siRNA Transfection and EMSA Analyses on Freshly Isolated Human Villous Cytotrophoblasts
Summary
This protocol describes a method for efficiently transfecting siRNA in freshly isolated human villous cytotrophoblasts using microporation and identifying DNA-protein complexes in these cells. Transfected cells can be monitored by Western blot and EMSA analyses during the 4-day culture time.
Abstract
Human primary villous cytotrophoblasts are a very useful source of primary cells to study placental functions and regulatory mechanisms, and to comprehend diseases related to pregnancy. In this protocol, human primary villous cytotrophoblasts freshly isolated from placentas through a standard DNase/trypsin protocol are microporated with small interfering RNA (siRNA). This approach provided greater efficiency for siRNA transfection when compared to a lipofection-based method. Transfected cells can subsequently be analyzed by standard Western blot within a time frame of 3-4 days post-transfection. In addition, using cultured primary villous cytotrophoblasts, Electrophoretic Mobility Shift Assay (EMSA) analysis was optimized and performed on extracts from days 1 to 4. The use of these cultured primary cells and the protocol described allow for an evaluation of the implication of specific genes and transcription factors in the process of villous cytotrophoblast differentiation into a syncytiotrophoblast-like cell layer. However, the limited time span allowable in culture precludes the use of methods requiring more time, such as generation of a stable cell population. Therefore testing of this cell population requires highly optimized gene transfer protocols.
Introduction
Human placental dysfunction is associated with the development of several pregnancy-associated diseases like preeclampsia and intrauterine growth restriction 1. An important cell constituent of the placenta is the trophoblasts, which can be classified as either extravillous or villous cytotrophoblasts. Upon fusion, villous cytotrophoblasts further differentiate into the syncytiotrophoblast layer, a multinuclear cell structure with an important role in feto-maternal exchange and hormone production 2. Human primary villous cytotrophoblasts and their differentiated counterparts represent important biological samples and allow researchers to study a number of placenta-related processes, such as cell fusion, in culture. Furthermore, substantial efforts are ongoing to identify markers that will facilitate appropriate management and improve preventive therapies specific to pregnancy-related diseases. Laboratories routinely isolate primary human villous cytotrophoblasts from fresh placentas, using a standard isolation procedure based on trypsin digestion of placenta villi 3. As cultured cytotrophoblasts lose their capacity to proliferate and quickly differentiate in a syncytiotrophoblast-like layer upon culture 4, very efficient transfection methods and optimized analysis approaches are needed. Previous studies have determined optimal conditions of transfection of these primary cytotrophoblasts 5. Herein, a different method of siRNA transfection, which has been previously tested in this cell type 6, is presented. In comparison to a lipofection-based approach, this microporation method improves transfection, as assessed by the extent of silencing of specific genes.
Promoter and gene expression studies also provide a better understanding of placental function. Although more difficult to use owing to the short time frame for which primary villous cytotrophoblasts can be cultured, promoter analyses using standard protocols can nonetheless be addressed, as previously published 7. Electrophoretic Mobility Shift Assay (EMSA) is one of these commonly used in vitro methods, allowing for fast and easy monitoring of DNA-protein interactions. Nuclear extracts from these primary trophoblasts were used to test a region of the Syncytin-2 promoter for specific interactions. Results revealed that bound factors could be detected at different time points of culture and in a specific and reproducible manner.
Data presented in this protocol confirm that our transfection approach and the EMSA protocol can be used for isolated primary villous cytotrophoblasts and will be of great use to study the diverse functions of villous cytotrophoblasts in normal or pathological conditions.
Protocol
The UQAM ethics committee has approved these protocols, which are in accordance with the guidelines of the ethics committee of St-Luc Hospital of the Centre Hospitalier Universitaire de Montréal (Montréal, Canada). Participants signed an informed consent form.
1. Medium Preparation and Isolation of Primary Villous Cytotrophoblasts
- Prepare culture medium for human primary villous cytotrophoblasts by supplementing Dulbecco’s Modified Eagle’s Medium (DMEM) with 25 mM HEPES, 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin. Filter the prepared medium through sterile 0.22 µm filter and store in a -20 °C freezer in aliquots of 50 ml.
- Isolate primary villous cytotrophoblasts from fresh placentas following standard protocols 3.
- Using pliers, remove the fetal membrane from the tissue by gently scratching it off, while being careful not to remove villous tissue. Cut the remaining placental villi in cubes of about 3 x 3 cm using scalpel and scissors. Thoroughly wash with water containing 0.9% NaCl to remove maternal blood.
