Determining Viral Disinfection Efficacy of Hot Water Laundering
Summary
In response to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, a laboratory protocol was developed to test the viral disinfection efficacy of hot water laundering of cloth face coverings, cotton scrubs, and denim pants. The Phi6 virus (bacteriophage) was used as the organism to test disinfection efficacy.
Abstract
This protocol provides an example of a laboratory process for conducting laundering studies that generate data on viral disinfection. While the protocol was developed for research during the coronavirus disease 2019 (COVID-19) pandemic, it is intended to be a framework, adaptable to other virus disinfection studies; it demonstrates the steps for preparing the test virus, inoculating the test material, assessing visual and integrity changes to the washed items due to the laundering process, and quantifying the reduction in viral load. Additionally, the protocol outlines the necessary quality control samples for ensuring the experiments are not biased by contamination and measurements/observations that should be recorded to track the material integrity of the personal protective equipment (PPE) items after multiple laundering cycles. The representative results presented with the protocol use the Phi6 bacteriophage inoculated onto cotton scrub, denim, and cotton face-covering materials and indicate that the hot water laundering and drying process achieved over a 3-log (99.9%) reduction in viral load for all samples (a 3-log reduction is the disinfectant performance metric in U.S. Environmental Protection Agency’s Product Performance Test Guideline 810.2200). The reduction in viral load was uniform across different locations on the PPE items. The results of this viral disinfection efficacy testing protocol should help the scientific community explore the effectiveness of home laundering for other types of test viruses and laundering procedures.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused unprecedented global supply chain disruption and led to a critical shortage of many items, including essential personal protective equipment (PPE)1,2,3. Those in high-risk occupations had to adapt using recommended crisis capacity strategies and the public adopted the use of non-specialized items such as cloth material face coverings primarily for source control, but also to provide some respiratory protection for wearers. In the United States, specialized respiratory protection (i.e., filtering facepiece respirators (FFRs) such as N95s) was reserved for some of these high-risk occupations (e.g., healthcare) during supply shortages4. When little was known about severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) transmission, a variety of other types of clothing materials were also considered as barrier protection early in the pandemic5. With the diversity of fabrics being used for wearer protection, questions arose about the use, reuse, and disinfection/decontamination of these items. While in the United States it was generally accepted that routine home machine laundering of face coverings and other clothing items rendered viruses on those surfaces non-infectious, little data existed to validate this claim, and there was a lack of published laboratory protocols for testing. The purpose of the research protocol presented here is to provide an example of a laboratory process for conducting laundering studies that generate data on viral disinfection. While the protocol was developed for research during the COVID-19 pandemic, it is intended to be a framework adaptable to other virus disinfection studies.
The role of clothing in disease transmission is a difficult concept to quantify. The International Scientific Forum on Home Hygiene attempted this challenging task by conducting a review of the role of clothing in the spread of infectious disease coupled with a risk assessment of home hygiene practices6. Included in this work was the review of several scientific studies that examined the survival of different viral strains on different types of fabrics such as wool and cotton7,8,9,10,11. Each study focused on a different type of virus including vaccinia, poliovirus, respiratory syncytial virus, herpesvirus, and influenza virus. The survival times of the different viruses on the fabrics ranged from 30 min to 5 months depending on the virus-material combination. Several of the studies also demonstrated the transfer of viral contamination from the material onto hands. As part of the publication, effective laundering was discussed as an important management technique to reduce transmission but recognized that the magnitude of the impact of laundering on reducing disease burden was dependent on the specific viral contaminant and difficult to quantify7,8,9,10,11.
The laundering process destroys microorganisms using chemical, physical, and thermal treatment processes. For example, soaps and detergents can separate soils and may impart some chemically mediated antimicrobial action. Physically, dilution and agitation may aid in the reduction in viral loads. A study examining the persistence of human coronavirus HCoV-OC43 on cotton swatches using industrial and domestic wash cycles with and without temperature and detergent found no detectable virus when washing in unheated water without detergent, but that in the presence of a soil load (artificial saliva), domestic wash cycles required detergent for samples to have non-detect virus loadings12. Hot water itself can also provide an effective means of destroying some microorganisms13,14.
