Microdissection and Dissociation of the Murine Oviduct: Individual Segment Identification and Single Cell Isolation
Summary
A method for microdissection of the mouse oviduct that allows collection of the individual segments while maintaining RNA integrity is presented. In addition, non-enzymatic oviductal cell dissociation procedure is described. The methods are appropriate for subsequent gene and protein analysis of the functionally different oviductal segments and dissociated oviductal cells.
Abstract
Mouse model systems are unmatched for the analysis of disease processes because of their genetic manipulability and the low cost of experimental treatments. However, because of their small body size, some structures, such as the oviduct with a diameter of 200-400 μm, have proven to be relatively difficult to study except by immunohistochemistry. Recently, immunohistochemical studies have uncovered more complex differences in oviduct segments than were previously recognized; thus, the oviduct is divided into four functional segments with different ratios of seven distinct epithelial cell types. The different embryological origins and ratios of the epithelial cell types likely make the four functional regions differentially susceptible to disease. For example, precursor lesions to serous intraepithelial carcinomas arise from the infundibulum in mouse models and from the corresponding fimbrial region in the human fallopian tube. The protocol described here details a method for microdissection to subdivide the oviduct in such a way to yield a sufficient amount and purity of RNA necessary for downstream analysis such as reverse transcription-quantitative PCR (RT-qPCR) and RNA sequencing (RNAseq). Also described is a mostly non-enzymatic tissue dissociation method appropriate for flow cytometry or single cell RNAseq analysis of fully differentiated oviductal cells. The methods described will facilitate further research utilizing the murine oviduct in the field of reproduction, fertility, cancer, and immunology.
Introduction
The murine oviduct is similar in function and morphology to the human fallopian tube1. Both consist of a pseudostratified ciliated epithelium, consisting of two historically described epithelial resident cells: ciliated cells and secretory cells1,2. The oviduct has three classically recognized segments: the infundibulum, the ampulla, and the isthmus. In a recent study, Harwalkar et al.3 investigated oviduct morphology and gene expression leading to the expansion of the categorization of resident epithelial cells to seven distinct populations. In addition, they established the ampullary-isthmus junction as a distinct segment of the oviduct3. The method described herein, which focuses on the infundibulum, ampulla, and isthmus, could easily be extended to include the ampullary-isthmic junction as well2,3. The infundibular region contains the ostium, or opening of the oviduct, and includes the fimbrial region as well as the proximal stalk. Moving toward the uterus, next is the ampulla, and then the isthmus. Ciliated cells are most prominent in the distal end of the region, proximal to the ovary, or infundibulum, while secretory cells are most prominent in the proximal end or isthmus segment1. Unlike the human fallopian tube, the murine oviduct is a coiled structure supported by the mesosalpinx, an extension of the broad ligament peritoneum1,4. In addition, the mouse oviduct is encased in a bursal sac that increases the likelihood of oocyte transfer into the oviduct4. The ampulla is identified as the location of fertilization, from which developing embryos pass into the isthmus before entering the uterus5. Tubal segments are 200-400 μm in diameter and the longer ampullary and isthmus regions are approximately 0.5-1.0 cm in length4. The oviduct distends during the estrous cycle and the ampulla and infundibulum are more distensible than the isthmus1.
Over proliferation of cells, especially secretory cells, characterize precursor lesions to serous tumors found in the pelvic cavity6. These precursor serous intraepithelial lesions arise in the oviduct epithelium solely in the fimbrial region; it is unknown why lesion formation is restricted to this region where normally the predominant cell type is ciliated, not secretory2,7,8. The regionality in terms of normal physiological function, as well as heightened interest in the oviductal origin of ovarian cancer9,10,11,12,13, underscores the importance of separate evaluation of the oviduct segments.
The method described here details the collection of separate oviductal segments for subsequent downstream analyses of segment-specific gene expression and function of dissociated cells. Traditionally, many tissues are processed for whole RNA extraction following either the phenol: chloroform method or an on-column complete extraction method; however, we found that RNA quality was maintained while producing sufficient yield with the described combination method. Utilizing this method, very small functional segments of the oviduct can be processed for downstream analyses rather than investigating the oviduct as a whole, which can mask results representative of the different segments14.
Dissociated murine oviductal cells have rarely been investigated by flow cytometry, most likely due to the limiting cell yield from this tissue. One approach to overcome this problem has been to dissociate cells, grow them in culture, and then stimulate re-differentiation in vitro to obtain appropriate cell numbers for downstream cell analysis15,16,17,18. A limitation to this approach is the time ex vivo and altered microenvironment in culture, both of which likely change gene expression. There is also an assumption that morphological re-differentiation has the same transcriptional and proteomic signature as was present in the intact animal. The current dissociation method was designed to achieve the highest number of epithelial cells in a heterogenous oviductal cell population while maintaining single cell differentiation. Further, the mostly non-enzymatic approach likely limits the loss of cell surface proteins.
