Early Detection of Cyanobacterial Blooms and Associated Cyanotoxins using Fast Detection Strategy
Summary
A fast-multidisciplinarystrategy for early detection of cyanobacterial blooms and associated cyanotoxins is described here. It allows for the detection of cyanobacteria and related cyanotoxins in water samples and in organic matrices, such as bivalve samples, in 24 h.
Abstract
Fast detection of cyanobacteria and cyanotoxins is achieved using a Fast Detection Strategy (FDS). Only 24 h are needed to unravel the presence of cyanobacteria and related cyanotoxins in water samples and in an organic matrix, such as bivalve extracts. FDS combines remote/proximal sensing techniques with analytical/bioinformatics analyses. Sampling spots are chosen through multi-disciplinary, multi-scale, and multi-parametric monitoring in a three-dimensional physical space, including remote sensing. Microscopic observation and taxonomic analysis of the samples are performed in the laboratory setting, which allows for the identification of cyanobacterial species. Samples are then extracted with organic solvents and processed with LC-MS/MS. Data obtained by MS/MS are analyzed using a bioinformatic approach using the online platform Global Natural Products Social (GNPS) to create a network of molecules. These networks are analyzed to detect and identify toxins, comparing data of the fragmentation spectra obtained by mass spectrometry with the GNPS library. This allows for the detection of known toxins and unknown analogues that appear related in the same molecular network.
Introduction
Cyanobacterial blooms have emerged as an environmental problem all over the world in the last 15 years1,2. Cyanobacterial blooms are due to the overgrowth of microorganisms named cyanobacteria. They are a conspicuous group of photosynthetic microorganisms that have adapted themselves to live in a large array of environments, including tropical areas and extremely cold waters. They are known for producing large blooms covering water surfaces, especially in response to a massive enrichment of nutrients, the so-called eutrophication process3.
Therefore, cyanobacteria are excellent bioindicators of water pollution4,5,6. They can also produce a wide array of natural compounds with interesting pharmacological properties7,8. The environmental problem related to cyanobacteria are the blooms themselves. Blooms can block sunlight to underwater grasses, consume oxygen in the water leading to fish kills, produce surface scum and odors, and interfere with the filter feeding of organisms9.
In addition, and even more seriously, in a specific combination of factors such as temperature, nutrients (phosphorus and nitrogen), sunlight (for the photosynthesis), and pH of the water, cyanobacterial blooms trigger toxin production; therefore, they become harmful to humans and animals. The most studied class of cyanotoxins is produced by the genera Microcystis. These are cyclic peptides known under the general name of microcystins (MCs): microcystin-LR being the most studied as being able to produce severe hepatoxicity10. Animals and humans may be exposed to MCs by ingestion of contaminated drinking water or food. The World Health Organization (WHO) suggested a total microcystin-LR value of 0.001 mg/L as a guideline11. However, this is related only to one variant (i.e., MC-LR) out of more than 100 microcystins that have been isolated so far.
Combined methods previously reported, such as remote sensing with MALDI-TOF MS analysis12,13,14,15, have focused on the concentration detection of MCs. The most recent methods use low-resolution sensors that are effective in detecting only wide bloom expanses; they are also capable of revealing only toxins for which standards are available. Moreover, most of these procedures are time-consuming, and time is a dramatic factor for early detection of the bloom to prevent or minimize safety problems. The multidisciplinary strategy proposed here provides rapid detection of cyanobacteria bloom and cyanotoxins, after only 24 h16.
In the frame of the program called MuM3, "Multi-disciplinary, Multi- scale and Multi-parametric Monitoring in the three-dimensional (3D) physical space"17,18, a Fast Detection Strategy (FDS) combines the advantages of several techniques: 1) remote sensing to detect the bloom; 2) microscopic observation to detect cyanobacteria species; and 3) analytical/bioinformatics analyses, namely, LC-HRMS-based molecular networking, to detect cyanotoxins. Results are obtained within 24 h.