In a recent publication summarizing the state of current laundry practices, many factors such as fabric composition, storage conditions, dirt load, wash temperature and time, and drying temperature was identified as varying in global practices of laundering15. While laundering is a common cleaning method for a large percentage of the population, this large variation in existing practices makes issuing detailed guidance for how to do this safely and effectively, when an item may be contaminated by a virus, challenging and sparse. During the COVID-19 pandemic, the United States Centers for Disease Control and Prevention (CDC) issued guidance on how to launder items for homeowners16,17. Much of this laundering guidance was based on several older studies on bacterial disinfection18,19 and supported by several benchtop studies that have found enveloped viruses inactivated in water with detergents20,21. The guidance can be summarized as 1) follow the manufacturer's instructions for the detergent, 2) use the warmest appropriate water setting, and 3) dry items completely. The rationale of these recommendations was that washing on the warmest possible cycle with detergent combined with drying completely (with heat if possible) will kill the SARS-CoV-2 virus.
The sheer number of possible variations in the laundering process necessitates a uniform protocol, such as presented here, to be able to isolate variables and test the viral disinfection efficacy of specific processes. The intent of this protocol coupled with an instructional video is to demonstrate a laboratory-based hot water laundering process for replication in other research studies. Additionally, the results of this viral disinfection efficacy testing should build consumer confidence in the effectiveness of home laundering during viral-based pandemics.
Protocol
Phi6 was received from a collaborator laboratory as a ~1 mL frozen aliquot and was stored at -80 °C until use. It was initially used to propagate more virus stocks which were subsequently stored at -80 °C until use. Phi6 was selected as the demonstration virus because it is commonly used as a model enveloped virus, can be propagated to high titers, and requires a low biosafety level laboratory to perform the testing22,23,24.
1. Prepare virus stock solution
- Propagate bacteriophage Phi6 in bacterial host Pseudomonas syringae using a modified Tryptic Soy Agar media preparation and soft agar overlay method as described below.
- Prepare modified Tryptic Soy Agar by weighing out and mixing the ingredients from Table 1 in deionized water.
- Prepare an overnight culture of P. syringae by adding a 1 mL aliquot of P. syringae with an optical density (OD600) between 0.9 to 1.5, to 100 mL of modified Tryptic Soy Broth (Table 1) and incubating in a shaking incubator at ~260 rpm at room temperature (20-26 °C).
- Prepare soft agar tubes by placing modified Tryptic Soy Agar (~6 mL) in test tubes and covering with an autoclavable cap. Store at 4 °C until use. Autoclave soft agar tubes at 121 °C for 15 min to melt agar. Hold at 48 °C until plating. Equilibration at 48 °C is important otherwise the virus can be inactivated in the assay.
- Add 1 mL of undiluted concentrated virus aliquot to soft agar and 100 µL of a log phase (OD600 between 0.9 to 1.5) P. syringae culture. Pour soft agar onto the surface of a solidified 100 mm diameter modified Tryptic Soy agar plate. To prevent bubbles and/or spilling, swirl plates gently to evenly distribute the soft agar over the solid agar surface and incubate overnight at room temperature.
- Gently scrape25 the contents of the three plates with a sterile cell spreader into a sterile 50 mL conical tube containing 15 mL of SM buffer. Vortex tubes at maximum setting for 1-2 min to break up agar and then centrifuge at 7,000 x g for 15 min.
- Remove supernatant and filter through a 0.2 µm syringe filter. Store 1 mL aliquots in cryovials at -80 °C until use.
2. Perform pre-test visual assessment of PPE item
- Place each PPE item on a clean, smooth surface (e.g., a laboratory bench covered with a disposable paper liner). Assess each PPE item in triplicate.
- Ensure proper lighting during the PPE examination. Measure and record the length and width of the unwashed items at various locations (Figure 1).
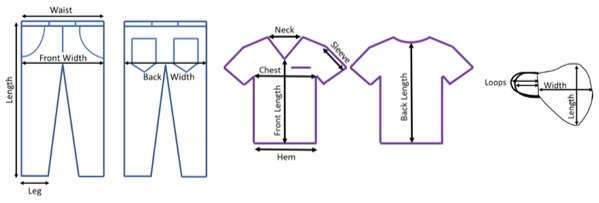
Figure 1. PPE pre-test assessment measurement locations. Denim, scrub, and face-covering locations where the length was recorded for tracking material changes from the laundering process. Please click here to view a larger version of this figure.
3. Prepare coupons
- Make 2 cm x 4 cm test coupons by cutting PPE, prepare two coupons per face covering (three face coverings were tested), five coupons per denim pant (three denim pants were tested), and three coupons per scrub shirt (three scrubs were tested).
- Prepare a set of 2 cm x 4 cm procedural blank coupons (a set for one full-sized PPE item) for each material type that will not be inoculated but will be laundered. Prepare two coupons for every day of face-covering experiments, five coupons for every day of denim experiments, and three coupons for every day of scrub experiments.