Protocol
All animal handling and procedures were approved by the University of California, Riverside institutional animal care and use committee and were in accordance with guidelines from the American Association for Laboratory Animal Care, the United States Department of Agriculture, and the National Institutes of Health. The described method utilized C57BL/6 adult, female mice. All animals were euthanized by decapitation prior to tissue harvesting.
NOTE: An overview of the protocol, which uses a blue dye to assist in efficient dissection and uncoiling of the oviduct, is shown in the first figure (Figure 1).
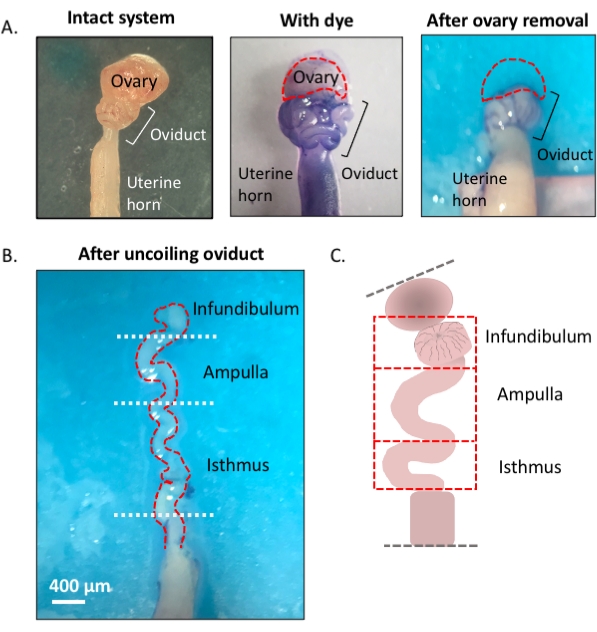
Figure 1: Overview of the microdissection method. (A) Left panel shows the intact structure from which most of the ovarian fat pad and remnants of connective tissue surrounding the oviduct have been removed. After this, the addition of toluidine blue helps distinguish the coils of the oviduct (middle panel). Right panel shows structures after the removal of the ovary. (B) An uncoiled oviduct with internal turns for determining where each segment begins and ends. (C) Cartoon of the segmentation used (not drawn to scale). Please click here to view a larger version of this figure.
1. Preparation of the dissection surface
- Affix a piece of dental wax to a Petri dish with adhesive and let it dry (Figure 2A). Sanitize the dish with 70% ethanol. The surface is ready for dissection.
2. Macrodissection of the ovary, oviduct, and uterus
- Immobilize the Petri dish containing the dental wax on a cold platform of choice (e.g., an ice pack kept at -20 °C prior to use) under a dissection microscope when ready for dissection (Figure 2A).
- Disinfect the ventral surface of the animal with 70% ethanol. Open the abdominal and pelvic cavity with sterile dissection scissors and move the gastrointestinal tract to one side to locate the dorsal bihornal uterus. Follow each uterine horn rostrally to locate each oviduct and ovary enveloped in the uterine fat pad, found just below the kidney (Figure 2B,C).
- Excise both lateral oviducts with the ovaries and a portion of the distal uterine horn still attached using dissection scissors. Cut each lateral uterine horn with approximately 1-2 cm of the horn still attached to the coiled oviduct and ovary (Figure 2D). Submerge the tissue immediately in 2 mL of cold dissection medium (see Table of materials).
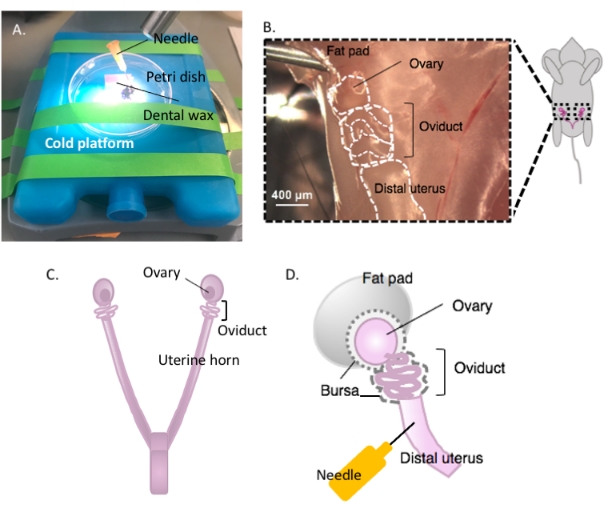
Figure 2: Macrodissection of the ovary and the dissection setup. The oviduct is located dorsally in the pelvic cavity at the base of the kidney within the ovarian fat pad (B). Locate the uterine bifurcation in the pelvic cavity (C) and follow the lateral uterine horn to locate the uterus-oviduct-ovary continuum (B). Remove these structures by cutting through one uterine horn and coarse dissection. Place onto the dissecting platform (A) and affix the uterine portion to dental wax with a needle (A and D). Please click here to view a larger version of this figure.