- Holding each cube with pliers, carefully remove the villous tissue from remaining vessels and fibrous tissue using scissors.
- For 15 to 30 mg of villous tissue, digest four times in Hank's balanced salt solution (HBSS) containing trypsin (from 9.6 x 105 to 1.8 x 106 U; final concentration 1.2 mg/ml) and deoxyribonuclease I (DNase I) (final concentration 200 µg/ml) for 30 min at 37 °C in a water bath with continuous shaking.
Note: Use 150 ml total volume for the first digestion, 100 ml for the second digestion and 75 ml for the two remaining digestions.
- Collect the supernatant containing dispersed cells and centrifuge at 1,250 x g for 15 min without brake. Remove the supernatant and the resuspend cells in 1 ml of prewarmed DMEM containing 1% penicillin/streptomycin.
- Layer the dispersed cells on top of a discontinuous 5% – 70% polyvinylpyrrolidone-coated silica gradient (see Table 1 for gradient preparation) and centrifuge for 23 min at 507 x g without brake. Remove the density layer between 1.017 to 1.033 by aspiration and collect the layers between 40% and 50% in the gradient with a new pipette. Wash cells extensively with 10 ml of prewarmed DMEM containing 1% penicillin/streptomycin.
Note: The collected fraction corresponds to densities between 1.048 and 1.062, which harbors trophoblasts. - Count the cells using a hemocytometer. Briefly, mix cells and add 10 µl in a new microcentrifuge tube. Add 10 µl of trypan blue and mix gently again. Draw up the cell suspension and fill the hemocytometer chambers. Count the cells under 10X objective.
- Place 1 x 106 and 4.5 x 106 cells into separate tubes for use in sections 1.3 and 2, respectively. Seed the remaining cells at a density of 1.5 x 106 cells/well in supplemented DMEM medium and culture at 37 °C and 5% CO2 for a maximum of 4 days for other experiments.
- Using pliers, remove the fetal membrane from the tissue by gently scratching it off, while being careful not to remove villous tissue. Cut the remaining placental villi in cubes of about 3 x 3 cm using scalpel and scissors. Thoroughly wash with water containing 0.9% NaCl to remove maternal blood.
- Evaluation of the purity of isolated primary villous cytotrophoblasts
- Spin 1 x 106 of the freshly isolated cytotrophoblasts in microcentrifuge tubes at 300 x g. Discard the supernatant and add 1 ml of prewarmed phosphate-buffered saline (PBS). Centrifuge cells at 300 x g and remove PBS by aspiration.
- Add 1 ml of cold methanol to cells and incubate at -20 °C for 20 min. Centrifuge cells at 300 x g for 5 min and remove methanol by aspiration.
Caution: Use canister mask, safety goggles and rubber gloves while handling methanol. - Add 1 ml of PBS at room temperature to rehydrate cells, centrifuge at 300 x g and remove PBS by aspiration.
- Incubate cells in PBS containing FBS (1:50 [v/v] dilution) and a FcR blocking reagent (1:10) for 45 min at room temperature to eliminate nonspecific binding.
- Wash twice with 500 µl of room temperature PBS, centrifuge cells at 300 x g for 5 min and remove PBS by aspiration. Resuspend cells in 100 µl room temperature PBS.
- Incubate cells with a mouse monoclonal fluorescein isothiocyanate (FITC)-conjugated anti-human cytokeratin-7 antibody clone LP5K (1:200) or a control isotype-matched non-specific antibody in PBS containing 0.2% BSA for 45 min at room temperature in the dark. Wash cells in PBS.
- Analyze the cells by flow cytometry 3. Use cell preparations, which demonstrate a minimum of 96% CK-7-positive cells by flow cytometry for further experiments.
2. siRNA Transfection of Human Primary Villous Cytotrophoblasts
- Transfection of human primary villous cytotrophoblasts using a microporation device
- Prepare 6-well plates containing 2 ml of supplemented DMEM medium for the incubation of microporated cells.
- After isolation of primary cytotrophoblasts (see section 1.2), take an aliquot of cells in suspension and determine cell number using a hemocytometer.
- Transfer 1.5 x 106 cells to a microcentrifuge tube and pellet cells by centrifugation at 300 x g for 5 min at room temperature. Wash the cells with 1 ml Dulbecco's PBS (DPBS) (Mg2+, Ca2+-free) and pellet the cells by centrifugation at 300 x g for 5 min at room temperature.
