Histological Quantification to Determine Lung Fungal Burden in Experimental Aspergillosis
Summary
Here we describe a protocol to determine pulmonary fungal burden in mice with invasive aspergillosis by quantification of Gomori's modified methanamine silver staining in histological sections. Use of this method resulted in comparable results with less animals compared to assessment of fungal burden by quantitative PCR of lung fungal DNA.
Abstract
The quantification of lung fungal burden is critical for the determination of the relative levels of immune protection and fungal virulence in mouse models of pulmonary fungal infection. Although multiple methods are used to assess fungal burden, quantitative polymerase chain reaction (qPCR) of fungal DNA has emerged as a technique with several advantages over previous culture-based methods. Currently, a comprehensive assessment of lung pathology, leukocyte recruitment, fungal burden, and gene expression in mice with invasive aspergillosis (IA) necessitates the use of a significant number of experimental and control animals. Here the quantification of lung histological staining to determine fungal burden using a reduced number of animals was examined in detail. Lung sections were stained to identify fungal structures with Gomori's modified methanamine silver (GMS) staining. Images were taken from the GMS-stained sections from 4 discrete fields of each formalin-fixed paraffin-embedded lung. The GMS stained areas within each image were quantified using an image analysis program, and from this quantification, the mean percentage of stained area was determined for each sample. Using this strategy, eosinophil-deficient mice exhibited decreased fungal burden and disease with caspofungin therapy, while wild-type mice with IA did not improve with caspofungin. Similarly, fungal burden in mice lacking γδ T cells were also improved by caspofungin, as measured by qPCR and GMS quantification. GMS quantification is therefore introduced as a method for the determination of relative lung fungal burden that may ultimately reduce the quantity of experimental animals required for comprehensive studies of invasive aspergillosis.
Introduction
IA is an opportunistic infection that may develop in susceptible individuals with congenital or acquired immune deficiencies due to immune suppressive therapy or chronic infection1,2. Primary infection often occurs in the lungs, although in some instances dissemination of Aspergillus fumigatus to the liver, kidneys, heart, and brain may occur, resulting in extensive tissue invasion of hyphae accompanied by severe disease and high rates of mortality1,2. Furthermore, the efficacy of existing pharmacotherapies is limited, and may be further weakened by the emergence of antifungal-resistant strains in the environment3. It is therefore important to understand the mechanisms of fungal virulence and host pathology that promote development or exacerbation of invasive fungal disease.
Murine models remain important for mechanistic IA studies, as they allow researchers to assess the roles of fungal virulence genes and host immune effectors for the establishment and growth of A. fumigatus in vivo4,5. Consequently, multiple strategies have been devised in order to effectively quantify or compare fungal burden in groups of experimental animals6,7. These strategies involve culture-based, biochemical, immunoassay, or qPCR methods, each with distinct advantages and disadvantages. Furthermore, each of these methods involve the dedication of a subset of animals in addition to those sacrificed for assessments of immune effector function, gene expression analysis, and comparative histopathology7. Thus, comprehensive IA studies often require significant numbers of research animals at a significant cost. Effective strategies that reduce experimental time, animal costs, and ethical considerations by utilizing animal tissues for multiple analyses are therefore extremely valuable7.
In this report, a method describing the quantification of GMS staining in histological sections for comparison of the relative fungal burden between experimental groups of mice with IA is introduced. Each step from fungal culture to infection, tissue harvest and processing, and image acquisition and data analysis, is described in detail. Fungal burdens obtained by GMS quantification were compared with qPCR in neutropenic models of IA, and in caspofungin-treated wild-type or eosinophil-deficient mice with IA8. The results show similarity with GMS quantification and qPCR of fungal DNA. This suggests that GMS quantification may be useful to researchers engaged in histological analyses as a supplemental or alternative method of comparison of relative fungal burden in mice with IA, and may ultimately reduce the cost and use of research animals in complex, mechanistic studies.
Protocol
All animal procedures were approved by the Animal Care and Use Committee of Indiana State University, the host campus of Indiana University School of Medicine — Terre Haute.
1. Preparation of A. fumigatus Conidia for Infection
- While working in a well-ventilated fume hood, add 125 µL of A. fumigatus 293 stock solution to 900 µL cell culture grade water (CCGW). Then transfer the solution to malt extract agar (MEA) plate via pipette and spread evenly with an inoculating loop.
- Store the fungal plate in the dark at 24 °C for at least 14 days and no more than 28 days.
- Harvest conidia using glass beads as previously described9.
- Pour 1.5 g of 0.5 mm glass beads onto the plate and gently tilt it back and forth until the beads are coated in order to extract conidia. Collect the bead/conidia mixture in a 15-mL conical tube. Suspend in 5 mL Dulbecco's phosphate-buffered saline (DPBS) and vortex.
- Count the conidia in a 50x dilution of supernatant in DPBS using a hemocytometer. Count the number of conidia in the area shown in Figure 1 and use Equation 1 to calculate the concentration.
#Conidia × Dilution Factor × 104=Conidia/mL (Equation 1)
NOTE: Additional 4 x 4 square corner areas may be counted, with the mean of these areas used as #Conidia in Equation 1.