- Prepare a set of 2 cm x 4 cm positive control coupons that will be inoculated, but not laundered. Prepare two coupons for every day of face-covering experiments (three face coverings were tested), five coupons for every day of denim experiments (three denim pants were tested), and three coupons for every day of scrub experiments (three scrubs were tested), and three stainless steel coupons.
NOTE: Different numbers of replicates were selected based on the size of the item. For example, it is physically difficult to adhere five coupons on the face covering, and two coupons would represent a limited area of the denim pants. The locations were selected to maximize coverage and in zones that might get folded during laundering and be more difficult to clean.
4. Perform inoculation
- Prepare a 10% beef extract solution by dissolving 1 g of beef extract in a total volume of 10 mL of 1x phosphate buffered saline. Filter sterilize the entire volume using a 0.2 µm syringe filter.
- Thaw the virus stock solution prepared in section 1 at room temperature. On the day of use, add 100 µL of the thawed Phi6 stock to 900 µL of the 10% beef extract solution.
- Inoculate test coupons and positive control coupons with ~107 PFU/sample by pipetting a 10 µL droplet of solution onto the PPE item and spreading the droplet using the tip of the pipette. Depending on the PPE material, the droplets will separate and reaggregate differently.
- Allow inoculated coupons to dry in a biosafety cabinet. Determined dry time(s) via observation for your specific materials. For the results presented here, the following times were used: scrubs = 30 min dry time; face-covering = 60 min dry time; denim = 30 min dry time; stainless steel = 120 min dry time.
- Attach inoculated coupons to full-sized PPE items according to Figure 2 using safety pins and aseptic techniques.
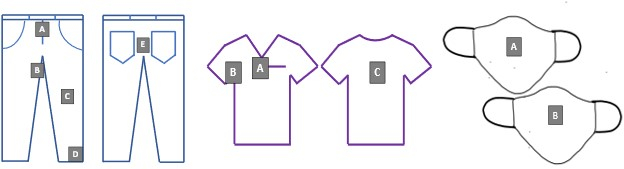
Figure 2. Test coupon locations on denim, scrubs, and face coverings. Letters A-D corresponds to the unique coupon identifiers for all laundering experiments. Please click here to view a larger version of this figure.
5. Perform laundering
- Prepare laundering detergent as follows.
- Sterilize tap water that will be used in the washing machine and collect 10 mL of autoclaved water for a sterility check. For this protocol, autoclave 7 L of tap water on a 60 min liquid cycle.
- Prepare laundering solution according to manufacturer's dilution directions. For this protocol, dissolve 1.54 mL of detergent in 3.5 L of sterile tap water. Warm the laundering solution to 50 °C using a hot plate and stir bar. Measure and record the pH and temperature of the laundering solution. Collect 10 mL of solution for sterility check.
- Pour laundering solution into a sterilized washer (3.25 L). Pre-sterilize washers using a 250 ppm-4 h cycle of vaporous hydrogen peroxide between tests.
- Place PPE item(s) in a sterilized washer. Add one denim pant and one scrub shirt per washer. Add one inoculated face covering and four non-contaminated fill masks per washer; fill masks did not have coupons attached.
- Wash PPE item for 18 min (two 9 min wash cycles with normal agitation). Drain the washer and triple rinse with room temperature tap water (5 L each time) to remove foam. Add room temperature sterilized tap water into the washer (3.25 L) and perform a 9 min long rinse cycle.
- Move the PPE item(s) into the washer's spin side and spin for 5 min. Move the PPE item(s) to the dryer and dry for 80 min on the high heat setting (93 °C).
- Move PPE from dryer to sterile workspace and aseptically remove the coupons from each item and place them in conical tubes. Pre-fill the tubes with 10 mL of 10% Dey-Engley broth extraction buffer and cover with aluminum foil.
6. Extract and enumerate viral loads on coupons
- Extract coupons in 10 mL of 10% Dey-Engley neutralizing broth by vortexing for 2 min at the maximum setting of your equipment.
- Plate extracts using a conventional soft top agar overlay method26.
- Prepare tubes of soft modified Tryptic Soy Agar and a P. syringae culture as described in section 1. On the day of testing, autoclave the soft agar tubes at 121 °C for 15 min to melt the agar. Hold the soft agar at 48 °C until plating. Equilibration at 48 °C is important otherwise the virus can be thermally inactivated in the assay.