NOTE: Leave enough uterine horn intact and attached to the oviduct to allow affixation to the dissection platform (Figure 2D).
- Affix the uterine tissue to the dental wax with a sterile 25 G needle to secure tissue for dissection (Figure 2A,D, and Figure 3A). Working under the dissection microscope, clean and dissect away the ovarian fat pad and connective tissue to allow clear visualization of the oviduct (Figure 3B).
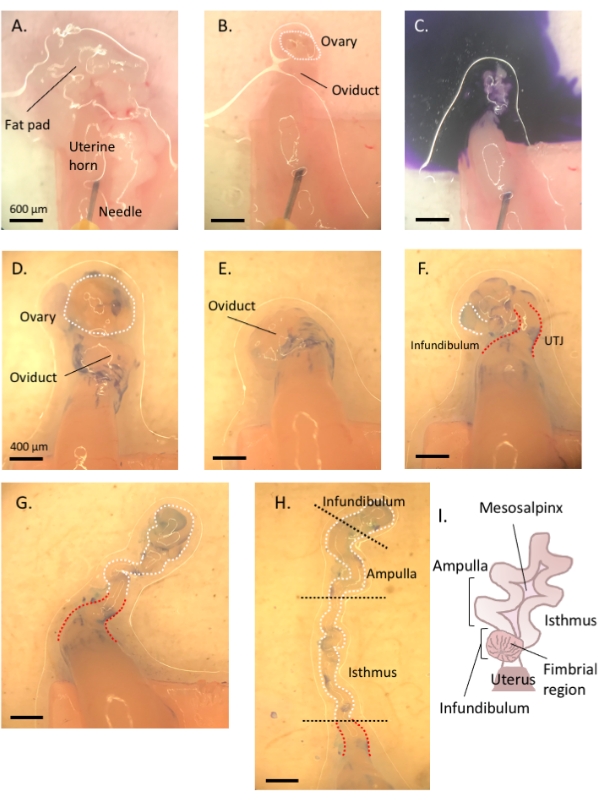
Figure 3: Step by step oviduct microdissection. Following removal of remnant fat and connective tissue (A,B), add toluidine blue to the tissue (C), and then wash away to facilitate visualization and subsequent removal of the ovary and uncoiling of the tube (D–H). The infundibulum can be found beside the proximal isthmus and uterine-tubal junction (UTJ). Lightly pull the distal end while cutting at the mesosalpinx to uncoil and reveal the endogenous turns of the oviduct (H) to orient for appropriate segmentation. Figure A-C scale bar = 500 μm, D–H scale bar = 400 μm (I) Cartoon of the various segments (not drawn to scale). Please click here to view a larger version of this figure.
- Using a 1 mL syringe, dispense and incubate the oviduct affixed to the uterus in 1-2 drops of sterile 1% toluidine blue dye solution for 30 s-1 min. Rinse with cold Dulbecco's phosphate-buffered saline (DPBS) and remove all the liquid (Figure 3C).
- Lightly pull the ovary from the coiled oviduct and cut away at the bursal membrane and broad ligament to remove the ovary from the oviduct without compromising the tubal tissue (Figure 3D,E).
NOTE: The ovary is not attached to the oviduct but encased in the bursal membrane with the oviduct (Figure 2D) and connected to the lateral side of the uterus via the broad ligament.
3. Microdissection and segmentation of the oviduct
- Locate the distal, fimbrial end of the oviduct found lateral to the uterine-tubal junction (UTJ). The oviduct coils back on itself; hence the fimbrial end can be located lateral to the proximal end and is an appropriate starting point to orient for uncoiling of the oviduct (Figure 3F,I).
- While gently pulling the tube, cut away at the mesosalpinx (Figure 3I) with spring-form micro-scissors to uncoil the oviduct (Figure 3G).
- Once uncoiled, cut the tube to produce the infundibular, ampullary, and isthmic regions (Figure 3H).
- Following excision of the infundibulum (fimbrial end and proximal stalk), excise the ampullary region by cutting after the second prominent turn of the tube (cut between turns two and three, approximately 1 cm in length). Finally, cut the remaining portion to the UTJ, which is the isthmic region (Figure 3H).
NOTE: The oviduct will not completely uncoil into a straight structure. The tube will maintain its turns (Figure 3H)3. Based on the subsequent turns following the ampullary region, one may further segment the remaining tube into the ampullary-isthmic junction and isthmus3. Following ampulla excision, cut between turns five and six to obtain the ampullary-isthmic junction; the remaining tube (turn six to the start of the UTJ) is the isthmus3.
