellular Na+ removal attenuates rundown of the epithelial Na+-channel (ENaC) by reducing the rate of channel retrieval. Pflügers Archiv: European Journal of Physiology. 447, 884-894 (2004).

