A Microfluidic Platform to Study Bioclogging in Porous Media
Summary
The present protocol describes a microfluidic platform to study biofilm development in quasi-2D porous media by combining high-resolution microscopy imaging with simultaneous pressure difference measurements. The platform quantifies the influence of pore size and fluid flow rates in porous media on bioclogging.
Abstract
Bacterial biofilms are found in several environmental and industrial porous media, including soils and filtration membranes. Biofilms grow under certain flow conditions and can clog pores, thereby redirecting the local fluid flow. The ability of biofilms to clog pores, the so-called bioclogging, can have a tremendous effect on the local permeability of the porous medium, creating a pressure buildup in the system, and impacting the mass flow through it. To understand the interplay between biofilm growth and fluid flow under different physical conditions (e.g., at different flow velocities and pore sizes), in the present study, a microfluidic platform is developed to visualize biofilm development using a microscope under externally-imposed, controlled physical conditions. The biofilm-induced pressure buildup in the porous medium can be measured simultaneously using pressure sensors and, later, correlated with the surface coverage of the biofilm. The presented platform provides a baseline for a systematic approach to investigate bioclogging caused by biofilms in porous media under flow conditions and can be adapted to studying environmental isolates or multispecies biofilms.
Introduction
Biofilms – bacterial colonies embedded in a self-secreted matrix of extra-polymeric substances (EPS) – are ubiquitous in natural porous media, such as soils and aquifers1, and technical and medical applications, like bioremediation2, water filtration3 and medical devices4. The biofilm matrix is comprised of polysaccharides, protein fibers, and extracellular DNA5,6, and strongly depends on the microorganisms, the availability of nutrients, as well as the environmental conditions7. Yet, the functions of the matrix are universal; it forms the scaffold of the biofilm structure, protects the microbial community from mechanical and chemical stresses, and is largely responsible for the biofilms' rheological properties5.
In porous media, the growth of biofilms can clog pores, causing the so-called bioclogging. Biofilm development is controlled by the fluid flow and pore size, defined as the distance separating two pillars, of the porous medium8,9,10. Both the pore size and the fluid flow control the nutrient transport and local shear forces. In turn, the growing biofilm clogs pores, affecting the velocity distribution of the fluid11,12,13, the mass transport, and the hydraulic conductivity of the porous medium14,15. The changes in hydraulic conductivity are reflected through increased pressure in confined systems16,17,18,19. Current microfluidic studies in biofilm development and bioclogging focus on studying the impact of flow velocities in homogeneous geometries16,20 (i.e., with a singular pore size) or heterogeneous porous media12,21,22. However, to disentangle the effects of flow rates and pore size on biofilm development and the resulting pressure changes in the bioclogged porous medium, a highly controllable and versatile experimental platform allowing the study of different porous media geometries and environmental conditions in parallel is required.
The present study introduces a microfluidic platform that combines pressure measurements with simultaneous imaging of the evolving biofilm within the porous medium. Because of its gas-permeability, bio-compatibility, and flexibility in the channel geometry design, a microfluidic device made of polydimethylsiloxane (PDMS) is a suitable tool for studying biofilm development in porous media. Microfluidics allow the control of physical and chemical conditions (e.g., fluid flow and nutrient concentration) with high precision to mimic the environment of microbial habitats23. Further, microfluidic devices can easily be imaged with micrometric resolution using an optical microscope and coupled with online measurements (e.g., the local pressure).
In this work, the experiments focus on studying the impact of pore size in a homogeneous porous medium analog under controlled imposed flow conditions. The flow of a culture medium is imposed using a syringe pump, and the pressure difference through the microfluidic channel is measured simultaneously with pressure sensors. Biofilm development is initiated by seeding a planktonic culture of Bacillus subtilis in the microfluidic channel. Regular imaging of the evolving biofilm and image analysis allows one to obtain pore scale resolved information on the surface coverage under various experimental conditions. The correlated information of pressure change and the extent of bioclogging provides crucial input for permeability estimations of bioclogged porous media.