- Following excision of the infundibulum (fimbrial end and proximal stalk), excise the ampullary region by cutting after the second prominent turn of the tube (cut between turns two and three, approximately 1 cm in length). Finally, cut the remaining portion to the UTJ, which is the isthmic region (Figure 3H).
- Immediately snap freeze dissected tissue segments in liquid nitrogen for RNA isolation or fix and process each segment for oriented embedding and immunohistological analysis.
- Add 200 μL of cold RNA extraction reagent and 1:1 homogenization bead mix (see Table of materials) to the tissue if proceeding for RNA extraction (see step 5.1).
4. Oviduct dissociation
- Continued from step 3.1.1 above) For optimal dissociation of the epithelium, slit open the oviduct in the areas where possible (i.e., infundibulum and ampulla) to expose the luminal epithelium (Figure 4A).
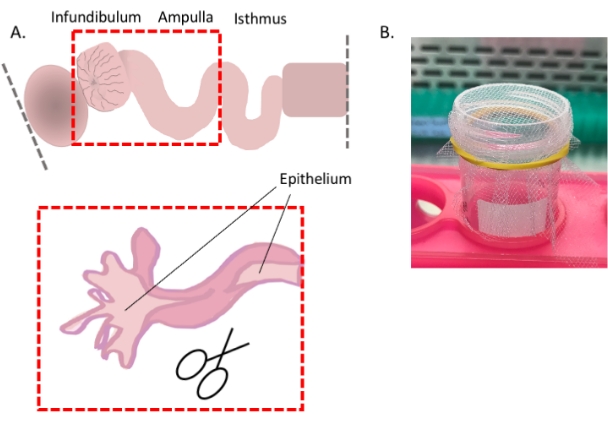
Figure 4: Oviduct cell dissociation, part 1. Cartoon showing how to expose the distal epithelium for optimal cell dissociation (A) and an image showing the easiest way to use the 1 mm2 mesh (B). Please click here to view a larger version of this figure.
- Starting at the fimbrial region, using forceps as leverage, slit the tube longitudinally with spring-form scissors.
- Mince the slit-open portions and the remaining oviduct into segments (~1-2 mm in size) and add 5 mL of warm, non-enzymatic dissociation buffer (see Table of Materials) and incubate at 37 °C.
- Resuspend the tissue pieces and dissociated cells with a bulb pipettor every 3 min for 9-10 min. Dilute the dissociation buffer 1:1 with cold dissection medium.
- Collect the cells by centrifugation (350 x g, 3 min, 4 °C). Resuspend the cells in 2 mL of RBC lysis buffer for 2 min at room temperature, and then dilute 1:1 with cold dissection medium.
- Collect the cells by centrifugation (350 x g, 3 min, 4 °C) and resuspend in 5 mL of cold pronase solution.
- Incubate in pronase digestion medium and resuspend the cell clumps with a bulb pipettor every 3-5 min until a single cell suspension is achieved (~25-30 min).
NOTE: This step is best performed in a tissue culture plate or flask to allow constant observation preventing unnecessary overexposure to pronase.
- Incubate in pronase digestion medium and resuspend the cell clumps with a bulb pipettor every 3-5 min until a single cell suspension is achieved (~25-30 min).
- Pass the cells through a sterile macro-mesh (1 mm x 1 mm mesh affixed to a conical tube with a rubber band (Figure 4B)) to remove any remaining undissociated tissue pieces. Collect the cells by centrifugation (350 x g, 3 min, 4 °C) and resuspend them in a cold medium appropriate for downstream analysis (e.g., FACS buffer if proceeding with staining for flow cytometry).
5. RNA extraction of oviductal segments
- Homogenize snap-frozen samples in 200 µL of RNA extraction reagent in a bead bullet blender tissue homogenizer on level 12 for two intervals (3 min each) at 4 °C using the stainless-steel bead mix.
NOTE: To preserve RNA integrity, samples should be immediately moved from snap-frozen state into RNA extraction reagent and not weighed. - Briefly centrifuge homogenate at 100-200 x g in a tabletop centrifuge for approximately 5 s at room temperature to separate the liquid from the beads. Transfer the supernatant to a fresh tube.
- Add another 200 µL of RNA extraction reagent to the beads to recover more samples, followed by centrifugation (100-200 x g, 5 s, room temperature). Combine the supernatants.
NOTE: Use a small pipette tip (most p200 tips are suitable) to prevent bead carryover when removing the supernatant. - Follow manufacturer's guidelines to phase separate and precipitate RNA. Representative samples were precipitated overnight at -20 °C with 100% isopropanol (approximately 2:1 volume, isopropanol: recovered aqueous phase).