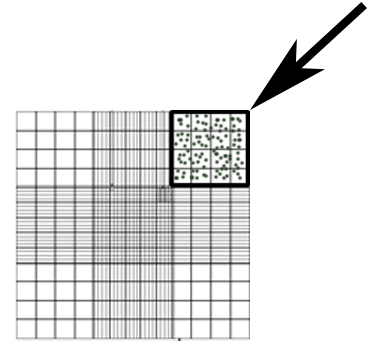
Figure 1: Hemocytometer representative area used for conidia counting (box, arrow).
- Dilute the conidia suspension with sterile saline (DPBS) to obtain the desired concentration, 1.0 x 108 conidia/mL for infection with 5 x 106 conidia/50 µL. Prepare 50 µL of conidia solution per 'n + 2' mice to be infected to account for pipetting error.
NOTE: Standard fungus harvest and preparation/filtration may be alternatively used10. The glass bead method is best used to ensure that conidia samples with minimal small/soluble hyphal antigens are obtained for aspiration.
2. Immunosuppression and Infection of Murine Model
- Use 7-10-week-old mice.
NOTE: BALB/c, C57BL/6 (B6), eosinophil-deficient (ΔdblGATA, BALB/c background), and γδ T cell-deficient (TCRδ-/-, B6 background) mice were used for this study. For each experiment, age and gender-matched (both males and females) mice were used for each group. - Deplete neutrophils via intraperitoneal (IP) injection of 0.1 mL (0.5 mg) anti-mouse-Ly-6G antibody in sterile saline at 45° in the lateral lower right quadrant (For an experimental schedule, see Figure 2).
- 24 h later, infect mice by aspiration with 5.0 x 106A. fumigatus conidia suspended in 50 µL sterile saline (see steps 2.4-2.10) as previously described11. Following infection, inject a subset of animals with antifungal agent such as 5 mg/kg caspofungin diacetate in DPBS used in this study (Daily, see Figure 2).
- Anesthetize mice with 99.9% isoflurane using a veterinary anesthesia machine.
- Place a paper towel on the floor of the induction chamber to catch feces and urine.
- Place the mouse in the induction chamber, secure the lid, and set the vaporizer to 5% isoflurane for 2 min and 30 s.
NOTE: Vet ointment is not used because the total time under anesthesia is less than 5 min. - Reduce the vaporizer to half of the maximum setting for 30 s.
- Confirm that the mouse is properly anesthetized by observing slowed breathing, lack of movement, and lack of response to toe-pinch stimulus.
- Remove the mouse from the induction chamber and place on a slant board. Place the mouse with its back against the board. Ensure that its upper incisors are gently restrained with a rubber band and the lower incisors are gently restrained with a metal wire.
- Carefully hold the tongue at full extension with small forceps and pipette 50 µL of conidia/PBS suspension at the base of the tongue.
- Hold the tongue at full extension; listen for rapid breathing and the distinct clicking noise of aspiration. Hold for 20 s or until suspension is fully aspirated.
- Remove the mouse from the slant board and gently place in a cage for observation.
- Following aspiration, the mouse should regain consciousness within 1 min. Monitor the mouse until fully conscious and able to walk around the cage. The animals should be housed at 21 °C and monitored twice daily until the lungs are harvested.
- Repeat the depletion of neutrophils (step 2.2) 24 h post-infection (Figure 2).
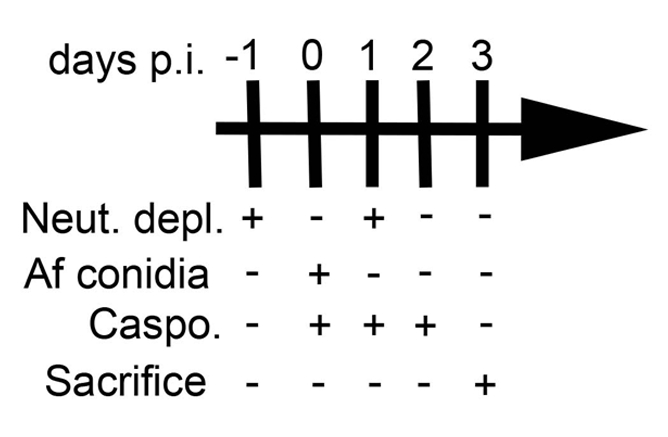
Figure 2: Experimental schedule. This figure was modified from a previous publication8.
3. Harvest and Preservation of Mouse Lungs for Histological Analysis
- Deeply anesthetize the animal with a fatal dose of 0.1 ml of 390 mg/ml pentobarbital sodium solution IP.
- Confirm euthanasia by observing lack of spontaneous breathing and absence of response to limb stimulation with tweezers. Once euthanized, spray the carcass with 70% ethanol to sterilize.
- Place the carcass on a foam board covered with an absorbent pad.
NOTE: The carcass should be placed on its back with pins securing all four limbs to the foam board. - Open the upper chest cavity using scissors with caution. Cut the rib cage away in order to expose the lungs and heart. Cut the inferior vena cava to allow for perfusion. Avoid cutting the blood vessels and puncturing organs.
- Slowly perfuse with 5 mL of cold DPBS in the right ventricle of the heart at a 45° angle with a 25-G needle. Repeat the perfusion with 5 mL of 10% formalin.