- Prepare a tenfold dilution series in 1x phosphate-buffered saline for each test sample used in the laundering study. Use both serially diluted (100 µL) and undiluted (1 mL and 100 µL) aliquots for plating.
- Add test sample aliquots to the soft agar tube containing 6 mL of soft agar and 100 µL of the log phase P. syringae culture (OD600 between 0.9-1.5). Pour the soft agar onto the surface of a solidified modified Tryptic Soy agar plate. Evenly distribute the soft agar over the solid agar surface by swirling the plate.
- Incubate plates overnight at room temperature and manually enumerate the following day by counting the plaque-forming units on each plate.
7. Perform post-test visual assessment of PPE item(s)
- Document the following in the various PPE items used for testing: signs of fading, discoloration, and/or damage (e.g., tearing, stretching); odors; small holes, cuts, or tears (use a small flashlight to look for damage); separation of layers, missing threads, areas where binding is damaged; damage to seams or zippers; measure and record the stretching of elastic.
Representative Results
Several different types of quality control data and results are generated after completion of this protocol. Plaque forming unit (PFU) plate counts along with the extracted sample volume enable the calculation of the number of PFU per test coupon. Table 2 is an example of a data recording sheet for serial dilution/plating results. Using the dilution factor, sample volume, and plate count from Table 1, Table 3 shows representative viral recovery results for a face covering test. Note that these data include the test coupons and the quality control samples for the inoculum, coupons, and wash water (with and without detergent). The procedural blank and sterility quality control samples are important for confirming that the water solutions and PPE materials were not contaminated with Phi6. Indication of contamination would cause erroneous calculations of disinfection efficacy and require the test to be repeated. The positive control samples are intended to verify that the virus stock solution did not environmentally/naturally degrade during the experiments, thereby inflating the effect of the laundering process in viral load reduction. These samples should remain within 1 log PFU of the inoculum controls to accept the test coupon results. A large reduction in PFU of the positive control samples also indicates that all steps of the coupon inoculation should be carefully reviewed to ensure that the analyst is executing the protocol with proper pipetting and spreading techniques.
This protocol also provides information for assessing changes to the material properties of clothing items due to laundering and quality control information pertaining to the protocol (Table 4 and Figure 3). These data are useful for several reasons. Recording the trends in measurements of the PPE items allows for the identification of an item with a manufacturing defect. This identification may help to explain outlier microbial data and contextualize the variation in product behavior. Taking notes of odors or damage may also provide an indication if the washer or dryer was operating sub-optimally during an experiment and if the tests should be repeated or the equipment serviced. Additionally, if the test plan calls for multiple laundering cycles of the same PPE item, the data may help determine how long the PPE items maintain their integrity for use when laundering. Keeping a record of the pH of the detergent solution provides an alert to changes in the water source or the detergent product. Maintaining a time log of the laundering steps ensures that the timer on the washer and dryer does not cause variations to the experimental protocol.
Ultimately, these data are used to report the disinfection efficacy of the hot water laundering procedure against a surrogate for viral pathogens. Laundering disinfection efficacy (Eqn. 1) is calculated by subtraction of the average log10 virus recoveries from PPE test coupon from the average log10 virus recoveries from PPE positive control results (Figure 4). For test coupon results that are non-detect, the log10 of the detection limit is used in the disinfection efficacy calculation. It is common to report disinfection efficacy as log values for comparison with other viral disinfection techniques and standards.
Disinfection Efficacy = Average log10 (Positive Controls) – Average log10 (Test Coupons) (Eqn. 1)
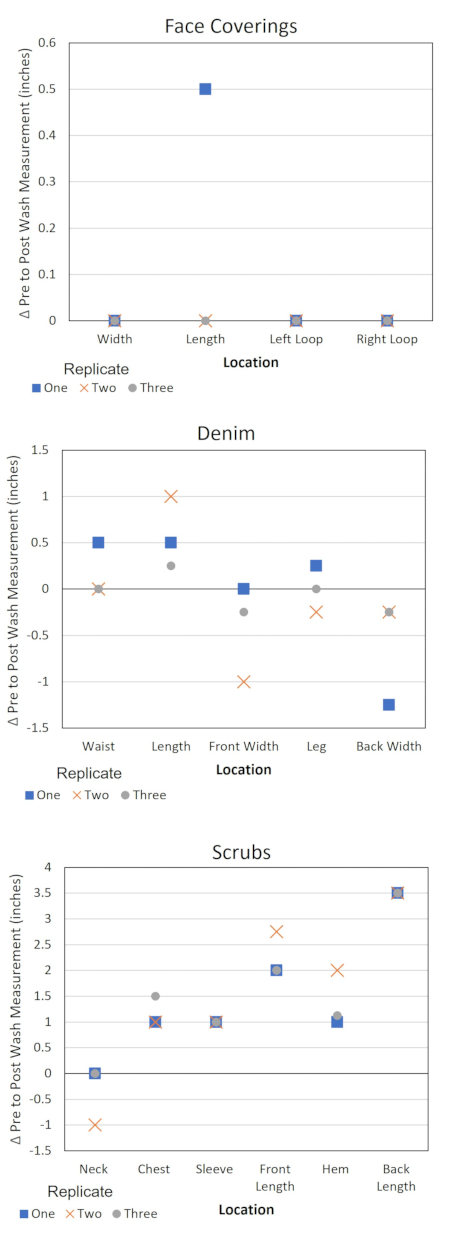
Figure 3. Change in PPE dimensions by location. Δ = Pre-test measurement – post-test measurement. A negative Δ value corresponds to the stretching of the item at the specified location and a positive value corresponds to shrinkage. Please click here to view a larger version of this figure.