Protocol
1. Silicon wafer preparation
- Design the geometries of the microfluidic channel in computer-aided design (CAD; see Table of Materials) software and print it onto a transparent film to create the photomask (Figure 1A).
- Fabricate the master mold by soft lithography (under clean-room conditions) following the steps below.
- Bake the silicon wafer at 200 °C for 2 h.
- Place the wafer at the center of a spin-coater and pour SU8 3050 photoresist (see Table of Materials) onto the wafer. Spin coat at 1,700 rpm for 40 s with a 10 s/100 rpm ramp time.
NOTE: The spin-coating parameters were set to obtain a target thickness of 100 µm for the SU8 3050. - After the spin-coating process, soft bake the silicon wafer at 65 °C for 600 s and 95 °C for 2,700 s. Let the wafer cool at room temperature overnight.
NOTE: The overnight cooling enhances the adhesion of the SU8 to the wafer. - Place the photomask (step 1.1) onto the wafer and expose it to UV light, with an exposure energy of 250 mJ/cm2 and at a wavelength of 350 nm.
- Post-exposure, bake the exposed substrate at 65 °C for 60 s and 95 °C for 300 s.
- Develop the silicon wafer to obtain the master mold by immersing it in a beaker filled with mrDev600 developer (see Table of Materials). Gently shake the beaker for 1,800 s to wash out the unpolymerized resist. Then, splash wash by spraying isopropanol on the silicon wafer and air dry.
- Hard bake the silicon wafer at 200 °C for 1,800 s.
- Silanize the master mold through vapor deposition of 20 µL of Trichloro (1H, 1H, 2H, 2H-perfluorooctyl) silane (see Table of Materials) placed on a glass slide next to the mold for 40 min in a vacuum desiccator, creating a gauge pressure of 100 mbar.
2. Fabrication of the microfluidic device
NOTE: The fabrication procedure described here is for a microfluidic device with one microfluidic channel. However, the same method can be applied to fabricate a microfluidic device with multiple microfluidic channels in parallel.
- Mix the elastomer with its crosslinker at a ratio of 10:1 (see Table of Materials) to prepare a PDMS mixture. Stir the mixture until it gets uniformly mixed and turns opaque due to the enclosed air bubbles.
- Degas the mixture in a vacuum desiccator, creating a gauge pressure of 100 mbar until the entrapped air bubbles are removed and it looks transparent. The time required for degassing is typically 30 min.
- Place the master mold (step 1) in a cell culture dish (see Table of Materials). Pour 20 g of the PDMS mixture on the master mold to produce channels with a final thickness of 5 mm.
- Bake the master mold at 70 °C for 2 h.
- Cut the cured PDMS around the microfluidic channel (at a distance of approximately 3 mm) using a blade, and then peel the PDMS microfluidic channel off the master mold.
- To create the microfluidic channels' inlet and outlet, punch holes with a biopsy punch (diameter of 1.5 mm) at its extremities (top of the triangles, see Figure 1A). Punch one additional hole at the center of the inlet triangle to install the pressure sensor later.
- Wash a glass slide and the microfluidic channel with a commercially available 1% detergent solution (see Table of Materials) for 5 min, then rinse them with deionized water. Thereafter, wash the PDMS microfluidic channel and the glass slide with isopropanol. Then, rinse them again with deionized water. Dry the PDMS microfluidic channel and the glass slide with compressed air at 1 bar for 1 min.
NOTE: The porous structure of the PDMS must be completely dry for the bonding to be effective. - Place the glass slide and the microfluidic channel in a plasma cleaner (see Table of Materials) and ensure the surfaces to be bonded are facing up. Turn on the plasma cleaner and treat the microfluidic channel and glass slide with air plasma at an airflow of 1 SL/h (standard liter per hour) for 1 min. Bond the microfluidic channel to the glass slide immediately after taking them out of the cleaner by putting them in contact with each other.