The new approach is useful to monitor wide coastal areas in a short time, avoiding numerous sampling and analyses, and reducing detection-time and costs. This strategy is the result of the study and application of different approaches to the monitoring of cyanobacteria and their toxins and combines the advantages of each of them. Specifically, the analysis of the results, coming from the use of different platforms (satellite, aircraft, drones) and sensors (MODIS, thermal infrared) for remote sensing analysis, such as of diverse methodological approaches for the identification of cyanobacterial species (microscope, UV-Vis spectroscopy, 16S analysis) and toxins (LC-MS analysis, molecular networking), allowed the selection of the most appropriate method both for the specific and general purposes. The new methodology was experimented and validated in subsequent monitoring campaigns on Campania coasts (Italy), in the frame of Campania environmental protection agency monitoring program.
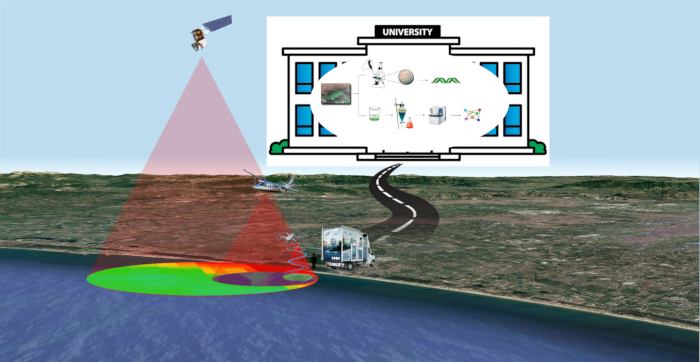
Figure 1: FDS strategy. An overview of Fast Detection Strategy for cyanobacteria and cyanotoxins. Please click here to view a larger version of this figure.
Protocol
1. Remote and proximal sensing: data acquisition and analysis
NOTE: In this case, remote/proximal sensing data are used for a first macro-area survey and to select specific spots of coastal areas to be sampled. In the MuM3 framework17 scheme the logic flow is based on a hierarchical monitoring model that includes several levels named information layers. The information of each level is based on data acquired using one or more sensors carried onboard platforms located at different altitudes. Each level defines a spatial scale depending on the altitude of the measurement19. There is the potential for multiple sensors at each level. Some examples are: visible near infrared (VNIR) and thermal infrared (TIR) imaging20 on satellites, aircraft, helicopters, UAV21 and at the surface; physical, chemical, and biological analyses, etc.22 at the surface and in fast response using the mobile lab. The data acquired by each sensor is processed and combined to calculate multispectral indexes (e.g., normalized difference vegetation index (NDVI), Normalized Difference Water Index (NDWI), Chlorophyll Index, etc.), so the raw data is converted into more useful parameters and formats (e.g., thematic map).
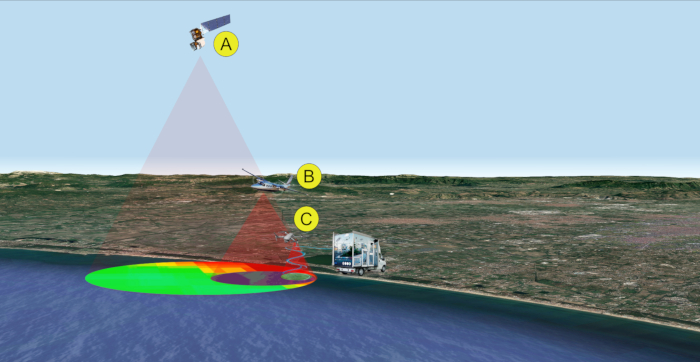
Figure 2: Remote/proximal sensing analyses for sampling (steps 1-2). Multi-level and multi-sensor approach for the detection of cyanobacterial bloom. Data acquisition is performed by satellite (A), aircraft (B), and/or drone (C). Please click here to view a larger version of this figure.
- Remote and proximal sensing data retrieval
- Retrieve data from the various public and private remote sensing datasets, produced especially for content-based retrieval and scene classification. Typical dataset sources used in this step include: Landsat products provided by U.S. Geological Survey23, Sentinel-2,3 products provided by NASA24, and Copernicus Open Access Hub25, MODIS-Aqua26.