- Carefully expose the trachea and detach the surrounding fat pads. Use caution and avoid puncturing the trachea. Ensure the excess connective tissue is removed in order to fully visualize the trachea before proceeding. Once the trachea is exposed, place forceps under the trachea to elevate it.
- Poke a small hole in the upper trachea using a 25-G needle.
NOTE: The hole should be positioned so that the entire cannula fits in the trachea. Use caution to avoid rupturing the trachea. - Insert a cannula through the hole and towards the lungs. Secure the cannula with a surgical thread tied loosely around the trachea.
NOTE: If the trachea is ruptured, the cannula may be inserted past the trachea as far as possible in the airway. - Using a syringe, inflate the lungs with 1 mL of 10% formalin buffer through the cannula. Ensure that the inflation of the lungs is visible. If not, adjust the catheter and repeat. Once the lungs are inflated, quickly remove the cannula with caution and tighten the thread securely to seal off the trachea.
NOTE: If the trachea is ruptured and the lungs cannot be inflated, the lungs may still be harvested but this may prevent proper fixation. - Gently remove the trachea and lungs from the chest by carefully disconnecting the connective tissue of the trachea with forceps while gently pulling up on the thread. Place the lungs and trachea in a 50-mL conical tube with 15 mL of 10% formalin buffer. Place one end of the thread outside of the tube and seal the cap. Invert the tube to completely submerge the sample in fixative.
- Store the samples at room temperature for at least 24 h or until further processing.
CAUTION: Formalin is hazardous. Wear appropriate Personal Protective Equipment including goggles and a face mask. Work under a chemical hood when processing samples.
4. Harvest and Preservation of Mouse Lungs for DNA Fungal Burden
- Follow steps 3.1-3.5 as described above on a separate mouse.
- Slowly perfuse with 10 mL of cold DPBS in the right ventricle of the heart at a 45° angle with a 25-G needle.
- Excise the lungs with scissors and place in labeled 1.5 mL tubes.
- Store the lungs at -80 °C until further processing.
- Use standard techniques as described (embedding and sectioning) and according to manufacturer's protocols (see Table of Materials).
5. Microscopy and Imaging
- Open the imaging quantification program (see Table of Materials).
- Using a light microscope, obtain at least 4 distinct fields of the GMS stained slides (10X objective). Ensure that the images are representative of the infected airways within the lung with similar tissue density.
NOTE: If the density and distribution of hyphal foci appear markedly different between control and experimental groups, additional fields may be taken, or the GMS area of the whole lung section may be imaged (4X objective). Obtain images of the same areas of the lung (identified by equivalent tissue morphology) on a serial lung section stained with hematoxylin and eosin, if desired. - Add scale bars to the images where necessary and save the images to be quantified. Select, Edit > Add/Edit Calibration Marks > Menu: choose line thickness, color and scale bar size > click on the image for where the scale bar is to be placed. Close the Menu and choose, Edit > Merge > Merge Calibration Marks > choose File Save As > name the file and click on Save.
6. Using an Image Processing Program to Calculate the Fungal Burden (Figure 3)
- Open an image processing program. Select File > folder with samples to quantify > image (Figure 3A). Select Image > Type > convert to "RGB Stack" (Figure 3B).
- Use the right arrow key to select the second of the three given images (Figure 3B). Hit "control + shift + T" to bring up the "Threshold" menu settings adjustor (Figure 3C). Locate the original .tiff or .jpg image as a reference and open with an image viewing program (Figure 3C).
- Pull the top slider all the way to the left and adjust the bottom slider until the area selected (in red) is representative of the microscopy image (typically ranging from 130-160 as indicated to the right of the slider (Figure 3C).
- Once selected, press "Set" > "OK" (Figure 3D). Select "Analyze" > "Set Measurements" > check "Area, Area Fraction, Limit to Threshold, Display Label" and leave the bottom settings (the drop-down menu and decimal places) as default > "OK" (Figure 3D).
- Significant areas of white space or background staining may be excluded by selection of the appropriate tissue with an area or polygon tool.
- Finally select, "Analyze" > "Measure" (a window with data should appear, or a tab on the windows taskbar will appear labeled, "Results") (Figure 3E). Transfer the data to a data analysis program for graphing and statistical analysis.
- Enter the mean of the four area percentages for each lung sample. 5-8 samples are generally sufficient to detect significant differences between groups. If comparing two experimental groups, perform an unpaired student's t-test to determine significance.
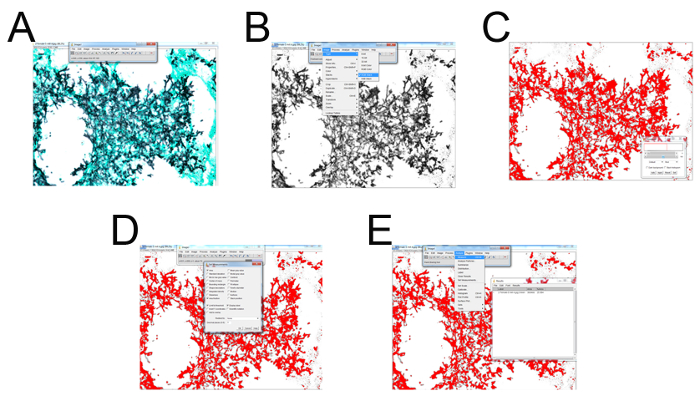
Figure 3: Representative screenshots for each step of GMS histological field quantification. (A) Select File > folder with samples to quantify > select image. (B) Select Image > Type > convert to "RGB Stack". Use the right arrow key to select the second of the three given images. (C) Hit "control + shift + T" to bring up the "Threshold" menu settings adjustor. Locate the original .tiff or .jpg image as a reference and open with photo viewer program. Pull the top slider all the way left and adjust the bottom slider until the area selected (in red) is representative of the microscopy image (typically ranging from 130-160 as indicated to the right of the slider. (D) Once selected press "Set" > "OK". Select "Analyze" > "Set Measurements" > check "Area, Area Fraction, Limit to Threshold, Display Label" and leave the bottom settings (the drop-down menu and decimal places) as default > "OK". (E) Finally select "Analyze" > "Measure" (a window with data should appear, or a tab on the windows taskbar will appear labeled "Results"). Please click here to view a larger version of this figure.