NOTE: Ensure not to touch the treated surfaces, as this might affect the bonding. When fabricating a microfluidic device with multiple microfluidic channels, expose the microfluidic channels simultaneously and bond them in a single step. - Place the bonded microfluidic device on an 80 °C hot plate for at least 15 min.
- Store the microfluidic device in a clean cell culture dish until the experiment starts.
3. Preparation of the bacterial suspension
- Grow a population of Bacillus subtilis NCIB 3610 20 h prior to the start of the experiment by directly inoculating 3 mL of nutrient broth no. 3 culture medium (see Table of Materials) from a frozen glycerol stock in a 15 mL culture tube. Incubate in a shaking incubator at 30 °C and 200 rpm overnight (for 16 h).
- Make a subculture from the overnight culture 4 h prior to the start of the experiment by adding 3 µL of the overnight culture in 3 mL of fresh culture medium (1:1,000 dilution) in a 15 mL culture tube. Incubate the subculture in a shaking incubator at 30 °C at 200 rpm for 3.5-4 h to obtain an optical density at 600 nm (OD600) of 0.1.
4. Biofilm growth experiment
- Turn on the box incubator of the microscope 3 h before the experiment to ensure a stable temperature of 25 °C. Mount the syringe pump and the pressure sensors (see Table of Materials).
- Connect the inlet and outlet tubing to the microfluidic device. Directly insert a needle (with an outer diameter of 0.6 mm) into the inlet tubing to secure the connection between the tubing and the syringe.
- Place the microfluidic device, 30 mL of deionized water, and 30 mL of culture medium in a vacuum desiccator and degas them for at least 1 h. Then, slowly pull the culture medium and the deionized water into two separate 30 mL syringes.
NOTE: This step is crucial to prevent bubble formation in the channel while flushing with the culture medium. - Mount the microfluidic device on the microscope and place the outlet tubing in a waste container.
- Connect the syringe filled with deionized water to the microfluidic channel through the microfluidic tubing and slowly inject the water until it exits from the pressure sensor outlet. Fill the pressure sensor with water and flush all the bubbles from the tubing connecting the microfluidic channel and the pressure sensor. Close the outlet of the pressure sensor with the screws dedicated to the pressure sensor.
NOTE: The described filling procedure ensures that the pressure changes at the microfluidic channels' inlet will be precisely recorded. When running an experiment with multiple microfluidic channels, connect each channel to a separate syringe to ensure equal flow conditions in all channels.
- Connect the syringe filled with deionized water to the microfluidic channel through the microfluidic tubing and slowly inject the water until it exits from the pressure sensor outlet. Fill the pressure sensor with water and flush all the bubbles from the tubing connecting the microfluidic channel and the pressure sensor. Close the outlet of the pressure sensor with the screws dedicated to the pressure sensor.
- Fill the rest of the microfluidic channel with the deionized water.
- Place a 1.2 µm filter (see Table of Materials) on the culture media syringe. Then, remove the water syringe and carefully connect the culture media syringe to the inlet microfluidic tubing. Mount the syringe on the syringe pump and flush the channel with the culture medium at a flow rate of 2 mL/h for 1 h.
NOTE: The filter prevents bacterial cells from entering the syringe during loading. Flushing the microfluidic channel with the culture media will remove the remaining bubbles in the porous structure. - Set the syringe pump at the desired flow rate (here 1 mL/h) during the experiment and set the pressure reading of the pressure sensors to zero.
NOTE: By setting the initial pressure reading to zero, only the pressure difference caused by biofilm development during the experiment will be measured. - Pipette 1 mL of the bacterial culture at an OD600 of 0.1 in a 1.5 mL centrifuge vial. Load the bacterial culture into the microfluidic channel by placing the outlet tube into the centrifuge vial. After waiting for 5 min to remove any potential air bubbles from the tubes' outlet, withdraw 150 µL of bacterial solution at a flow rate of 1 mL/h, until the microfluidic channel is filled with the bacterial culture.