- Identify the products available for the specific area, dates range and consider the limit derived by the cloud cover. Define: A) the region of interest using graphical user interface and polygon selection; B) the dates range and the cloud coverage using the text query filter fields.
NOTE: Even if many scientific reports cite that an acceptable cloud cover value is <20%, in this type of research, it is important to pay attention to the real cloud cover only on the water surface, so an effectiveness value for this region is <5%. - Choose products derived from the platforms best fitting the specific needs of the mission. From each combination of satellite and payload technical specifications, it is possible to obtain several collections of products. For example, the NASA EOSDIS24 data products are processed at various levels ranging from Level 0 to Level 4: the Level 0 products are raw data at full instrument resolution while at higher levels the data is converted into more useful parameters and formats. Usually, the Levels 0-1 are available, while the products in Levels 2-4 need specific processing related to specific research goals, so their availability is limited in time and the region covered.
- Download the chosen dataset of satellite observations. In detail, selecting among the list of the results from the previous action, download a complete dataset product (e.g., a group of geo-tiff files, one for each band, plus infos, and other metadata) or only a specific single band.
NOTE: For the specific description of each basic action included in the procedure of step 1, please also refer to Huan-Huan Chen et al. 202027.
- Data pre-processing and processing for the analysis and transformation into more useful parameters and formats to permit a first screening of the macro-area.
NOTE: The following actions (step 1.2) are dedicated to data processing finalized to transform raw data coming from lower levels into information and then into useful information (higher levels). This step consists of processing of the raw data from each sensor (e.g., product Levels 0-1) and transformation into higher-level products (e.g., Levels 2-4) and, subsequently, in information layers to generate more useful parameters and formats (e.g., thematic map).- Pre-process raw data involving geometric and radiometric calibration. This action could be performed using specific software that, in an automatic or semi-automatic way (unsupervised or supervised), provides a set of connected tools for raster processing in order to perform an easy classification respecting a correct workflow.
NOTE: In this research, two free open source tools are used: Q-GIS 3.1428 combined with Semi-Automatic Classification Plugin (SCP). The SCP plugin29 allows the semi-automatic classification of remote sensing images, providing a complete tool set for the download of free images, the pre-processing, the post-processing, and the raster calculation. - Process calibrated data calculating multispectral indexes. Typically, this action is performed using a raster calculator tool. The calculation of a multispectral index defines a correlation between different bands/layers that as a result generates a new layer that could be managed on a GIS platform. For example, starting from Sentinel and Landsat operational land imager sensor (OLI), it is possible to calculate multispectral indexes such asNormalized Difference Vegetation Index (NDVI). Vegetation index is then used to estimate chlorophyll content30,31.
- Analyze the results obtained from the estimation of chlorophyll content index represented as in false color thematic maps and define critical area with high chlorophyll concentration. In this study, chlorophyll-a map is generated using dataset of MODIS sensor32, mounted on Terra and Aqua satellite platforms. The sampling spots are localized where abnormal values are registered.
NOTE: The algal bloom flag (that define each specific spot) is raised in the remote sensing domain when the Chl-a concentration exceeds 20 mg/L33.
- Pre-process raw data involving geometric and radiometric calibration. This action could be performed using specific software that, in an automatic or semi-automatic way (unsupervised or supervised), provides a set of connected tools for raster processing in order to perform an easy classification respecting a correct workflow.
2. Guided samplings
NOTE: The choice of sampling spots is driven by remote/proximal sensing layer analysis that allows to select spots in large coastal areas. Taking account that sampling could be dangerous for operators due to microcystins' toxicity, safety sampling procedures are needed. Particularly, it is needed to protect operators from aerosol inhalation and skin contact. Then, perform sampling following the procedure detailed below.
- Wear goggles for eye protection, FFP2 mask to prevent aerosol inhalation, and safety gloves to prevent skin contact.
- Collect 0.5 L of water in triplicate in each site.