- Remove the supernatant and resuspend the cells gently in 100 µl of resuspension buffer.
Note: Avoid keeping cell suspension for longer than 15-30 min at room temperature, to maximize cell viability and transfection efficiency. - Add 300 ng of Syncytin-2 siRNA (or control-scrambled siRNA) to the 1.5 ml microcentrifuge tube and aspirate the cell-siRNA mixture using a 100 µl transfection pipette. Insert the pipette into the station (see List of Materials) and subject the cells to a 1,300 V single pulse of 30 msec.
- Immediately transfer the cells to 6-well plates (prepared in step 2.1.1) containing 37 °C pre-warmed supplemented medium to avoid cell damage.
- Repeat steps 2.1.5 and 2.1.6 for the remaining samples.
- Gently rock the plate for 30 sec to ensure even distribution of the cells. Incubate the plate at 37 °C in a humidified CO2 incubator for 24 to 48 hr. Refresh culture medium 24 hr after transfection.
- Evaluate transfection efficiency by Western blot analysis (see steps 3 to 6).
- Lipofection of siRNA in human primary cytotrophoblasts
- After isolation of villous cytotrophoblasts, seed the cells at a density of 1.5 x 106 cells per well in 6-well plates in 2 ml of culture medium and incubate for 18 hr.
- Dilute 300 ng of Syncytin-2-specific siRNA (or control-scrambled siRNA) and 5 µl of lipid-based transfection reagent in separate 1.5 ml tubes in a total volume of 10 µl of antibiotic- and serum-free medium. Incubate for 5 min.
- Mix the diluted reagents and incubate for an additional 20 min at room temperature to form the transfection complex. Add 90 µl of antibiotic- and serum-free medium to the mix to a final volume of 100 µl.
- Remove culture medium from the 6-well plates (step 2.2.1). Add 2 ml of fresh medium and the 100 µl transfection mix to each well. Incubate the cells at 37 °C in a CO2 incubator for 24 to 48 hr. Refresh culture medium 24 hr after transfection.
- Evaluate transfection efficiency by Western blot analysis (see steps 3 to 6).
3. Preparation of Lysates from Whole Cell Culture
- Remove the media and rinse each well (from steps 2.1.8 and 2.2.4) using pre-warmed DPBS (Mg2+, Ca2+ free). Aspirate the DPBS and add 500 µl of 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA). Incubate for 1-3 min at 37 °C in a CO2 incubator. Stop trypsin treatment by adding 500 µl of serum containing growth media.
Note: Use appropriate media volume according to the type of flask/dish: 1 ml per 107 cells/100 mm dish/150 cm2 flask; 0.5 ml per 5 x 106 cells /60 mm dish /75 cm2 flask. - Transfer cells to a 1.5 ml microcentrifuge tube, pellet cells by centrifugation at 300 x g for 2 min at room temperature. Wash the cells with DPBS (Mg2+, Ca2+-free) and pellet cells by centrifugation at 300 x g for 2 min at room temperature.
- Carefully remove supernatant without disturbing the pellet. Place the tube on ice. Resuspend the pellet in 50-200 µl ice-cold Radio-Immunoprecipitation Assay (RIPA) buffer (see Table 2 for buffer composition) containing freshly added protease and phosphatase inhibitors at a 1x final concentration. Briefly, vortex the tube and leave on ice for 30 min.
- Spin at 16,000 x g for 20 min at 4 °C. Carefully place the tube on ice. Using a pipette, transfer the supernatant to a fresh pre-cooled microcentrifuge tube kept on ice. Discard the pellet.
4. Preparation of Nuclear Extracts from Cultured Cells
- For each well (step 1.2.5), wash cultured cytotrophoblasts twice with 1 ml of pre-warmed DPBS (Mg2+, Ca2+ free). Aspirate DPBS and add 500 µl of 0.25% trypsin/EDTA. Incubate for 1-3 min at 37 °C in a CO2 incubator and stop trypsin treatment by adding 500 µl of serum containing growth media.
- Transfer cells to a 1.5 ml microcentrifuge tube, pellet cells by centrifugation at 300 x g for 2 min at room temperature. Wash the cells with DPBS (Mg2+, Ca2+-free) and pellet cells by centrifugation at 300 x g for 2 min at room temperature. Carefully remove all supernatant without disrupting the pellet and place the tube on ice.