- Carefully remove the culture media syringe filter and place the outlet into the waste container. Leave the bacterial cells at zero-flow conditions in the microfluidic channel for 3 h to allow their surface attachment in the porous medium.
NOTE: Leaving the bacterial cells at zero-flow conditions for 3 h was optimized for the attachment of the bacterial strain used while assuring a well-oxygenated bacterial culture. Other bacterial strains might require more or less time. - To start the experiment, start the flow by setting the syringe pump to the desired flow rate (here 1 mL/h) and start the pressure reading at 1 Hz.
- Acquire images of the growing biofilm at the desired time interval, optical configuration, and magnification.
NOTE: In the present study, images at 4x magnification in the bright-field mode in 18 positions spanning the entire domain of the porous medium were acquired every 6 min for 24 h.
5. Image analysis
- Reconstruct the entire porous medium from the image sequences recorded by stitching the images from the 18 positions using image analysis software (see Table of Materials) and a stitching algorithm24.
- Save the stitched image sequences as a sequence of singular images.
NOTE: If the files are too big, at this point, the images can be rescaled and binned to a reasonable size for further processing. - Create a mask of the pillars of the porous medium to remove them from the analysis.
- Remove the background of the images by dividing them by their background with the image taken at t = 0 h (Figure 2A) and binarize the images at a threshold suitable to segment the biofilm (Figure 2B).
- Compute the saturation of the biofilm by calculating the area covered by the biofilm (number of pixels attributed to the biofilm) compared to the void area of the porous domain (Figure 3).
Representative Results
For the present study, a microfluidic device with three parallel microfluidic channels with different pore sizes was used (Figure 1) to study biofilm formation in porous media systematically. The biofilm formation process was visualized using bright-field microscopy. The bacterial cells and the biofilm appeared in the images as darker pixels (Figure 2). In addition, a gradual clogging process was observed; during a 24 h experiment, the initially randomly growing biofilm colonized almost the entire porous medium.
The surface coverage of the biofilm in time grown at a flow rate of Q = 1 mL/h, which corresponds to a mean initial fluid flow velocity of 0.96 mm/s, was quantified for three different pore sizes (75 µm, 150 µm, and 300 µm) (Figure 3, black lines). It was found that the surface coverage, which was used as a proxy for the bioclogging degree, occurred 10% faster at the smallest pore size of 75 µm than at the biggest pore size (300 µm) when comparing the surface coverage at t = 20 h. Then, the surface coverage was correlated to the pressure buildup caused by the biofilm (Figure 3, blue lines). The clogging in the smaller pore size microfluidic channel led to a higher pressure difference between the inlet and the outlet than in the larger pore size microfluidic channels, indicating that smaller-sized porous media will develop higher pressure buildup when subjected to bioclogging.
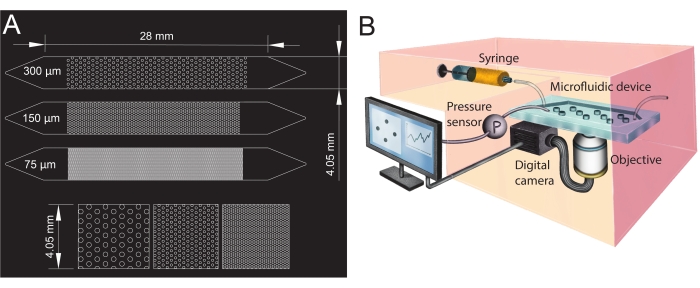
Figure 1: Microfluidic channel design and experimental setup. (A) Photomask of the microfluidic channels with different pore sizes (75 µm, 150 µm, and 300 µm) used as porous media analogs and a zoomed-in view of the pillars' arrangement (bottom row). The circles show the location of the pillars (impermeable obstacles), representing the porous media's solid phase. (B) Schematic of the experimental setup showing the syringe, the pressure sensor, the microfluidic device (with a single microfluidic channel), and the digital camera setup with the objective (i.e., the microscope). Please click here to view a larger version of this figure.