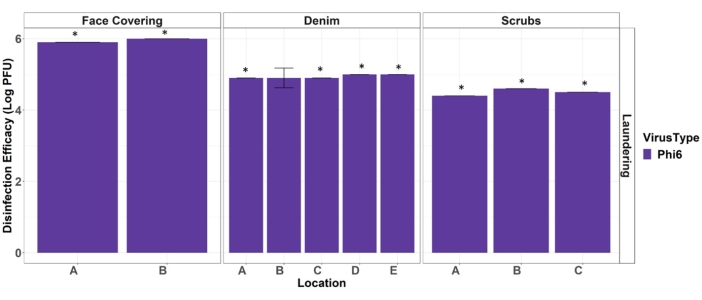
Figure 4. Efficacy of hot water laundering at disinfecting face coverings, denim, and scrub PPE materials from Phi6. Stars denote full disinfection occurred (non-detects on the test coupons). Error bars indicate standard deviation (n = 3). Location letters correspond to the placement depicted in Figure 2. Please click here to view a larger version of this figure.
Table 1. Solution recipes. Ingredients and amounts necessary to prepare tryptic soy agar, tryptic soy broth, and beef extract solutions. Please click here to download this Table.
Table 2. An example portion of a serial dilution/plating results sheet. Template for reporting raw microbial data. Please click here to download this Table.
Table 3. Face covering microbial results. Example summary sheet for processed plaque-forming unit (PFU) data. Please click here to download this Table.
Table 4. Scrubs quality control and material assessment log. Template for reporting calibration of pH probe, pH of detergent solution, pre-and post-wash measurements, and laundering cycle times. Please click here to download this Table.
Discussion
This protocol was developed to execute systematic laboratory testing to assess the laundering effectiveness of viral disinfection from full-sized PPE/clothing items. The procedures outline the critical steps for preparing the virus, inoculating the test material, assessing the changes to the items due to the laundering process, and quantifying the reduction in viral load as a result of the laundering (machine washing and drying) process. Additionally, the protocol outlines the necessary quality control samples for ensuring the experiments are not biased by contamination and measurements/observations that should be recorded to track the material integrity of the PPE items after multiple laundering cycles. The results using Phi6 indicate that the hot water laundering process used in this protocol achieved a greater than 3-log reduction in viral load for all samples (face covering, scrubs, and denim pants). The viral load reduction was also uniform across different locations on the PPE/clothing items. To demonstrate 3-log reduction, this protocol requires the use of a high viral load and a stabilizing agent (beef extract) that may not be representative of the soil load for all situations.
Mini washers and compact dryers were selected to optimize the number of replicate experiments that could be conducted in a space-constrained setting and to keep the sterilization of equipment and water volume used during the experiments manageable for laboratory staff. As a result of using the mini washer, the rinsing steps were manual as compared to most home laundering applications that are fully automated. It is also important to remember that machine washing predominates in developed countries, but handwashing is still practiced all over the world15. Additionally, some may not have access to hot water for washing, and others manually air-dry clothes rather than machine drying. These differences in laundering practices were not addressed in this current protocol but could easily be investigated with minor modifications such as substituting the washing and drying steps with using a bucket and a close line
There has been minimal focus on the cleaning/disinfecting of virally contaminated face coverings and street clothing in the scientific literature at the full scale. More commonly, studies assess the filtration performance of face coverings after repeated washing and drying but do not evaluate viral disinfection efficacy27,28. For example, Clapp et al. evaluated fitted filtration efficiency of cloth masks and modified procedure masks and found wide variation in performance, with simple modifications providing increased fit and filtration efficiency29. Another study looked at the filtration efficiency of four cloth masks of different materials30, again focusing on source control or personal protection. This may be due to a lack of specialization for both the microbial portion and mechanical testing in the same laboratory. The protocol presented here provides an evaluation of disinfection efficacy as well as material degradation.