- Extract nuclear protein according to manufacturer's instructions. Determine protein concentration of each sample using protein quantification assay (Bradford or bicinchoninic acid assay (BCA)).
5. Sample Preparation and Electrophoresis
- Determine the protein concentration for each cell extract using a protein quantification assay (for example, Bradford 8 or BCA 9).
- As per manufacturer's instructions (if antibodies could be used in reduction and denaturation conditions), add an equal volume of 2x Laemmli Sample Buffer (Table 2) to 20-30 µg of total protein.
- Boil cell lysates at 100 °C for 5 min. Aliquot 50-100 µl of lysate to avoid repeated freeze/thaw cycles and store the remaining tubes at -20 °C for future use.
- While the samples are heating, prepare 1 L of 1x running buffer (Table 2).
- Spin the samples at 16,000 x g for a few seconds and place them on ice.
- Load equal amount of protein into each well of a 12% SDS-PAGE gel (see Table 2 for gel composition), along with a molecular weight marker. Run the gel for 1 to 2 hr at 100 V.
6. Transferring the Protein from the Gel to the Membrane
- Activate polyvinylidene fluoride (PVDF) membrane with methanol for 1 min and rinse with 1x transfer buffer/10% methanol solution. According to manufacturer's instructions, transfer for 30 min to 1 hr depending on the protein size.
Note: Transfer efficiency can be assessed using Ponceau Red staining before the blocking step.
7. Antibody Staining
- Block the membrane for 1 hr at room temperature or overnight at 4 °C using 5% blocking solution.
- Incubate the membrane with anti-Syncytin-2 antibodies (4 µg/ml; 1:5,000) 6 or with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 0.4 µg/ml; 1:500) in 5% blocking solution overnight at 4 °C (anti-Syncytin-2) or for 45 min at room temperature. Wash the membrane three times in Tris-buffered saline Tween (TBST; see Table 2 for buffer composition) for 5 min.
- Incubate the membrane with appropriate horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse antibody (1:10,000) in 5% blocking buffer in TBST at room temperature for 1 hr. Wash the membrane three times in TBST for 5 min and then rinse in TBS. Detect signals using the chemiluminescence-based blotting substrate.
8. Radiolabeling of Single Stranded Oligonucleotide
- Phosphorylation reaction
- In a microcentrifuge tube, add 50 ng of a forward oligonucleotide along with 1x T4 polynucleotide kinase buffer, 1 U T4 polynucleotide kinase and 20 µCi (γ-32P) ATP and make up the reaction volume to 20 µl with nuclease-free water.
- Incubate at 37 °C for 30 min. Stop the reaction by adding 5 µl of 0.5 M EDTA. Make up the volume to 50 µl with nuclease-free water.
- Removal of unincorporated nucleotides from oligonucleotides
- Prepare a G-25 column equilibrated with TE buffer (Table 2) following manufacturer's instructions. Place the column in a fresh DNase-free 1.5 ml microcentrifuge tube.
- Slowly add 50 µl of probe (from step 8.1.2) over the resin, being careful not to disturb the resin bed. Spin for 2 min at 350 x g to collect the purified sample in the 1.5 ml microcentrifuge tube. Wash the G-25 column with 36 µl of nuclease-free water and spin for 2 min at 350 x g to collect it in the microcentrifuge tube.
- Hybridization with the complementary strand oligonucleotide
- Transfer the radiolabeled probe (86 µl) to a 1.5 ml microcentrifuge tube and add 200 ng of the reverse oligonucleotide and 1x of annealing buffer (Table 2). Heat 5 min at 95 °C and leave overnight at room temperature for cooling.
- Prepare non-radioactive double-stranded oligonucleotides by adding 50 ng of unlabeled forward and reverse oligonucleotides and 1x of annealing buffer in a 1.5 ml microcentrifuge tube. Make up the volume to 100 µl with nuclease-free water. Heat and keep at room temperature as described in step 8.3.1.
9. Preparation of a Non-denaturing Polyacrylamide Gel for EMSA
- Prepare a 4% native gel (10 x 12 cm) with a 1.5 mm space-comb (see Table 2 for gel composition). Clean all glass plates using distilled water.
Note: It is critical that the plates be completely free of ionic detergent, like SDS. - Immediately pour the acrylamide solution in between the plates and allow polymerization to proceed (approximately 30 min).
- Assemble the electrophoresis apparatus and fill the tank with 0.5x TBE. Pre-run the gel for 30 min to 1 hr at 150 V before sample loading.