7. DNA Fungal Burden
- Remove the lungs from -80 °C and lyophilize for at least 4 h (overnight is optimal) or until completely dried by a freeze dry system.
- While the lungs are drying, prepare DNA extraction buffer (0.1 M NaCl, 10 mM EDTA pH 8.0, 10 mM Tris HCl pH 8.0, and 0.5% SDS).
- Place the prepared DNA extraction buffer in a 60 °C water bath to pre-warm. Also, place phenol: chloroform: isoamyl alcohol (25:24:1) in a 50-mL conical tube and allow for separation.
- Place the lyophilized lungs in a 2-mL screw cap tube with 0.2 mL of glass beads. Using a bead beater, grind dry lung tissue for 15-30 s until it forms a fine powder.
- Add 0.8 mL warm DNA Extraction buffer to each tube and vortex well. Incubate each tube for 15 min in a 65 °C bead bath and then vortex before incubating for an additional 15 min. After incubating, vortex and then centrifuge tubes at 25,000 x g for 10 min.
- Label 3 sets of 1.5 mL tubes for each of the samples and transfer the top layer of the supernatant into one set of tubes. Use caution while pipetting out the aqueous top layer to avoid disturbing the middle layer.
- Add equal volume of phenol:chloroform:isoamyl alcohol and mix well. Then centrifuge at 25,000 x g for 15 min before pipetting out the upper layer into the second set of tubes.
- Add equal volume of chloroform:isoamyl chloroform, mix well, and centrifuge at 25,000 x g for 10 min. Carefully pipette out the upper layer into the third set of tubes.
- Add 500 µL of isopropanol and 50 µL NaoAc (pH 5.2), mix well, and allow samples to incubate at room temperature for 10 min. Then centrifuge at 25,000 x g for 30 min and decant the supernatant.
- Wash the DNA pellet with 750 µL of ice cold 70% ethanol and centrifuge for 5 min at 25,000 x g. Decant the ethanol and allow the pellets to air dry in the tubes in a fume hoods for 40 min.
- Resuspend the dry pellets in 300 µL of TE buffer.
NOTE: The DNA may be stored at -20 °C until quantified. - Quantify DNA using a spectrophotometer and prepare 100 µL of 0.2 µg/µL of each DNA sample for qPCR analysis.
- Perform a standard qPCR in triplicate as previously described with 1 µg of DNA sample, fungal 18S rDNA primer, and probe sets with a modified probe quencher (5'-/56-FAM/AGC CAG CGG/ZEN/CCC GCA AAT G/3IABkFQ/-3')12,13.
- Prepare a standard curve from A. fumigatus genomic DNA and calculate the concentration of fungal DNA in pg in each sample using the average reporter dye Ct values.
- Plot the pg/µg of fungal DNA with standard error of the mean. Perform a student's t-test analysis to determine significance.
NOTE: As a less toxic alternative to the phenol-chloroform DNA extraction used herein, column based DNA extraction kits can be used.
Representative Results
Figure 4 includes a graph of survival of wild type or eosinophil-deficient mice with IA treated with caspofungin. The results show that eosinophil-deficient mice exhibited increased survival when compared to wild-type mice (50% mortality versus 100% mortality, respectively). Figure 5 shows representative GMS staining from neutropenic wild-type and eosinophil-deficient mice with IA treated or untreated with caspofungin. Caspofungin treatment resulted in relative fungal clearance in eosinophil-deficient, but not wild-type mice (right panels), while both groups that did not receive caspofungin were similar (left panels). Figure 6 shows the comparison of fungal burden in caspofungin-treated and control untreated wild-type or eosinophil-deficient mice, using both qPCR of fungal DNA (Figure 6A) and representative GMS quantification of 4 (10X objective) fields (Figure 6B). The results generated from both techniques show that caspofungin treatment results in the most significant fungal burden decrease between wild-type and eosinophil-deficient mice (Figure 6A). However, in untreated mice, only GMS quantification resulted in a significant decrease in mice lacking eosinophils (Figure 6B). When the mean area of whole-lung GMS-stained sections were calculated (4X objective), the differences were similar to the results obtained with 4 representative 10X fields, though with less statistical significance (Figure 6C). Figure 7 shows survival, images of representative GMS staining (4 10X fields), and quantification of fungal burden by both methods in γδ T cell-deficient (TCRδKO) mice that were treated with caspofungin in comparison to control, untreated mice. Caspofungin treatment improved survival (Figure 7A) and fungal burden (Figure 7B-D) in TCRδKO mice. Similar to the results of Figure 6, the results of fungal burden as measured by qPCR (Figure 7B) and representative GMS quantification (Figure 7C) were comparable.