- Measure the salinity for each sample using a refractometer. Put a drop of the sample on the refractometer and read the salinity value in terms of parts per thousand (ppt – ‰).
- At the end of sampling, first wash the hands; then in turn remove the gloves, mask, and goggles taking care not to touch the external surface of the personal protection equipment.
- Carry the sample to the lab at room temperature.
3. Identification of cyanobacteria species by microscopic observations and taxonomic identification
- Centrifuge each sample (≈0.5 L) at 11,200 x g for 5 min.
- Extract supernatants as follows: pour 500 mL of butanol in each sample and using a funnel, transfer the mixture to be extracted into the separatory funnel. Place the separatory funnel upright in the ring clamp to allow both the layers to separate. Let the aqueous phase to drain in an Erlenmeyer flask. Repeat this step three times. Then, concentrate the organic phases under vacuum and weigh them.
- Collect pellets from step 3.1 and analyze them at 400x and 1,000x magnification with an optical microscope equipped with an 18 MP digital camera for microscope. Look for the presence of cyanobacteria on the basis of their morphological features: blue-green color, cell shape, and size allow to recognize cyanobacteria among other microorganisms.
- Identify the species through taxonomic analysis by microscopic observation, according to the procedure described in Komarék et al. 201434.
NOTE: Complementary 16S metagenomic analysis can be performed in order to identify cyanobacterial taxa3. - Dilute an aliquot of the pellets with 10 mL of seawater/freshwater BG11 media, according to its salinity, for cultivation.
4. Identification of cyanotoxins
- Extraction of samples with organic solvents
- Put each sample of pellet in a flask and sonicate for 5 min using an ice-bath. Then, add 50 mL of fresh MeOH and gently shake. Filter the solution using a paper filter and collect the filtrate in a round bottom flask. Repeat this step twice. Then, extract the pellets adding 50 mL mixture of MeOH/DCM twice (1:1, 50 mL), and twice using 100% DCM (x2, 50 mL)35. Label each round bottom flask respectively, as "MeOH extract", "MeOH/DCM extract", and "DCM extract", and concentrate under vacuum.
- Analyze each organic extract (MeOH, MeOH/DCM, DCM) by LC-HRMS/MS.
- LC-HRMS/MS analysis
- LC-HRMS and LC-HRMS/MS samples preparation: dissolve each sample using MeOH ≥ 99.9% to get a final concentration of 10 mg/mL.
- Analyze each sample using a high-resolution ESI mass spectrometer coupled with an HPLC system (LC-HRMS/MS system). Work on the HPLC with a 5-µm C18 column (100 x 2.10 mm), at room temperature. Use a gradient elution with H2O (supplemented with 0.1% HCOOH) and MeOHat 200 µL min-1. The gradient program is: 10% MeOH for 3 min, 10% to 100% MeOH for 30 min, 100% MeOH for 7 min. Use MeOH as the control.
- Data acquisition. Collect data in the data-dependent acquisition mode (DDA): the 10 most intensive ions of a full-scan mass spectrum have to be subjected to high-resolution tandem mass spectrometry (HRMS/MS) analysis. Acquire HRMS/MS scans for selected ions with CID fragmentation, an isolation width of 2.0, normalized collision energy of 35, Activation Q of 0.250, and an activation time of 30 ms.
- Bioinformatic analyses and molecular networking
- Analyze fragmentation patterns for each intense ion using MS specific software.
- Use MS-Cluster to cluster the data (parent mass tolerance of 1.0 Da and MS/MS fragment ion tolerance of 0.5 Da). Consensus spectra containing less than 2 spectra are eliminated.
- Analyze MS/MS data by GNPS (Global Natural Products Social36) for molecular networking.
- Pairwise compare consensus spectra and also with those reported in the GNPS-Mass-Ive libraries.
- Visualize the obtained network37.
NOTE: For molecular networking please refer to Sigrist et al. 202038.