10. DNA Binding Reaction
- In a 1.5 ml microcentrifuge tube, add 10 µg of nuclear extract (step 4.3) in 1x Gel Shift Binding buffer (Table 2) and make up the final volume to 10 µl with nuclease-free water. As a control, prepare a tube with no nuclear extract. For the competition reaction, add 1 µl of cold non-specific or specific double-stranded oligonucleotide (step 8.3.2).
- Add 1 µl of radioactive probe from step 8.3.1 to each reaction. Incubate the reaction at room temperature for 20 min. Add 1 µl of 10x gel loading buffer per reaction and load each sample on the gel prepared in section 9.
- Run the gel for 1 1/2 to 2 hr at 150 V. After migration, wrap the gel and the glass in a plastic wrap and position in an exposure cassette below a phosphor screen. Leave cassette at 4 °C for 24 hr.
- Remove the screen from the exposure cassette and insert in the phosphor imaging device for scanning.
Representative Results
Fresh placentas from term pregnancies were used to isolate human primary villous cytotrophoblasts to conduct the set of experiments presented in the Protocol section. Following their isolation, we first analyzed the purity of cytotrophoblasts through the use of the cytokeratin-7 marker (Figure 1). Cell preparations were thus stained using a monoclonal anti-cytokeratin-7 antibody. Figure 1 represents results from a typical experiment following purification of primary villous cytotrophoblasts, analyzed by standard flow cytometry. Following the density gradient step, >97% cells (in this case, 99%) are positively stained for cytokeratin-7 in standard experiments, thereby demonstrating a very high degree of efficiency in the purification of this cell type.
After their isolation, human primary cytotrophoblasts can be directly used for transfection. Two different protocols were evaluated, i.e., microporation and lipid-based transfection. Both protocols were tested with siRNA specific to Syncytin-2 mRNA and compared to a scrambled siRNA (negative control). Transfection efficiency was next evaluated by Western blot analysis. Results confirmed that Syncytin-2 expression was significantly silenced in cytotrophoblast-derived extracts upon microporation (Figure 2). On the other hand, no significant silencing was noted when siRNAs were transfected by the lipid-based transfection reagent.
We also conducted EMSA experiments on freshly isolated cytotrophoblasts. A radiolabeled probe that originated from a Syncytin-2 promoter region was synthesized. The synthesized probe (termed WT; 5'-CTCTAGGAACACCTGACTGATAAGGGAAAAATGTC-3') has been previously shown to bind CREB-related transcription factors 7. In the presence of nuclear extracts from 24 and 48 hr cultured villous cytotrophoblasts, the Syncytin-2-promoter-derived probe showed the presence of two DNA-protein complexes. Addition of 100x cold WT oligonucleotide competed for the formation of the complex, while no similar competition with excess of a cold unrelated oligonucleotide (mouse Neural Crest Enhancer 2; 5′-GATCCTGTGATTTTCGTCTTGGGTGGTCTCCCTCG-3') was observed, confirming the specificity of both signals (Figure 3).
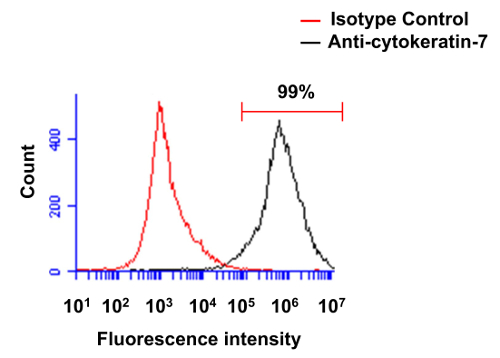
Figure 1. Evaluation of the purity of primary villous cytotrophoblasts through flow cytometry. Freshly isolated preparations of villous cytotrophoblasts were first permeabilized and then labeled with isotype control antibody (red line) or monoclonal antibody specific for cytokeratin-7 (black line). Cells were analyzed by flow cytometry. Please click here to view a larger version of this figure.