- Transfer the complete RNA precipitate along with the solution to an RNA-binding spin column and centrifuge for 30 s at 9,500 x g. Purify the RNA on-column as per the manufacturer's guidelines, and then elute the RNA in 20 µL of nuclease-free water. Assess the quality/quantity on a microvolume spectrophotometer and/or bioanalyzer.
Representative Results
The described dissociation protocol yields 100,000-120,000 cells per mouse with pooling of both oviducts. The method is gentle enough to leave multi-ciliated cell borders intact, allowing for a distinction between multi-ciliated cells and secretory cells, and verifying that the digestion method is gentle enough to prevent de-differentiation. Representative immunofluorescence images in Figure 5 show small cell clumps following step 4.2.1, fixed for 3 min in 4% paraformaldehyde (PFA)/ DPBS, washed with 1% bovine serum albumin (BSA)/ Tris-buffered saline-tween (BTBST) and stained in suspension. Briefly, cells were permeabilized for 15 min at room temperature in 0.5% TritonX-100/BTBST, washed in BTBST, quenched for background fluorescence for 2 h at room temperature in 20 mM glycine/ DPBS, washed in BTBST, and then blocked for 1 h at room temperature in 5% BSA/ 0.1% TritonX-100/ TBST. Cells were then incubated in the primary antibody at 4 °C overnight in BTBST (mouse anti-mouse occludin (1:2000); rabbit anti-mouse acetylated tubulin (1:1000)) followed by several washes in BTBST. Secondary antibody was added for 1 h at room temperature in BTBST (goat anti-mouse IgG Alexa Fluor 555 (1:1000); goat anti-rabbit IgG Alexa Fluor 488 (1:1000)), washed several times in BTBST, once in DPBS, and then incubated in nuclear stain for 20 min at room temperature (1:2000, Hoescht 33342) followed by a wash in DPBS. Cells were resuspended in 80 µL of antifade mounting medium and left overnight in the dark before imaging (Figure 5A). Figure 5B shows cells at the end of the dissociation protocol. Viability assessments were made throughout the protocol, and representative images are shown in Figure 5B–D. Propidium iodide was added at 1:100 dilution, and cells were immediately observed on an inverted compound fluorescence microscope (Figure 5C, red stain). Cilia remained intact in small cell clusters (Figure 5A) and single cell suspensions (Figure 5E) with many cells observed to have cilia still beating (Suppl. Video 1).
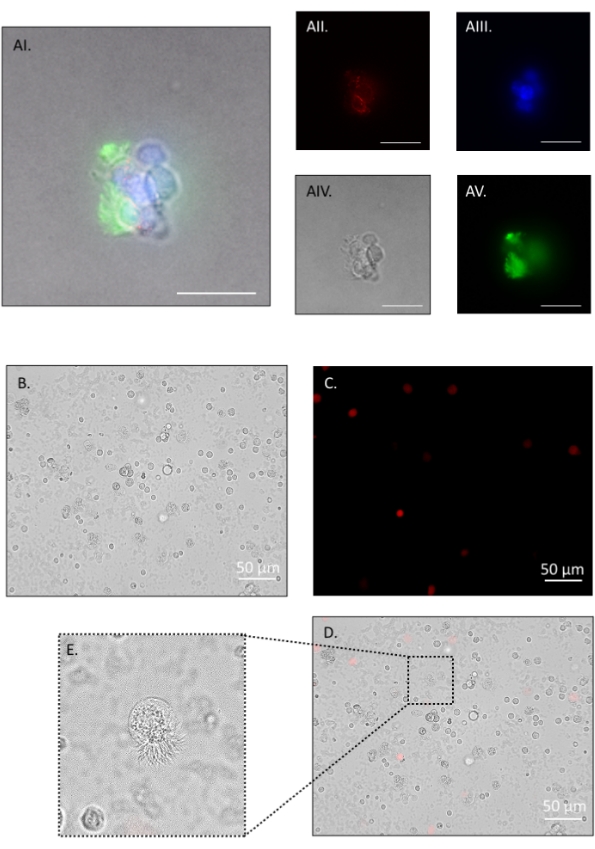
Figure 5: Oviduct cell dissociation, part 2. Fluorescent and phase-contrast images of a cell cluster positive for occludin (red), nuclei (blue), and acetylated tubulin (green). The ciliated apical border (green in AI and AV) remains intact throughout the protocol and withstands further digestion to single cells (E). AI: merge, AII: red channel, AIII: blue channel, AIV: phase contrast, AV: green channel. After a brief pronase incubation, the majority of cells are single. Propidium iodide (PI) staining shows approximately 93% viability. (B) Bright field, (C) red channel PI positive, (D) merge. (E) inset on merge shows an intact ciliated border on a single dissociated cell. Such cells had actively beating cilia (see Suppl. Video 1). Images were taken on an inverted compound fluorescence microscope. Figure AI-V scale bar = 20 μm, B-D scale bar = 50 μm. Please click here to view a larger version of this figure.