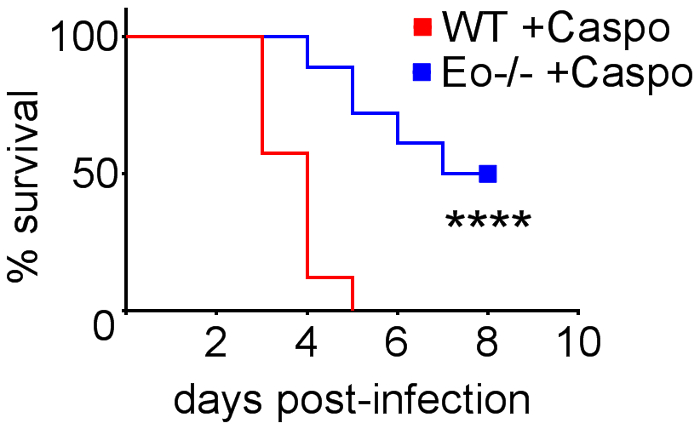
Figure 4: Increased survival in eosinophil-deficient (ΔdblGATA) mice with IA after treatment with caspofungin. 15-30 mice/group. Summary of 3-6 experiments. **** p < 0.0001. This figure was modified from a previous publication8. Please click here to view a larger version of this figure.
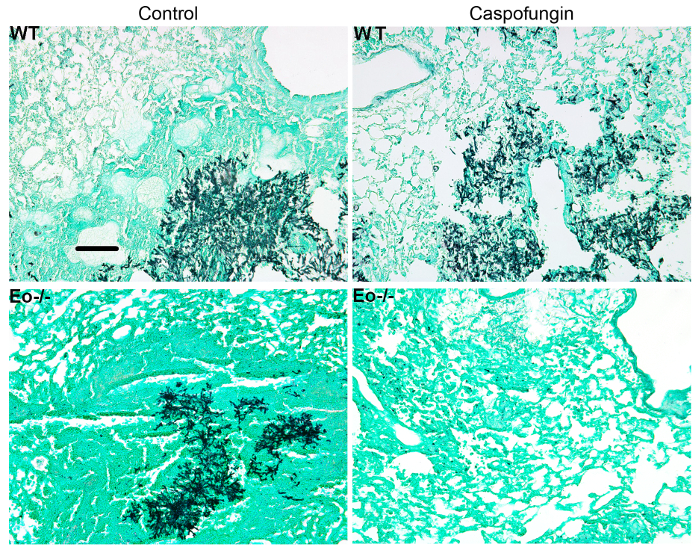
Figure 5: Representative images of GMS-stained lung sections from wild-type, eosinophil deficient, and caspofungin-treated or control untreated mice with IA. Representative of 3-4 mice/group. Scale bar is equivalent to 100 µm. This figure was modified from a previous publication8. Please click here to view a larger version of this figure.
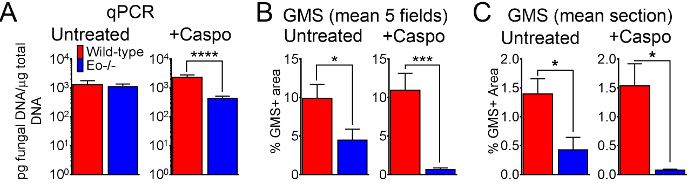
Figure 6: Fungal burden in wild-type, eosinophil-deficient mice with IA treated or control treated with caspofungin. (A) qPCR of fungal DNA. 15-30 mice/group, summary of 3-6 experiments. (B) GMS quantification of histological sections, mean% of GMS staining from 4 fields (10x objective). (C) GMS quantification, mean% of whole lung section (4X objective). B and C are a summary of 5 mice/group. * p < 0.05. *** p < 0.001. **** p < 0.0001. This figure was generated using data that were reported in a previous publication8. Please click here to view a larger version of this figure.
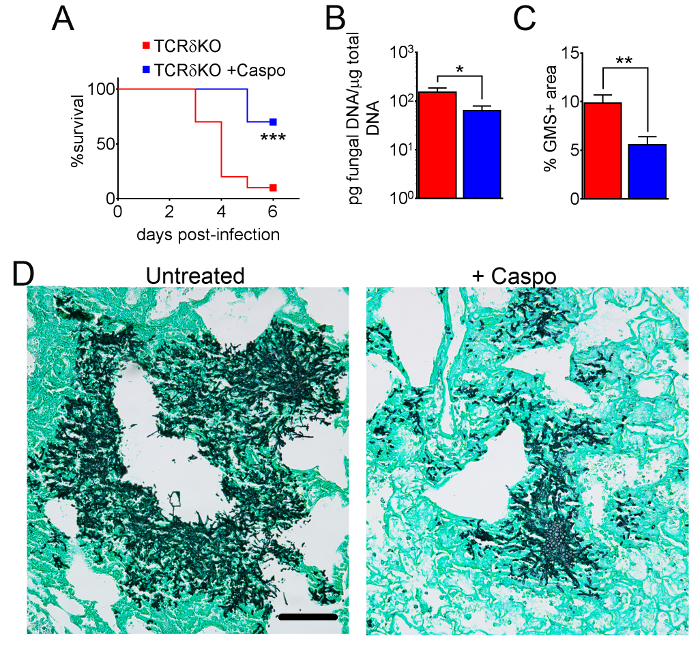
Figure 7: Decreased severity of IA in γδ T cell-deficient mice after caspofungin treatment. (A) Survival. (B) qPCR of fungal DNA. (C) GMS quantification of histology. (D) Representative images of GMS-stained sections from γδ T cells with IA, treated or untreated with caspofungin. A and B are a summary of two experiments with 7-10 mice/group. C and D are a summary of 4 mice/group. * p < 0.05. ** p < 0.01. *** p < 0.001. This figure was modified from a previous publication8. Please click here to view a larger version of this figure.