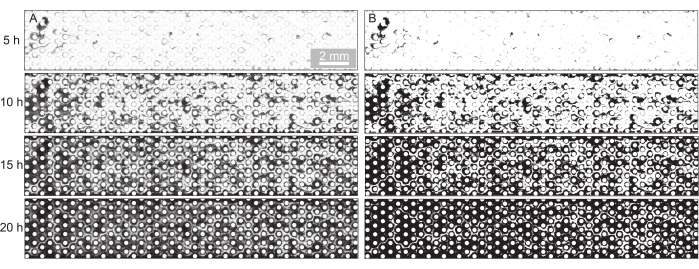
Figure 2: Visualization and quantification of biofilm development in the porous medium. (A) Representative image sequence of the biofilm development at the imposed flow rate of Q = 1 mL/h (corresponds to a mean initial fluid flow velocity of 0.96 mm/s) and a pore size of d = 300 µm shown for the experimental time points t = 5 h, t = 10 h, t = 15 h, and t = 20 h. The bright-field images were stitched, and the background was removed. (B) The binarization of these images and quantification of the area occupied by biofilm (dark pixels) led to the quantification of surface coverage in Figure 3. Please click here to view a larger version of this figure.
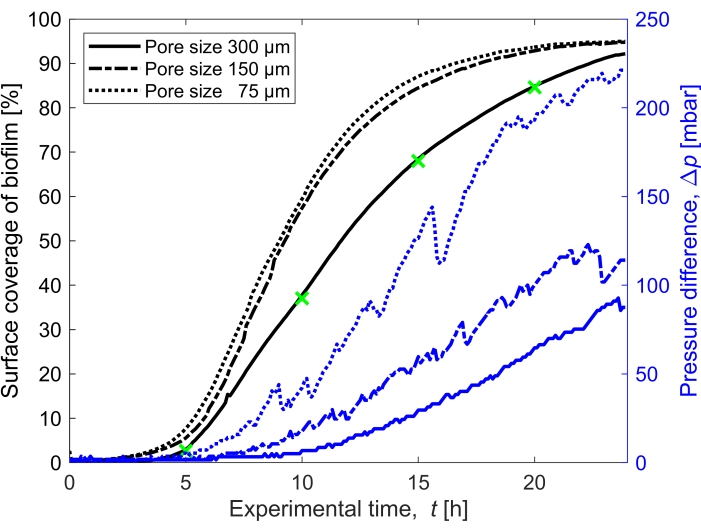
Figure 3: Temporal evolution of biofilm coverage and impact on pressure. Biofilm coverage with simultaneous pressure reading for the three pore sizes (300 µm, 150 µm, and 75 µm) in the same experimental conditions as Figure 2. The pressure difference caused by the biofilm in the porous medium microfluidic channel, Δp, (blue lines) shown on the right y-axis, increases with an increased surface coverage of the biofilm (black lines). The green markers correspond to the data points of the images shown in Figure 2. Please click here to view a larger version of this figure.
Discussion
Microfluidic porous media analogs coupled with pressure sensors provide a suitable tool to study biofilm development in porous media. The versatility in the design of the microfluidic porous medium, specifically the arrangement of the pillars, including diameter, irregular shapes, and pore size, allows the investigation of many geometries. These geometries range from single pores to highly complex, irregularly arranged obstacles mimicking different natural (e.g., soils) and industrial (e.g., membranes and filters) porous media. In the present microfluidic platform, three porous media geometries were created with regularly arranged cylindrical pillars (pore sizes: 75 µm, 150 µm, and 300 µm), where the fluid flow rate could be chosen per experiment. The presented platform can be easily adapted to study bioclogging with a fixed pressure head rather than an imposed fluid flow rate. In this case, the flow control device should be a pressure controller with a culture medium reservoir instead of a syringe pump. The resulting changes in flow rate due to bioclogging could be monitored by measuring the outflow over time using a flow rate sensor.