The RNA isolation protocol described gives sufficient yield and purity necessary for segmental RT-qPCR and RNAseq. Results show that this method yields 800-1200 ng RNA per segment (Figure 6A) except for infundibular samples where pooling from two animals may be necessary for appropriate yield for downstream assays. The presented results for infundibulum are pooled from two animals, totaling four infundibular regions (Figure 6). This protocol is successful in achieving pure RNA, initially analyzed by microvolume spectrophotometer and further by bioanalyzer (Figure 6B,C). Samples have a 260/280 nm absorption ratio between 1.8 and 2.0 (Figure 6B) typical of pure RNA nucleotide species19. Absorption ratios (260/230 nm) were taken for each sample with an inclusion criterion between 1 and 2.2, representative of limited contamination from isolation reagents (data not shown)19. Bioanalyzing the samples showed high integrity of the RNA, with assessed RNA integrity number (RIN) above seven (Figure 6C), appropriate for downstream expression and sequencing analysis20.

Figure 6: Quality assessment of segmental RNA. Whole RNA was assessed for yield (A) and quality (B,C) by microvolume spectrophotometer and bioanalyzer. RIN: RNA integrity number, as assessed by the bioanalyzer. Bars are the mean ± standard deviation (SD), N = 12 samples per segment from two separate RNA extractions. Please click here to view a larger version of this figure.
Supplemental Video 1: Live cell imaging of beating cilia on dissociated cells. Beating cilia following pronase digestion. Videos were taken on an inverted compound microscope. Please click here to download this Video.
Discussion
The three segments of the oviduct are histologically, morphologically, and functionally distinct1,2,3. The epithelium varies greatly from one end of the oviduct to the other. Ciliated cells dominate at the fimbrial/infundibular end, while secretory cells dominate in the isthmic region1. While this overall gradient has been recognized for some time, recent work has uncovered more distinctions among the oviductal segments. Thus, while ciliated cells become less frequent toward the proximal end of the tube, there is a sharp drop in their number (from approximately 60% to 10% ciliation) transitioning from the ampulla to the ampullary-isthmic junction3. Further, epithelial cell populations in the proximal and distal segments are derived from developmentally distinct lineages1,2,3. Distal epithelial cells of the infundibulum and ampulla do not contribute to proximal epithelial cell populations in the isthmus, as shown by lineage tracing of epithelial markers. At embryonic day 12.5 (E12.5), oviductal epithelial cells have a distinct lineage independent of the proximal epithelial cell marker, PAX22. These discoveries, in addition to the diverse cell population among the various segments, further emphasize the necessity to investigate the segments independently. The described method details a step-by-step unwinding and segment identification protocol as per the three historically described segments (infundibulum, ampulla, and isthmus). Due to the recent establishment of the ampullary-isthmic junction as a definitive oviduct segment, priority was given to the identification and collection of the three classically described areas. However, the method can be extended to include a separate collection of the ampullary-isthmic junction, as described above3.
The use of the blue dye and location of the infundibulum is a critical step to unwind the oviduct in a fast and efficient way. The blue dye highlights the ostium and coils of the tube, allowing a faster and more efficient de-coiling and collection. The initial location of the ostium orients one to the distal (infundibular) and proximal (isthmic) areas of the oviduct throughout uncoiling. In addition, keeping the proximal uterine horn portion affixed to the dissection platform during uncoiling allows the surgeon to use both hands for the delicate dissection steps. Initial removal of the coiled oviduct and attempts to uncoil this structure without rendering the tube immobile or without the use of the dye can easily lead to confusion regarding the distal and proximal ends or cause breakage of the tube. Tube breakage is easy, so care must be exercised when uncoiling since breakage will make reproducible segmentation unachievable. The use of high-quality, spring-form micro-scissors is highly recommended to achieve the best results.
Evaluating the gene expression signature in the diverse segments of the oviduct, for example, during the estrous cycle and in response to administered hormones and cytokines, will most likely produce a much better understanding of how the oviduct changes in response to these stimuli. These data may also shed light on what factors contribute to epithelial transformation. The RNA extraction protocol herein was developed to obtain the highest quantity and quality possible for this tissue of limited size and resilient tubal structure. The described homogenization, RNA precipitation, and conjunctional use of the phenol: chloroform and on-column purification methods were found to be most efficient in achieving appropriate quality and quantity of RNA for downstream analysis. The oviductal segments were not weighed and entire segments were immediately processed for tissue homogenization in order to prevent any warming of the tissue leading to RNA degradation. In addition, because the small tubal segments require multiple homogenization steps, it is critical to ensure that homogenization is conducted at 4 °C to preserve sample integrity. Because this method is sufficient to produce appropriate yield for downstream analysis, a future application of this technique would be to perform segmental RNAseq, previously not reported, which would contribute greatly to our understanding of oviduct physiology.