Discussion
The purpose of this article was to introduce a method for determination of lung fungal burden in mice with IA by utilizing GMS-stained lung histological sections for image analysis and quantification. In this study, treatment with the β-glucan-synthesis-targeting antifungal drug caspofungin14 did not improve survival or fungal burden in neutropenic wild-type mice with IA8. However, in the absence of eosinophils or γδ T cells, survival and fungal burden improved. The results of our study also demonstrated that comparable results may be obtained by GMS fungal burden in comparison to the widely-used fungal DNA qPCR method6.
There are several benefits to utilizing GMS fungal burden quantification. First, the process may utilize existing histological samples, thus potentially reducing the number of experiments required to determine significant differences. Second, in this study, less animals were required for GMS fungal burden quantification to achieve significant differences in comparison to qPCR of fungal DNA (Figure 6, Figure 7). Third, comparison of fungal burden in different isolates by qPCR may be affected by isolate-dependent differences in ribosomal DNA copy number15. In contrast, GMS quantification is not affected by copy number, as fungal burden is determined by relative levels of lung fungal growth. Thus, the use of GMS quantification for fungal burden reduces the use of vertebrate animals and does not require pre-determination of rDNA copy number. Finally, in addition to modifying fungal morphology, caspofungin therapy increases fungal fragmentation, and thus may artificially increase fungal burden when measured by isolation of colony forming units from lung homogenates12,16,17. Thus, GMS quantification of fungal burden avoids several limitations inherent with other commonly used methods.
However, the limitations of GMS quantification and/or this study are important to note. First, the authors assumed a comparable distribution of hyphal growth throughout the lungs of each experimental group, and thus used quantification from 4 representative 10x objective fields as a representative measurement for the fungal burden in the entire lung (Figure 6B). It is possible that in some instances the relative distribution, size, and density of hyphal foci would sufficiently differ so that the fungal burden would appear different with this method and equivalent by the qPCR method. However, our additional results with whole lung section quantification using the 4X objective fields showed similar, albeit less statistically significant, differences between groups (Figure 6C). The standard error of this quantification was increased with this strategy, likely due to decreased hyphal resolution with the 4X objective and increased background in suboptimal fields. Therefore, a representative quantification of fewer fields at higher magnification is preferred. Second, only a single, central section was used for each sample. It is possible, based on the fungal isolates or mice strains used, that some studies may result in an uneven distribution of hyphal growth. In those instances, additional sections throughout each paraffin block should be quantified to obtain a more representative burden. Third, in experiments that induce substantial production of mucins (i.e., quantifying airway fungal growth in allergic bronchopulmonary aspergillosis (ABPA)18 or cystic fibrosis (CF)18), GMS reactivity with polysaccharide-rich mucins19 could yield non-specific GMS+ results and thus skew some samples in favor of higher fungal burden. Since only the neutropenic model of IA was used in this study, it is possible that the use of other immune competent or suppressive models could results in less comparable results. Despite these caveats, GMS quantification provides a comparable technique to determine fungal burden, and its continued use in additional studies may further validate the utility of this method as consistent, reliable, and cost-effective.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This study was supported in part by an Indiana University School of Medicine Research Enhancement Grant and by NIH-NIAID 1R03AI122127-01. N.A. was partly supported during this period by a Careers in Immunology Fellowship from the American Association of Immunologists.