Several critical points must be considered to run a successful microfluidic experiment with biofilm growth. To avoid air bubble formation in the microfluidic channel during the experiment, the microfluidic channel and the culture medium were degassed (step 4.3). Next, filling of the microfluidic channel with the degassed culture medium must be conducted rapidly but carefully to obtain a fully saturated channel without any air bubbles. In case air bubbles are trapped in the porous medium, flushing the microfluidic channel at a higher flow rate can remove the bubbles after a short time. The second crucial step is to ensure a constant temperature environment to reproduce biofilm growth consistently. The growth of microorganisms varies with temperature25, which might lead to non-reproducible results when not keeping the temperature stable during the experiment (in this case, 24 h). For the present platform, a box incubator was used around the microscope, though a smaller temperature-stable casing for the microfluidic device would likely be sufficient too. Finally, during the image acquisition, the positions of the individual images should be chosen with an overlap of at least 15% to obtain enough overlap for the stitching algorithm24.
The present microfluidic platform is limited to two-dimensional observation, whereas porous media applications like soil or membranes have a three-dimensional structure. However, advantages of the quasi-2D microfluidic platform compared to 3D porous media platforms to study bioclogging are the full optical access and the high time resolution, as 3D platforms usually perform endpoint imaging26,27. In addition, it is expected that the bioclogging process (i.e., the time evolution of surface coverage) persists in 3D systems26,27, as it also occurs for the cluster size distribution of an immiscible phase within porous media28, which presents the same scaling in 2D and 3D systems.
This method allows measuring the pressure response to biofilm growth in porous media while studying its spatio-temporal development at high temporal and spatial resolution and different pore sizes. The data sets obtained from such measurements bring insight into the correlation of pore-scale biofilm development with pressure responses of the biofilm-porous medium system, and can provide a benchmark for the numerical modeling of biofilms. These modeling efforts are especially relevant to extending the range of conditions (e.g., pore sizes, flow velocities, and biofilm properties for other species or multispecies biofilms) that exceed experimental capacities. The latter is highly relevant to understanding the mechanisms of bioclogging in the vicinity of wells, bioremediation applications, and biomineralization29,30,31. Overall, this method could easily be adapted to study biomineralization or track the biotransformation of contaminants by biofilms in porous media.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge support from SNSF PRIMA grant 179834 (to E.S.), discretionary funding from ETH (to R.S.), ETH Zurich Research Grant (to R.S. and J.J.M.), and discretionary funding from Eawag (to J.J.M.). The authors would like to thank Roberto Pioli for illustrating the experimental setup in Figure 1B and Ela Burmeister for the silicon wafer preparation.
Materials
| Acrodisc 25 mm Syringe Filter, 1.2 µm Versapor Membrane | Pall Corporation | PN4190 | 1.2 µm filters |
| BD 10 mL Syringe (Luer-Lock) | BD | 300912 | used to fill the channel with deionised water |
| Box Incubator | Life Imaging Services | used to have a stable temperature during the biofilm growth experiment | |
| Cell density meter CO8000 | WPA biowave | OD meter | |
| Centrifuge vial | Eppendorf | 30120086 | 1.5 mL |
| CETONI Base 120 | CETONI GmbH | syringe pump | |
| CorelCAD | CorelDRAW | software used to design the microfluidic channel geometries | |