There have been a number of decontamination/disinfection methods for disposable respiratory protection (primarily N95s) recently published in the scientific literature31,32,33. The primary focus on FFRs (e.g., N95s) is due to the critical respiratory protection they provide for healthcare workers and other front-line occupations. Primary technologies for respirator decontamination involved vaporized hydrogen peroxide (VHP), ultraviolet germicidal radiation (UVGI), and moist heat (steam) for virus inactivation. Viscusi et al. evaluated five decontamination methods for FFRs and UVGI; ethylene oxide and VHP were found to be the most promising decontamination methods31. Fischer et al. evaluated four different decontamination methods-UV light, dry heat, 70% ethanol, and VHP-for their ability to reduce contamination with SARS-CoV-2 and their effect on N95 respirator function32. There are many additional studies on effective decontamination technologies for FFRs which were summarized and published in 202033. However, these specialized methods aren't accessible or designed to be used safely by the average home or small business owner.
This protocol was developed using Phi6, an enveloped bacteriophage that is similar to SARS-CoV-2, has spike proteins, and is of similar size (80-100 nm)34, for all testing. Since Phi6 is not a known pathogen, it can be manipulated in a general microbiological Biosafety Level 1 (BSL-1) laboratory. Efficacy against Phi6 may indicate the efficacy of other enveloped viruses, however, empirical verification for each virus of interest is necessary35. By using a similar, nonpathogenic viral agent, it is hoped that this protocol can be repeated elsewhere and used for studying future viral epidemics/pandemics. Future research may include the use of disinfectants (e.g., bleach) in addition to detergents and a standardized protocol for hand washing and line drying.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development directed the research described herein under EP-C-15-008 with Jacobs Technology Inc. It has been reviewed by the Agency but does not necessarily reflect the Agency's views. No official endorsement should be inferred. EPA does not endorse the purchase or sale of any commercial products or services. The authors would like to thank EPA contractors Denise Aslett for the oversight of the EPA RTP microbiology, Brian Ford, Rachael Baartmans, and Lesley Mendez Sandoval for their work on this project in the EPA RTP microbiology lab, Ramona Sherman for providing the EPA quality assurance review, and Worth Calfee and Shannon Serre for providing EPA technical reviews.
Materials
| Freezer (- 80 °C) | ThermoFisher Scientific | FDE30086FA | |
| Hot Plate | VWR | 97042-714 | |
| Safety Pins (steel) | Singer | 319921 | |
| Shaker | Lab-Line Instruments, Inc. | 3525 | |
| SM buffer | Teknova, Hollister, CA | S0249 | |
| Syringe filter (0.2 μm) | Corning, Corning, NY | PES syringe filters, 431229 | |
| 1X Phosphate Buffered Saline | Teknova, Hollister, CA | P0196, 10X PBS solution | |
| Agar | Becton Dickinson | 214010 | |
| Autoclavable caps | DWK Life Sciences, Millville, NJ | KIM-KAP Caps, 73663-18 | |
| Autoclave | Steris | AMSCO 250LS Steam Sterilizer Model 20VS | |
| Beef Extract | Sigma-Aldrich, Millipore Sigma, St. Louis, MO, USA | P/N B4888-100g | |
| Calcium chloride | Sigma-Aldrich | 793639 | |
| Cell spreaders | Busse Hospital Disposables | 23600894 | |
| Centrifuge | ThermoFisher Scientific | 75004271 | Heraeus MegaFuge 16R Centrifuge |
| Certified Timer | https://nist.time.gov/ | Not Applicable | |
| Conical tubes (50 mL) | Corning Life Sciences | 352098 | Falcon 50-mL high-clarity polypropylene conical centrifuge tubes |
| Cryovials | Thermo Fisher Scientific, Waltham, MA | AY509X33 | |
| Denim | Wrangler | Rustler Regular Fit Straight Leg Jean Four Pocket Jean with Scoop Front Pockets, PN:87619PW | |
| Detergent | Proctor and Gamble | Tide Original Scent Liquid Laundry Detergent Product Number (PN): 003700023068 | |