During the development of the cell dissociation protocol, we found that robust and/or long enzymatic digestions caused demonstrable loss of cilia. We, therefore, concentrated on the development of, and achieved, a method that preserved cell morphology. Improvements in single cell yield, as described herein, will allow more robust and immediate multiplexed analyses of the different epithelial and stromal cell types by flow cytometry. The use of a non-enzymatic dissociation protocol followed by a short incubation in pronase is also likely to improve the preservation of cell surface antigens over dissociation methods utilizing long or robust digestion methods2,18. Single cell sequencing analysis requires far fewer cells compared to multiplexed flow cytometry panels2,21. So single cell sequencing of the three described segments of the oviduct may be possible with the described protocol.
Because the oviduct cannot be fileted along its entire length, a potential limitation of the current method could be that cells forming the epithelium of the narrower isthmus would have reduced exposure to the dissociation reagents and, therefore, may not be present in the final suspension in the same proportion as in vivo.
Due to the coiling of the oviduct, orientation for immunohistochemical analyses of all oviductal segments can be challenging3. However, the described segment dissection followed by oriented embedding of the oviductal segments allows for more comprehensive longitudinal and cross-sectional image analysis without having to serially section through the entire intact coiled tube. Further, the blue dye utilized for uncoiling, aids in correct orientation during embedment of the small tubal segments.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by a DoD Breakthrough Award to AMW (BCRP W81XWH-14-1-0425). KCR was partially supported by intramural fellowships: the Pease Cancer Pre-Doctoral Fellowship and the Mary Galvin Burden Pre-Doctoral Fellowship in Biomedical Sciences and University of California, Riverside, intramural awards: the Graduate Council Fellowship Committee Dissertation Research Grant and the Graduate Division Dissertation Year Program Award. The authors thank Gillian M. Wright and Alyssa M. Kumari for assistance in early troubleshooting of this method.
Materials
| 0.5 mm Stainless steel bead mix | Next Advance | SSB05 | Mix 1:1 with 1.4 mm SSB14B, sterilized |
| 1.4 mm Stainless steel bead mix | Next Advance | SSB14B | Mix 1:1 with 0.5 mm SSB05, sterilized |
| 1X Dulbecco's Phosphate Buffered Saline A, pH 7.4 (DPBS) | Gibco | 21600-010 | Cold, sterile |
| 25G needle | BD | 305122 | |
| 60 mm sterile petri dishes | Corning | 430166 | |
| 70 μm cell meshes | Fisherbrand | 22-636-548 | |
| Agilent Eukaryote Total RNA 6000 Pico Chip kit | Agilent 2100 Bioanalyzer | 5067-1513 | |
| Bead Bullet Blender Tissue Homogenizer | Next Advance | BBY24M | |
| Bioanlyzer | Agilent 2100 Bioanalyzer | ||
| Bovine serum albumin | Sigma Aldrich | A7906 | |
| Cold plate/pack/surface of choice | N/A | N/A | Kept at -20C for dissection |
| Dental wax | Polysciences Inc. | 403 | |
| Dulbecco’s Modified Eagle’s Medium (DMEM)/ Ham’s F12 | Corning | 10-090-CV | Prepare dissection medium: DMEM/ Ham’s F12, 10% FBS, 25 mM Hepes, 1% Pen-Strep |
| Fetal Bovine Serum | Corning | 35-015CV | |
| Fine point forceps of choice | N/A | N/A | |
| Glycine | Sigma Aldrich | G712b | Immunocytochemical validation images |
| Goat anti-mouse IgG Alexa Fluor 555 | Invitrogen | A-21422 | Immunocytochemical validation images |
| Goat anti-rabbit IgG Alexa Fluor 488 | Invitrogen | A-11001 | Immunocytochemical validation images |
| Hepes | Sigma Aldrich | H-3784 | |
| Hoescht 33342 | Cell Signaling Technologies | 4082S | Immunocytochemical validation images |
| Inverted compound microscope | Keyence BZ-X700 | ||
| Mouse anti-mouse Occludin | Invitrogen | 33-1500 | Immunocytochemical validation images |
| Non-enzymatic dissociation buffer | N/A | 5 mM EDTA, 1 g/L glucose, 0.4% BSA, 1X DPBS | |
| Nylon macro-mesh 1 mm x 1 mm | Thomas Scientific | 1210U04 | |
| Paraformaldehyde | Sigma Aldrich | P-6148 | Immunocytochemical validation images |
| Pen-Strep | MP Biomedicals | 10220-718 | |
| Prolong Gold Antifade Reagent | Cell Signaling Technologies | 9071S | Immunocytochemical validation images: antifade mounting medium |