Materials
| Aspergillus fumigatus 293 Stock Solution | Fungal Genetics Stock Center | FGSC #A1100 | |
| HyPure Cell Culture Grade Water | Thermo Fisher Scientific | SH30529.03 | |
| Malt Extract | MP Biomedicals | 2155315 | White Powder |
| BD BBL Acidicase Dehydrated Culture Media: Peptone | Fisher Scientific | L11843 | |
| Dextrose (D-Glucose) Anhydrous (Granular Powder/Certified ACS) | Fisher Scientific | D16-3 | |
| Fisher BioReagents Agar, Powder / Flakes, Fisher BioReagents | Fisher Scientific | BP1423-500 | |
| Auto Dry Cabinet | Shanghai Hasuc Instrument Manufacture Co.,LTD | HSFC160FD | |
| 1300 Series Class II, Type A2 Biological Safety Cabinet | Thermo Fisher Scientific | 1375 | |
| 0.5 mm Glass Beads | BioSpec Products | 11079105 | |
| 15 ml Conical Tubes | Thermo Fisher Scientific | 339650 | |
| Hemacytometer | Fisher Scientific | # 0267110 | |
| Leica Model DME Microscope | Leica | 13595XXX | |
| Dulbecco's Phosphate Buffered Saline | Sigma-Aldrich | D8662-500 ml | |
| 1.5 ml tubes | Fisher Scientific | # 05-408-129 | |
| BD Precisionglide syringe needles, gauge 27, L 1/2 in. | Sigma-Aldrich | Z192384 | |
| Anti-mouse-Ly-6G antibody | BioXCell | BP0075-1 | Clone 1A8 |
| Caspofungin diacetate | Sigma-Aldrich | SML0425 | |
| Isoflurane | Henry Schein Animal Health | 1169567762 | |
| Non-Rebreathing Table Top Veterinary Anesthesia Machine | Supera Anesthesia Innovations | M3000 | |
| Pureline Oxygen Concentrator | Supera Anesthesia Innovations | OC8000 | |
| Slant Board Restraint | Indiana State University Facilities Management | Custom made | |
| Gilson PIPETMAN Classic 200 ml Pipets | Fisher Scientific | F123601G | |
| Pentobarbital Sodium (Fatal-Plus) | Vortech Pharmaceuticals | 0298-9373-68 | |
| General-Purpose Broad-Tipped Forceps | Fisher Scientific | 10-300 | |
| Fisherbrand Standard Dissecting Scissors | Fisher Scientific | 08-951-20 | |
| Fisherbrand General-Purpose Curved Forceps | Fisher Scientific | 10-275 | |
| Ethyl Alcohol-200 Proof | PHARMCO-AAPER | 111000200 | |
| Fisherbrand Absorbent Underpads | Fisher Scientific | 14-206-63 | |
| BD Precisionglide syringe needles, gauge 25, L 1 in. | Fisher Scientific | Z192406 | |
| All-Plastic Norm-Ject Syringes | Fisher Scientific | 14-817-30 | |
| IV CATH ANGIOCATH 22GX1GIN 50B | Fisher Scientific | NC9742754 | |
| Formalin Solution, neutral buffered, 10% | Sigma-Aldrich | HT501128-4L | |
| 50 ml Conical Tubes | Thermo Fisher Scientific | 339652 | |
| Thermo Scientific Shandon Embedding Cassettes II in Tube Packs | Fisher Scientific | B1000729 | |
| Fisherfinest Histoplast Paraffin Wax | Fisher Scientific | 22-900-700 | |
| Disposable Base Molds | Fisher Scientific | 22-363-554 | |
| Reichert Jung Histocut 820 Microtome | Labequip.com | 31930 | |
| Water Bath | Precision Scientific | 66630-23 | |
| CO2 Incubator | Fisher Scientific | 116875H | |
| Silver Stain (Modified GMS) Kit | Sigma-Aldrich | HT100A-1KT | |
| Fast Green FCF | Fisher Scientific | AC410530250 | |
| Frosted Microscope Slides | Fisher Scientific | 12-550-343 | |
| Fisherfinest Superslip Cover Glass | Fisher Scientific | 12-545-89 | |
| Cytoseal XYL | Fisher Scientific | 22-050-262 | |
| Olympus Provis AX70 Microscope | Olympus | OLYMPUS-AX70 | |
| U-PHOTO Universal Photo System | Olympus | OLYMPUS-U-PHOTO | |
| U-MCB-2 MULTI CONTROL BOX | Olympus | OLYMPUS-U-MCB-2 | |
| U-PS POWER SUPPLY UNIT | Olympus | OLYMPUS-U-PS | |
| FreeZone 1 Liter Benchtop Freeze Dry System | LABCONCO | 7740020 | |
| Maxima C Plus Vacuum Pump | Fisher Scientific | 01-257-80 | Displacement- 6.1 cfm |
| 1-Butanol | Fisher Scientific | A383-4 | |
| AMRESCO PHENOL-CHLORFORM-OSOAMYL 100ML DFS | Fisher Scientific | NC9573988 | |
| Free-Standing Microcentrifuge Tubes with Screw Caps | Fisher Scientific | # 02-682-557 | |
| Mini-Beadbeater-24 | BioSpec Products | 112011 | |
| BioSpec ProductsSupplier Diversity Partner 2.3 MM ZIRCONIA BEADS | Fisher Scientific | NC0451999 | |
| Tris Base (White Crystals or Crystalline Powder/Molecular Biology) | Fisher Scientific | BP152-1 | |
| Hydrochloric Acid, Certified ACS Plus | Fisher Scientific | A144S | |
| Ethylenediaminetetraacetic Acid, Disodium Salt Dihydrate | Fisher Scientific | S311 | |
| Sodium Dodecyl Sulfate (SDS), White Powder, Electrophoresis | Fisher Scientific | BP166 | |
| Chloroform/isoamyl alcohol 24:1 | Fisher Scientific | AC327155000 | |
| Biotek Epoch Microplate Spectrophotometer | Fisher Scientific | 11-120-570 | |
| Thermo Scientific™ ABsolute Blue qPCR Mixes | Fisher Scientific | AB4137A | |