| Dextrose | Fisher | BP350 | |
| Dey-Engley neutralizing broth | Becton Dickinson | DF0819172 | |
| Dryer | Magic Chef | MCSDRY15W | |
| Face Coverings | Felina | Reusable Organic Cotton Face Masks, PN: 990121P4 | |
| Incubator (top agar) | Symphony | 414004-596 | |
| Laboratory Notebook | Scientific Notebook Company | 2001 | |
| Magnesium chloride | Sigma-Aldrich | M9272 | |
| Media sterilization and dispensing system | Integra | Media Clave/Media Jet | |
| Petri Dishes (100 mm) | VWR | 25384-342 | |
| pH Meter | Orion/Oakton | STARA1110/EW-35634-35 | |
| pH Probe | Orion | 8157BNUMD | |
| pH Standards | Oakton | 00654-(00/04/08) | |
| Phi 6 and Pseudomonas syringae | Battelle Memorial Institute, Columbus, OH | Not Applicable | |
| Pipette & Tips | Rainin | (Pipettes) 17014391, 17002921; (Pipette Tips) 30389239, 17014382 | |
| Refrigerator | True Manufacturing Co., Inc. | GDM-33 | |
| Scrubs | Gogreen cool | PN: WS19100PT | |
| Sodium chloride | Sigma-Aldrich | 57656 | |
| Stir Bar | Fisherbrand | 16-800-512 | |
| Tape Measure | Lufkin | PS3425 | |
| Test Tubes for Soft agar (14 mL) | Corning, Corning, NY | 352059 | |
| Thermometer | Fisherbrand | 14-983-19B | |
| Tryptone | Sigma-Aldrich | T9410 | |
| Vaporous hydrogen peroxide sterilization bags | STERIS | 62020TW | |
| Vortex (during the plating process) | Daigger Scientific, Inc | 3030A | Vortex Genie 2 |
| Vortex (for sample extraction) | Branson Ultrasonics | 58816-115 | Multi-Tube vortexer |
| Washer | Kuppet | KP1040600A | |
| Washer Sterilization | Steris | STERIS VHP ED1000 generator | |
| Yeast extract | Gibco | 212750 |
Referencias
- Emanuel, E. J., et al. Fair allocation of scarce medical resources in the time of Covid-19. The New England Journal of Medicine. 382 (21), 2049-2055 (2020).
- Cohen, J., van der Meulen Rodgers, Y. Contributing factors to personal protective equipment shortages during the COVID-19 pandemic. Preventive Medicine. 141, 106263 (2020).
- Burki, T. Global shortage of personal protective equipment. The Lancet Infectious Diseases. 20 (7), 785-786 (2020).
- Optimizing Personal Protective Equipment (PPE) Supplies. CDC Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/ppe-strategy/index.html (2020)
- Livingston, E., Desai, A., Berkwits, M. Sourcing personal protective equipment during the COVID-19 pandemic. Jama. 323 (19), 1912-1914 (2020).
- Bloomfield, S. F., Exner, M., Signorelli, C., Nath, K. J., Scott, E. A. The infection risks associated with clothing and household linens in home and everyday life settings, and the role of laundry. International Scientific Forum on Home Hygiene. , (2011).
- Hall, C. B., Douglas, R. G., Geiman, J. M. Possible transmission by fomites of respiratory syncytial virus. Journal of Infectious Diseases. 141 (1), 98-102 (1980).
- Turner, R., Shehab, Z., Osborne, K., Hendley, J. O. Shedding and survival of herpes simplex virus from ‘fever blisters’. Pediatrics. 70 (4), 547-549 (1982).
- Bean, B., et al. Survival of influenza viruses on environmental surfaces. Journal of Infectious Diseases. 146 (1), 47-51 (1982).
- Brady, M. T., Evans, J., Cuartas, J. Survival and disinfection of parainfluenza viruses on environmental surfaces. American Journal of Infection Control. 18 (1), 18-23 (1990).
- Sidwell, R. W., Dixon, G. J., Mcneil, E. Quantitative studies on fabrics as disseminators of viruses: I. Persistence of vaccinia virus on cotton and wool fabrics. Applied Microbiology. 14 (1), 55-59 (1966).
- Owen, L., Shivkumar, M., Laird, K. The stability of model human coronaviruses on textiles in the environment and during health care laundering. Msphere. 6 (2), 00316-00321 (2021).
- Sehulster, L., et al. Guidelines for environmental infection control in health-care facilities. Recommendations from CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). American Society for Healthcare Engineering/American Hospital Association. , (2004).