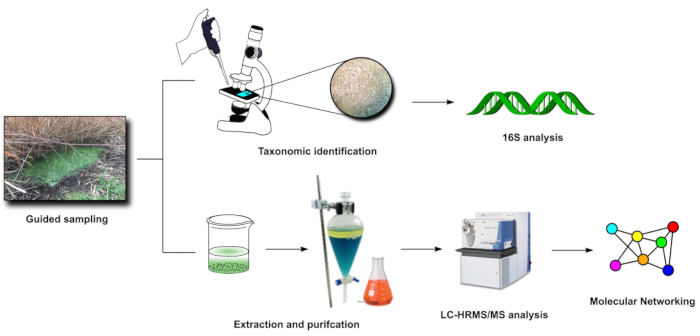
Figure 3: FDS in-lab steps (3-4). Visual representation of the main activities carried out in laboratory after sampling (steps 3 and 4). Please click here to view a larger version of this figure.
Representative Results
In a first study3, four anthropogenically-impacted sites along the Campania coast in SW Italy were observed using satellite Landsat 8 and aircraft during summer 2015. Landsat 8 operational land imager sensor (OLI) and the aircraft multispectral camera allowed to create Normalized Difference Water Index (NDWI) images for the areas, therefore, to reveal the presence of cyanobacterial communities. Cyanobacterial community composition was determined through spectrophotometric analyses for the detection of the cyanobacterial pigment phycocyanin (PC). Then, complementary 16S metagenomic analysis allowed to identify cyanobacterial taxa. The simplified multispectral image indexing and classification through satellite/aerial platforms in combination with metagenomic analyses were effective in detecting the presence of cyanobacteria belonging to genera associated with strong eutrophic conditions (such as Leptolyngbya sp., Pseudooscillatoria sp.), at an early stage of blooming.
In a second study14, FDS approach was tested during Spring/Summer 2017. Satellite data were used as the only remote sensing level. In detail, data acquired by MODerate Image Spectroradiometer (MODIS) sensor, mounted on Terra and Aqua satellite platforms, allowed quantification of chlorophyll-a (Chl-a) in the water bodies along Campania coasts and drove the choice of ten sampling spots. Samples were processed in lab by microscopic observation and taxonomic identification, then extracted with organic solvents. Organic extracts were processed by LC-MS-MS analysis. Data obtained by MS-MS were analyzed using a bioinformatic approach, using the GNPS platform to create a network of molecules. The network was analyzed to detect and identify toxins comparing data of the fragmentation spectra obtained by mass spectrometry with the GNPS library. This allowed to detect known toxins and unknown analogues that will appear related in the same molecular network. Specifically, Lyngbyatoxin A, a lipophilic dermatotoxins, was detected in all water-samples and bivalves' samples; in the Lyngbyatoxin A molecular cluster, nodes not related to any known compounds of lyngbyatoxins family were also present, suggesting the presence of unknown lyngbyatoxin analogues. No microcystins and other toxins were detected in the samples. All the results were obtained within 24 man-hours.
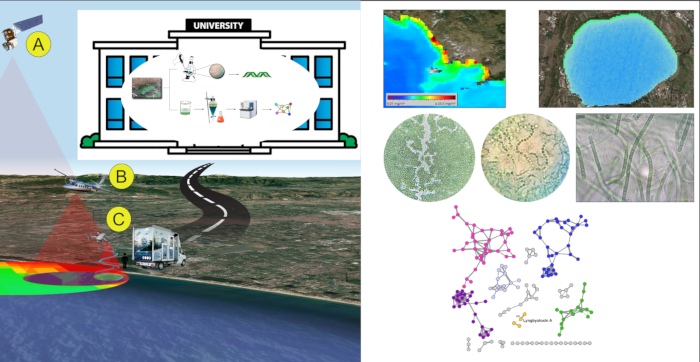
Figure 4: FDS representative results. An example of application of FDS strategy on Campania coast (Italy). Please click here to view a larger version of this figure.
Discussion
During the last years, our team tested and validated several different approaches that allowed unraveling the presence of cyanobacteria and cyanotoxins in water bodies and bivalves. The new developed strategy represents the result of these studies. The optimal techniques and technologies that fit the scope of fast detection, are gathered under the hat of a unique procedure that maximize the effectiveness of each single step. The target area, the bloom extension, and growing stage are the driving force to the choice of suitable methods and technologies to use.