| Culture tubes (14 mL, sterile) | greiner bio-one | Culture tubes | |
| Drying oven, VENTI-Line | VWR | Oven to cure the PDMS | |
| Handy | Migros | Detergent solution | |
| Hot plate with temperature control | VRW | to cure the PDMS-glass bonding after plasma treatment | |
| ImageJ | FIJI | Image analysis software | |
| Innova 42 Inc Shaker (New Brunswick) | Eppendorf | Incubator | |
| Isopropanol (> 99.8%) | Sigma Aldrich | 67-63-0 | |
| Masterflex transfer tubing | Masterflex | HV-06419-05 | 0.020'' ID, 0.06'' OD |
| Micro Slides, Plain, 75 x 60 mm | Corning | 2947-75X50 | Glass slides |
| Microfluidic pressure sensor (1 bar) | Elveflow | Pressure sensors | |
| Miltex Biopsy puncher, diameter 1.5 mm | Integra | Puncher to make the inlet and outlet holes of the microfluidic channel | |
| mrDev600 developer | Microresist | ||
| Nikon Eclipse Ti2 | Nikon Instruments | Microscope | |
| Nutrient broth n°3 | Sigma Aldrich | ||
| Omnifix Syringe with Luer-Lock | B.Braun | syringes of different volume | |
| Plasma chamber Zepto | Diener Electronic | ZEPTO-1 | used to plasma bond the PDMS and the glass slide |
| Precision wipes (Kimtech Science) | Kimberly Clark | KCP-7552 | to dry the glass slide |
| Scale | VWR-CH | 611-2605 | used to weigh the elastomer to crosslinking agent ratio |
| Silicon wafer (10 cm) | Silicon Materials Inc. | N//Phos <100> 1-10 Ω cm | |
| Spincoater, Spin module SM150 | Sawatec | ||
| SU8 3050 Photoresist | Kayakuam | ||
| Süss MA6 Mask aligner | SUSS MicroTec Group | used to align the chrome-glass mask | |
| Sylgard 184 | Dow Corning | silicone elastomer kit; curing agent | |
| Techni Etch Cr01 | Technic | Technic | |
| Tissue culture dish 150 | TPP | 93150 | |
| Trichloro (1H, 1H, 2H, 2H perfluorooctyl) silane | Sigma Aldrich | Sigma Aldrich | used to silanize the silicane wafer |
| Veeco Dektak 6 M | Veeco | Profilometer |
Referencias
- Flemming, H. C., Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nature Reviews Microbiology. 17 (4), 247-260 (2019).
- Cunningham, A. B., Sharp, R. R., Hiebert, R., James, G. Subsurface biofilm barriers for the containment and remediation of contaminated groundwater. Bioremediation Journal. 7 (3-4), 151-164 (2003).
- Pronk, W., et al. Gravity-driven membrane filtration for water and wastewater treatment: A review. Water Research. 149, 553-565 (2019).
- Caldara, M., Belgiovine, C., Secchi, E., Rusconi, R. Environmental, microbiological, and immunological features of bacterial biofilms associated with implanted medical devices. Clinical Microbiology and Infection. 35 (2), 00221 (2022).
- Flemming, H. C., Wingender, J. The biofilm matrix. Nature Reviews Microbiology. 8 (9), 623-633 (2010).
- Davey, M. E., O’Toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiology and Molecular Biology Reviews. 64 (4), 847-867 (2000).
- Stoodley, P., Dodds, I., Boyle, J. D., Lappin-Scott, H. M. Influence of hydrodynamics and nutrients on biofilm structure. Journal of Applied Microbiology. 85 (1), 19-28 (1998).
- Thomen, P., et al. Bacterial biofilm under flow: First a physical struggle to stay, then a matter of breathing. PLoS ONE. 12 (4), 0175197 (2017).
- Horn, H., Reiff, H., Morgenroth, E. Simulation of growth and detachment in biofilm systems under defined hydrodynamic conditions. Biotechnology and Bioengineering. 81 (5), 607-617 (2003).
- Thullner, M., Mauclaire, L., Schroth, M. H., Kinzelbach, W., Zeyer, J. Interaction between water flow and spatial distribution of microbial growth in a two-dimensional flow field in saturated porous media. Journal of Contaminant Hydrology. 58 (3-4), 169-189 (2002).
- Bottero, S., et al. Biofilm development and the dynamics of preferential flow paths in porous media. Biofouling. 29 (9), 1069-1086 (2013).
- Durham, W. M., Tranzer, O., Leombruni, A., Stocker, R. Division by fluid incision: Biofilm patch development in porous media. Physics of Fluids. 24 (9), 091107 (2012).