| Pronase | Sigma Aldrich | 10165921001 | Prepare pronase digestion medium: 0.15% Pronase in DMEM/Ham's F12, sterile |
| Propidium Iodide | Roche | 11 348 639 001 | Viability validation images |
| Rabbit anti-mouse Acetylated-Tubulin | Abcam | ab179484 | Immunocytochemical validation images |
| RBC lysis buffer | BD Biosciences | 555899 | |
| RNeasy Mini Kit | Qiagen | 74134 | Utilized for on-column purification in text. |
| Spring form microdissection scissors | Roboz Surgical | RS-5610 | |
| Sterile 3 mL bulb pipettors | Globe Scientific | 137135 | |
| Toluidine blue | Alfa Aesar | J66015 | Prepare toluidine blue solution: 1% in 1X DPBS, sterile |
| Tris-Buffered Saline-Tween (TBST) | N/A | N/A | Immunocytochemical validation images; 0.1 N NaCl, 10 mM Tris-Cl pH 7.5, 1% Tween 20 |
| Triton-X-100 | Mallinckrodt Inc. | 3555 | Immunocytochemical validation images |
| Trizol RNA Extraction Reagent | Invitrogen | 15596026 | Referred to as RNA extraction reagent. Nucelase-free water, chloroform and isoproponal are required in supplement of performing Trizol extraction per manufacturer's guidelines |
References
- Stewart, C. A., et al., Kubiak, J. Z., et al. . Mouse oviduct developmemt in Mouse Development: From Oocyte to Stem Cells. , 247-262 (2012).
- Ford, M. J., et al. Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the distal-proximal axis. Cell Reports. 36 (10), 109677 (2021).
- Harwalkar, K., et al. Anatomical and cellular heterogeneity in the mouse oviduct-its potential roles in reproduction and preimplantation development. Biology of Reproduction. 104 (6), 1249-1261 (2021).
- Agduhr, E. Studies on the structure and development of the bursa ovarica and the tuba uterina in the mouse. Acta Zoologica. 8 (1), 1 (1927).
- Pulkkinen, M. O. Oviductal function is critical for very early human life. Annals of Medicine. 27 (3), 307-310 (1995).
- Kindelberger, D. W., et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: Evidence for a causal relationship. American Journal of Surgical Pathology. 31 (2), 161-169 (2007).
- Ghosh, A., Syed, S. M., Tanwar, P. S. In vivo genetic cell lineage tracing reveals that oviductal secretory cells self-renew and give rise to ciliated cells. Development. 144 (17), 3031-3041 (2017).
- Lee, Y., et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. Journal of Pathology. 211 (1), 26-35 (2007).
- Kim, J., et al. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proceedings of the National Academy of Sciences of the United States of America. 109 (10), 3921-3926 (2012).
- Perets, R., et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 24 (6), 751-765 (2013).
- Sherman-Baust, C. A., et al. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. Journal of Pathology. 233 (3), 228-237 (2014).
- Zhai, Y. L., et al. High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. Journal of Pathology. 243 (1), 16-25 (2017).
- Karthikeyan, S., et al. Prolactin signaling drives tumorigenesis in human high grade serous ovarian cancer cells and in a spontaneous fallopian tube derived model. Cancer Letters. 433, 221-231 (2018).
- Shao, R., et al. Differences in prolactin receptor (PRLR) in mouse and human fallopian tubes: Evidence for multiple regulatory mechanisms controlling PRLR isoform expression in mice. Biology of Reproduction. 79 (4), 748-757 (2008).
- Alwosaibai, K., et al. PAX2 maintains the differentiation of mouse oviductal epithelium and inhibits the transition to a stem cell-like state. Oncotarget. 8 (44), 76881-76897 (2017).
- Peri, L. E., et al. A novel class of interstitial cells in the mouse and monkey female reproductive tracts. Biology of Reproduction. 92 (4), 102 (2015).
- Lõhmussaar, K., et al. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nature Communications. 11 (1), 2660 (2020).
- Chen, S., et al. An air-liquid interphase approach for modeling the early embryo-maternal contact zone. Scientific Reports. 7, 42298 (2017).
- Desjardins, P., Conklin, D. NanoDrop microvolume quantitation of nucleic acids. Journal of Visualized Experiments: JoVE. (45), e2565 (2010).
- Schroeder, A., et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 7, 3 (2006).
- McGlade, E. A., et al. Cell-type specific analysis of physiological action of estrogen in mouse oviducts. The FASEB Journal. 35 (5), 21563 (2021).