When cyanobacteria and cyanotoxins fast detection is the priority, the strategy is streamlined reducing the total number to four main steps: (1) Remote and Proximal sensing and data analysis for a first survey, localization of sites and definition of bloom pattern and extension; (2) Guided sampling; (3) Microscopic observation and taxonomic analysis; (4) Chemical analysis and molecular networking of LC-MS data for dereplication of the water samples and fast detection of cyanotoxins.
Regarding the first step, even if the availability of data acquired by a complete chain of platforms that cover all the layers of hierarchical monitoring approach would be the best solution to restitute a complete vision of the analyzed scenario, often just one information layer can drive the area survey action and effectively focus on the hot spots to perform in-situ sampling actions. According to the reported experiences in which data was acquired using satellites, aircrafts, helicopters, UAVs, the solution that totally matches the needs required by the fast detection strategy is the use of the only satellite products.
In addition, the information layers that derive from missions performed by platforms that fly at lower altitudes than satellites (e.g., aircrafts, helicopters, UAVs) restitute information with great resolution but these are very expensive and also require more time to complete the full acquisition process that also includes flight plan defining and approval.
Once the spots to be samples have been selected (step 2), analytical/bioinformatics analyses (Molecular networking of LC-MS data) is the tool for fast dereplication of the water samples and fast detection of cyanotoxins (steps 3 and 4). 16S metagenomic analysis takes at least 2 weeks of work. Moreover, even when cyanobacterial species that are generically toxic are identified, their toxin production is not demonstrated. For the same reason, microscopic observation is not itself sufficient to reveal the presence of toxic cyanobacteria. Of course, MS analysis and molecular networking have some limitations; they are quite effective if compounds of interest (e.g., toxins) are well ionized in the applied conditions, if they are in sufficient amount to be detected. For the purpose of the known cyanobacterial toxin detection and monitoring, MS-based molecular networking actually represents one of the more robust and reliable technologies.
Therefore, this approach proves to be quite useful when a fast detection of cyanobacteria and related cyanotoxins is needed; moreover quantification of both cyanobacterial bloom and toxin over space and time is also possible by this strategy to prevent health communities' problems that could arise by large cyanobacterial toxic blooms.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This research was funded by "Centro di Riferimento Regionale per la Sicurezza Sanitaria del Pescato (CRiSSaP)" in the frame of the project "Attività pilota di Monitoraggio di Cianobatteri nella fascia costiera della regione Campania", and performed in cooperation with the Campania Region Environmental Protection Agency, Italy (ARPAC), "Istituto Zooprofilattico Sperimentale del Mezzogiorno/Osservatorio Regionale per la Sicurezza Alimentare" (IZSM/ORSA), University of Naples "Federico II" – Department of Veterinary Medicine and Animal Production, ref. prof. A. Anastasio).