- Coyte, K. Z., Tabuteau, H., Gaffney, E. A., Foster, K. R., Durham, W. M. Microbial competition in porous environments can select against rapid biofilm growth. Proceedings of the National Academy of Sciences. 114 (2), 161-170 (2017).
- Taylor, S. W., Jaffé, P. R. Biofilm growth and the related changes in the physical properties of a porous medium: 1. Experimental investigation. Water Resources Research. 26 (9), 2153-2159 (1990).
- Cunningham, A. B., Characklls, W. G., Abedeen, F., Crawford, D. Influence of biofilm accumulation on porous media hydrodynamics. Environmental Science and Technology. 25 (7), 1305-1311 (1991).
- Valiei, A., Kumar, A., Mukherjee, P. P., Liu, Y., Thundat, T. A web of streamers: Biofilm formation in a porous microfluidic device. Lab on a Chip. 12 (24), 5133-5137 (2012).
- Biswas, I., Sadrzadeh, M., Kumar, A. Impact of bacterial streamers on biofouling of microfluidic filtration systems. Biomicrofluidics. 12 (4), 044116 (2018).
- Hassanpourfard, M., Ghosh, R., Thundat, T., Kumar, A. Dynamics of bacterial streamers induced clogging in microfluidic devices. Lab on a Chip. 16 (21), 4091-4096 (2016).
- Stewart, T. L., Scott Fogler, H. Pore-scale investigation of biomass plug development and propagation in porous media. Biotechnology and Bioengineering. 77 (5), 577-588 (2002).
- Hassanpourfard, M., Ghosh, R., Thundat, T., Kumar, A. Dynamics of bacterial streamers induced clogging in microfluidic devices. Lab on a Chip. 16 (21), 4091-4096 (2016).
- Aufrecht, J. A., et al. Pore-scale hydrodynamics influence the spatial evolution of bacterial biofilms in a microfluidic porous network. PLoS ONE. 14 (6), 0218316 (2019).
- Karimifard, S., Li, X., Elowsky, C., Li, Y. Modeling the impact of evolving biofilms on flow in porous media inside a microfluidic channel. Water Research. 188, 116536 (2021).
- Yawata, Y., Nguyen, J., Stocker, R., Rusconi, R. Microfluidic studies of biofilm formation in dynamic environments. Journal of Bacteriology. 198 (19), 2589-2595 (2016).
- Preibisch, S., Saalfeld, S., Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25 (11), 1463-1465 (2009).
- Ratkowsky, D. A., Olley, J., McMeekin, T. A., Ball, A. Relationship between temperature and growth rate of bacterial cultures. Journal of Bacteriology. 149 (1), 1-5 (1982).
- Ostvar, S., et al. Investigating the influence of flow rate on biofilm growth in three dimensions using microimaging. Advances in Water Resources. 117, 1-13 (2018).
- Carrel, M., et al. Biofilms in 3D porous media: Delineating the influence of the pore network geometry, flow and mass transfer on biofilm development. Water Research. 134, 280-291 (2018).
- Iglauer, S., Favretto, S., Spinelli, G., Schena, G., Blunt, M. J. X-ray tomography measurements of power-law cluster size distributions for the nonwetting phase in sandstones. Physical Review E. 82 (5), 10-12 (2010).
- Wu, C., Chu, J., Wu, S., Cheng, L., van Paassen, L. A. Microbially induced calcite precipitation along a circular flow channel under a constant flow condition. Acta Geotechnica. 14 (3), 673-683 (2019).
- Nassar, M. K., et al. Large-scale experiments in microbially induced calcite precipitation (MICP): reactive transport model development and prediction. Water Resources Research. 54 (1), 480-500 (2018).
- Jimenez-Martinez, J., Nguyen, J., Or, D. Controlling pore-scale processes to tame subsurface biomineralization. Reviews in Environmental Science and Biotechnology. 21 (1), 27-52 (2022).