- Chen, H., et al. Influence of different inactivation methods on severe acute respiratory syndrome coronavirus 2 RNA copy number. Journal of Clinical Microbiology. 58 (8), e00958 (2020).
- Abney, S. E., Ijaz, M. K., McKinney, J., Gerba, C. P. Laundry hygiene and odor control-state of the science. Applied and Environmental Microbiology. 87 (14), 0300220 (2021).
- COVID-19 Cleaning and Disinfecting Your Home. CDC Available from: https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/disinfecting-you-home.html (2021)
- COVID-19 Your Guide to Masks – How to select, properly wear, clean, and store masks. CDC Available from: https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/about-face-coverings.html (2021)
- Walter, W. G., Schillinger, J. E. Bacterial survival in laundered fabrics. Applied Microbiology. 29 (3), 368-373 (1975).
- Blaser, M. J., Smith, P. F., Cody, H. J., Wang, W. -. L. L., LaForce, F. M. Killing of fabric-associated bacteria in hospital laundry by low-temperature washing. Journal of Infectious Diseases. 149 (1), 48-57 (1984).
- Gerhardts, A., et al. Testing of the adhesion of herpes simplex virus on textile substrates and its inactivation by household laundry processes. Journal of Biosciences and Medicines. 4 (12), 111 (2016).
- Heinzel, M., Kyas, A., Weide, M., Breves, R., Bockmühl, D. P. Evaluation of the virucidal performance of domestic laundry procedures. International Journal of Hygiene and Environmental Health. 213 (5), 334-337 (2010).
- Casanova, L. M., Weaver, S. R. Inactivation of an enveloped surrogate virus in human sewage. Environmental Science & Technology Letters. 2 (3), 76-78 (2015).
- Aquino de Carvalho, N., Stachler, E. N., Cimabue, N., Bibby, K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environmental Science & Technology. 51 (15), 8692-8700 (2017).
- Ye, Y., Chang, P. H., Hartert, J., Wigginton, K. R. Reactivity of enveloped virus genome, proteins, and lipids with free chlorine and UV254. Environmental Science & Technology. 52 (14), 7698-7708 (2018).
- Bacteriology Culture Guide. ATCC Available from: https://www.atcc.org/resources/culture-guides/bacteriology-culture-guide (2022)
- Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., Johnson, R. P. . Bacteriophages. , 69-76 (2009).
- Sankhyan, S., et al. Filtration performance of layering masks and face coverings and the reusability of cotton masks after repeated washing and drying. Aerosol and Air Quality Research. 21 (11), 210117 (2021).
- Kumar, A., Bhattacharjee, B., Sangeetha, D., Subramanian, V., Venkatraman, B. Evaluation of filtration effectiveness of various types of facemasks following with different sterilization methods. Journal of Industrial Textiles. , (2021).
- Clapp, P. W., et al. Evaluation of cloth masks and modified procedure masks as personal protective equipment for the public during the COVID-19 pandemic. JAMA Internal Medicine. 181 (4), 463-469 (2021).
- Lu, H., Yao, D., Yip, J., Kan, C. -. W., Guo, H. Addressing COVID-19 spread: development of reliable testing system for mask reuse. Aerosol and air quality research. 20 (11), 2309-2317 (2020).
- Viscusi, D. J., Bergman, M. S., Eimer, B. C., Shaffer, R. E. Evaluation of five decontamination methods for filtering facepiece respirators. Annals of Occupational Hygiene. 53 (8), 815-827 (2009).
- Fischer, R. J., et al. Effectiveness of N95 respirator decontamination and reuse against SARS-CoV-2 virus. Emerging Infectious Diseases. 26 (9), 2253 (2020).
- Derraik, J. G., Anderson, W. A., Connelly, E. A., Anderson, Y. C. Rapid review of SARS-CoV-1 and SARS-CoV-2 viability, susceptibility to treatment, and the disinfection and reuse of PPE, particularly filtering facepiece respirators. International Journal of Environmental Research and Public Health. 17 (17), 6117 (2020).
- Fedorenko, A., Grinberg, M., Orevi, T., Kashtan, N. Survival of the enveloped bacteriophage Phi6 (a surrogate for SARS-CoV-2) in evaporated saliva microdroplets deposited on glass surfaces. Scientific Reports. 10 (1), 1-10 (2020).
- Calfee, M. W., et al. Virucidal efficacy of antimicrobial surface coatings against the enveloped bacteriophage Φ6. Journal of Applied Microbiology. 132 (3), 1813-1824 (2022).