Materials
| 10X Vitamin mix | Nicotinic acid 100 mg/100 mL; PABA 10 mg/100 mL; Biotin 1 mg/100 mL; Thiamine 200 mg/100 mL; B12 1 mg/100 mL; Folic Acid 1 mg/100 mL; i-inositol 1 mg/100 mL; Ca-pantothenate 100 mg/100 mL | ||
| 1-BuOH | Sigma-Aldrich | 33065.2.5L-R | |
| BG11 stock solution | Na2EDTA 20 mg/L; Ferric ammonium citrate 120 mg/L; Citric acid·1H2O 120 mg/L; CaCl2·2H2O 700 mg/L, MgSO4·7H2O 1.5 g/L, K2HPO4·3H2O 800 mg/L, NiSO4(NH4)2SO4·6H2O (0.1 mM stock) 5 mL; Na2SeO4 (0.1 mM stock) 2 mL, Nitsch's Solution 20 mL | ||
| Centrifuge | Hermle | Z36HK | |
| CHCl3 | Honeywell | 32211.2.5L | |
| H2O | Sigma-Aldrich | 34877.2.5L | |
| Kinetex C18 cloumn | Phenomenex | ||
| LTQ Orbitrap XL high-resolution ESI mass spectrometer coupled to a U3000 HPLC system | Thermo | ||
| MeOH | Honeywell | 32213.2.5L | |
| Microscope equipped with an OMAX 18 MP CMOS camera | Optech | Biostar B3 | |
| Multiband camera | Intergraph DMC | ||
| Nitsch's Solution | H3BO3 0.5 g/L MnSO4· H2O 2.28 g/L ZnSO4·7H2O 0.5 g/L CuSO4·5H2O 0.025 g/L COCl2·6H2O 0.135 g/L Na2MoO4·2H2O 0.025 g/L |
||
| Refractomer mr 100 ATC | AQL | ||
| SWBG11 medium | BG11 stock solution 50 mL/L; Instant Ocean 33 g/L; Water 950 mL/L 10X; Vitamin mix 100 µL/L |
Referencias
- Tamele, I. J., Silva, M., Vasconcelos, V. The incidence of marine toxins and the associated seafood poisoning episodes in the African countries of the Indian ocean and the Red sea. Toxins. 11 (1), 25-48 (2019).
- Lürling, M., Faassen, E. J. Dog poisonings associated with a Microcystis aeruginosa bloom in the Netherlands. Toxins. 5 (3), 556-567 (2013).
- O’Neil, J. M., Davis, T. W., Burford, M. A., Gobler, C. J. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae. 14, 313-334 (2012).
- Teta, R., et al. Cyanobacteria as indicators of water quality in Campania coasts, Italy: A monitoring strategy combining remote/proximal sensing and in situ data. Environmental Research Letters. 12 (2), (2017).
- Teta, R. Bioindicators as a tool in environmental impact assessment: Cyanobacteria as a sentinel of pollution. International Journal of Sustainable Development and Planning. 14 (1), 1-8 (2019).
- Teta, R., Della Sala, G., Mangoni, A., Lega, M., Costantino, V. Tracing cyanobacterial blooms to assess the impact of wastewaters discharges on coastal areas and lakes. International Journal of Sustainable Development and Planning. 11 (5), 804-811 (2016).
- Teta, R., et al. A joint molecular networking study of a: Smenospongia sponge and a cyanobacterial bloom revealed new antiproliferative chlorinated polyketides. Organic Chemistry Frontiers. 6 (11), 1762-1774 (2019).
- Singh, R. K., Tiwari, S. P., Rai, A. K., Mohapatra, T. M. Cyanobacteria: an emerging source for drug discovery. Journal of Antibiotics. 64 (6), 401-412 (2011).
- Huisman, J., et al. Cyanobacterial blooms. Nature Reviews Microbiology. 16 (8), 471-483 (2018).
- Gupta, N., Pant, S. C., Vijayaraghavan, R., Rao, P. V. L. Comparative toxicity evaluation of cyanobacterial cyclic peptide toxin microcystin variants (LR, RR, YR) in mice. Toxicology. 188 (2-3), 285-296 (2003).
- WHO. Cyanobacterial toxins: Microcystin-LR in drinking water. Background document for development of WHO Guidelines for Drinking-Water Quality. WHO. 2, (1998).
- Agha, R., Cirés, S., Wörmer, L., Domínguez, J. A., Quesada, A. Multi-scale strategies for the monitoring of freshwater cyanobacteria: Reducing the sources of uncertainty. Water Research. 46 (9), 3043-3053 (2012).
- Giménez-Campillo, C. Determination of cyanotoxins and phycotoxins in seawater and algae-based food supplements using ionic liquids and liquid chromatography with time-of-flight mass spectrometry. Toxins. 11 (10), 610 (2019).
- Sanseverino, I., António, D. C., Loos, R., Lettieri, T. Cyanotoxins: methods and approaches for their analysis and detection. JRC Technical Reports. , (2017).