| Hybridization Probe, 5′-FAM-AGCCAGCGGCCCGCAAATG-TAMRA-3′ | Integrated DNA Technologies | N/A | |
| Sense Amplification Primer, 5′-GGCCCTTAAATAGCCCGGT-3′ | Integrated DNA Technologies | N/A | |
| Antisense Amplification Primer, 5′-TGAGCCGATAGTCCCCCTAA-3′ | Integrated DNA Technologies | N/A | |
| Applied Biosystems™ MicroAmp™ Optical 8-Tube Strip, 0.2mL | Fisher Scientific | 43-165-67 | |
| Thermo Scientific Domed and Flat PCR Cap Strips | Fisher Scientific | AB-0386 | |
| Mx3005P QPCR System, 110 Volt | Agilent | 401443 | Stratagene is now owned by Agilent |
| BALB/c mice | The Jackson Laboratory | 000651 | |
| C57BL/6 (B6) mice | The Jackson Laboratory | 000664 | |
| Eosinophil-deficient (ΔdblGATA, BALB/c background) mice | The Jackson Laboratory | 005653 | |
| γδ T cell-deficient (TCRδ-/-, B6 background) mice | The Jackson Laboratory | 002120 | |
| ImageJ Software | National Institutes of Health | N/A | https://imagej.nih.gov/ij/ |
| Spot Advanced Software | Spot Imaging | SPOT53A | http://www.spotimaging.com/software/spot-advanced/ |
| GraphPad Prism 6 | GraphPad Software | N/A | https://www.graphpad.com/scientific-software/prism/ |
| Fisherbrand Absorbent Underpads | Fisher Scientific | 14-206-62 | |
| Step 4.5 | http://www.bdbiosciences.com/sg/resources/protocols/paraffin_sections.jsp ; http://www.jove.com/science-education/5039 ; http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/General_Information/1/ht100.pdf |
Referencias
- Hohl, T. M., Feldmesser, M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 6 (11), 1953-1963 (2007).
- Kwon-Chung, K. J., Sugui, J. A. Aspergillus fumigatus–what makes the species a ubiquitous human fungal pathogen?. PLoS Pathog. 9 (12), e1003743 (2013).
- Ostrosky-Zeichner, L., Casadevall, A., Galgiani, J. N., Odds, F. C., Rex, J. H. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov. 9 (9), 719-727 (2010).
- Desoubeaux, G., Cray, C. Rodent Models of Invasive Aspergillosis due to Aspergillus fumigatus: Still a Long Path toward Standardization. Front Microbiol. 8, 841 (2017).
- Hohl, T. M. Overview of vertebrate animal models of fungal infection. J Immunol Methods. 410, 100-112 (2014).
- Sheppard, D. C., et al. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin Microbiol Infect. 12 (4), 376-380 (2006).
- Patterson, T. F. The future of animal models of invasive aspergillosis. Med Mycol. 43 Suppl 1, S115-S119 (2005).
- Amarsaikhan, N., et al. Caspofungin Increases Fungal Chitin and Eosinophil and gammadelta T Cell-Dependent Pathology in Invasive Aspergillosis. J Immunol. 199 (2), 624-632 (2017).
- Templeton, S. P., Buskirk, A. D., Law, B., Green, B. J., Beezhold, D. H. Role of germination in murine airway CD8+ T-cell responses to Aspergillus conidia. PLoS One. 6 (4), e18777 (2011).
- Brunel, S. F., et al. Live Imaging of Antifungal Activity by Human Primary Neutrophils and Monocytes in Response to A. fumigatus. J Vis Exp. (122), (2017).
- Rao, G. V., et al. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J Toxicol Environ Health A. 66 (15), 1441-1452 (2003).
- Bowman, J. C., et al. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob Agents Chemother. 45 (12), 3474-3481 (2001).
- Li, H., et al. The small GTPase RacA mediates intracellular reactive oxygen species production, polarized growth, and virulence in the human fungal pathogen Aspergillus fumigatus. Eukaryot Cell. 10 (2), 174-186 (2011).
- Sucher, A. J., Chahine, E. B., Balcer, H. E. Echinocandins: the newest class of antifungals. Ann Pharmacother. 43 (10), 1647-1657 (2009).
- Herrera, M. L., Vallor, A. C., Gelfond, J. A., Patterson, T. F., Wickes, B. L. Strain-dependent variation in 18S ribosomal DNA Copy numbers in Aspergillus fumigatus. J Clin Microbiol. 47 (5), 1325-1332 (2009).
- Kirkpatrick, W. R., Perea, S., Coco, B. J., Patterson, T. F. Efficacy of caspofungin alone and in combination with voriconazole in a Guinea pig model of invasive aspergillosis. Antimicrob Agents Chemother. 46 (8), 2564-2568 (2002).
- Petraitiene, R., et al. Antifungal efficacy of caspofungin (MK-0991) in experimental pulmonary aspergillosis in persistently neutropenic rabbits: pharmacokinetics, drug disposition, and relationship to galactomannan antigenemia. Antimicrob Agents Chemother. 46 (1), 12-23 (2002).
- Cowley, A. C., Thornton, D. J., Denning, D. W., Horsley, A. Aspergillosis and the role of mucins in cystic fibrosis. Pediatr Pulmonol. 52 (4), 548-555 (2017).
- Taylor, M. J., et al. Detection of fungal organisms in eosinophilic mucin using a fluorescein-labeled chitin-specific binding protein. Otolaryngol Head Neck Surg. 127 (5), 377-383 (2002).