- Teta, R. Combined LC-MS/MS and molecular networking approach reveals new cyanotoxins from the 2014 cyanobacterial bloom in Green Lake, Seattle. Environmental Science and Technology. 49 (24), 14301-14310 (2015).
- Esposito, G., et al. A fast detection strategy for cyanobacterial blooms and associated cyanotoxins (FDSCC) reveals the occurrence of lyngbyatoxin A in campania (South Italy). Chemosphere. 225, 342-351 (2019).
- Lega, M., Casazza, M., Teta, R., Zappa, C. J. Environmental impact assessment: a multilevel, multi-parametric framework for coastal waters. International Journal of Sustainable Development and Planning. 13 (8), 1041-1049 (2018).
- Di Fiore, V. Integrated hierarchical geo-environmental survey strategy applied to the detection and investigation of an illegal landfill: A case study in the Campania Region (Southern Italy). Forensic Science International. 279, 96-105 (2017).
- Gargiulo, F., Persechino, G., Lega, M., Errico, A. IDES project: A new effective tool for safety and security in the environment. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). , (2013).
- Ferrara, C., Lega, M., Fusco, G., Bishop, P., Endreny, T. Characterization of terrestrial discharges into coastal waters with thermal imagery from a hierarchical monitoring program. Water. 9 (7), (2017).
- Alfeo, A. L., Cimino, M. G. C. A., De Francesco, N., Lega, M., Vaglini, G. Design and simulation of the emergent behavior of small drones swarming for distributed target localization. Journal of Computational Science. 29, 19-33 (2018).
- Casazza, M., Lega, M., Liu, G., Ulgiati, S., Endreny, T. A. Aerosol pollution, including eroded soils, intensifies cloud growth, precipitation, and soil erosion: a review. Journal of Cleaner Production. 189, 135-144 (2018).
- . U.S. Geological Survey Available from: https://ers.cr.usgs.gov (2020)
- . NASA Earthdata Available from: https://urs.earthdata.nasa.gov (2020)
- . Copernicus Opern Access Hub Available from: https://scihub.copernicus.eu/apihub (2020)
- Chen, H. -. H., Tang, R., Zhang, H. -. R., Yu, Y., Wang, Y. Investigating the relationship between sea surface chlorophyll and major features of the south china sea with satellite information. Journal of Visualized Experiments: JoVE. (160), (2020).
- . QGIS Available from: https://www.qgis.org/it/site/ (2020)
- . Semi-automatic classification plugin documentation release 4.8.0.1 Available from: https://manualzz.com/doc/7130012/semi-automatic-classification-plugin-documentation (2016)
- Rouse, J. W., Hass, R. H., Schell, J. A., Deering, D. W. Monitoring vegetation systems in the great plains with ERTS. Third Earth Resources Technology Satellite (ERTS) Symposium. 1, 309-317 (1973).
- Carmona, F., Rivas, R., Fonnegra, D. C. Vegetation index to estimate chlorophyll content from multispectral remote sensing data. European Journal of Remote Sensing. 48 (1), 319-326 (2015).
- Savtchenko, A., et al. MODIS data from terra and aqua satellites. International Geoscience and Remote Sensing Symposium (IGARSS). 5, 3028-3030 (2003).
- Binding, C. E., Greenberg, T. A., McCullough, G., Watson, S. B., Page, E. An analysis of satellite-derived chlorophyll and algal bloom indices on Lake Winnipeg. Journal of Great Lakes Research. 44 (3), 436-446 (2018).
- Komárek, J., Kaštovský, J., Mareš, J., Johansen, J. R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 86, 295-335 (2014).
- Esposito, G., et al. Chlorinated thiazole-containing polyketide-peptides from the caribbean sponge smenospongia conulosa: structure elucidation on microgram scale. European Journal of Organic Chemistry. 2016 (16), 2871-2875 (2016).
- Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 13 (11), 2498-2504 (2003).
- Sigrist, R., Paulo, B. S., Angolini, C. F. F., De Oliveira, L. G. Mass spectrometry-guided genome mining as a tool to uncover novel natural products. Journal of Visualized Experiments: JoVE. (157), (2020).