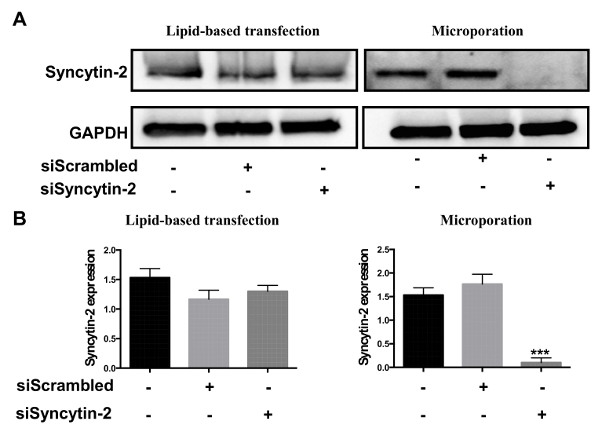
Figure 2. Comparison of siRNA transfection efficiencies between lipid-based transfection and microporation. Freshly isolated villous cytotrophoblasts were transfected with 300 ng of Syncytin-2-specific siRNA vs. a scrambled control siRNA using the lipid-based reagent or microporation. (A) Western blot analyses were performed on cellular extracts from each transfection condition using anti-Syncytin-2 and anti-GAPDH antibodies. (B) Syncytin-2 protein levels from Western blot analyses were quantified in terms of band intensity following normalization with corresponding GAPDH signals (Error bars are defined as SD, ***p<0.001). Please click here to view a larger version of this figure.
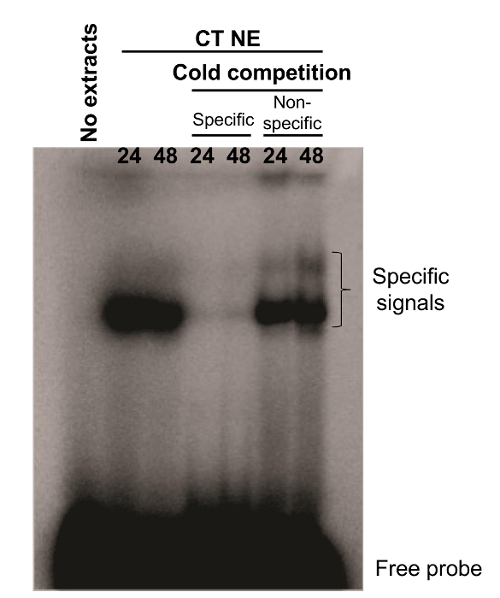
Figure 3. DNA-protein complexes identified by EMSA analysis. Nuclear extracts from primary villous cytotrophoblasts cultured for 24 or 48 hr were incubated with a WT probe corresponding to a promoter region of Syncytin-2 with or without excess (100x) of specific or nonspecific cold oligonucleotides. DNA-protein complexes were separated upon migration on a native gel. Specific signals and free probes are indicated on the right side of the gel. WT probe incubated in the absence of nuclear extract was also used as a negative control. CT NE = cytotrophoblast nuclear extracts. Please click here to view a larger version of this figure.
| % of Percoll final | Volume of Percoll (90%) (ml) | Volume of HBSS (ml) |
| 70 | 2.33 | 0.67 |
| 65 | 2.17 | 0.83 |
| 60 | 2.00 | 1.00 |
| 55 | 1.83 | 1.17 |
| 50 | 1.67 | 1.33 |
| 45 | 1.50 | 1.50 |
| 40 | 1.33 | 1.67 |
| 35 | 1.17 | 1.83 |
| 30 | 1.00 | 2.00 |
| 25 | 0.83 | 2.17 |
| 20 | 0.67 | 2.33 |
| 15 | 0.50 | 2.50 |
| 10 | 0.33 | 2.67 |
| 5 | 0.17 | 2.83 |
Table 1. 5%-70% polyvinylpyrrolidone-coated silica gradient setting.
| Buffer | Composition | Comments/Description |
| PBS 1x | 0.9 mM CaCl2, 2.7 mM KCl, 1.5 mM KH2PO4, 0.5 mM MgCl2, 136.9 mM NaCl and 0.8 mM Na2HPO4 | |
| RIPA Buffer | 150 mM NaCl, 1.0% NP-40 or 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS (sodium dodecyl sulfate), 50 mM Tris-HCl pH 8.0 | |
| Laemmli Sample Buffer | 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 M Tris-HCl, pH 6.8 | |
| 10x running buffer | 25 mM Tris base, 190 mM glycine, 0.1% SDS | |
| 10x transfer buffer | 25 mM Tris base, 192 mM Glycine | |
| TBST 1x buffer | 10 mM Tris-HCl, 15 mM NaCl, 0.05% Tween 20 at pH 7 | Mix well and filter. Failure to filter can lead to “spotting” of the membrane. |
| Blocking buffer | 5% milk or BSA (bovine serum albumin) added to TBST 1x buffer | |
| TE Buffer | 10 mM Tris-HCl (pH 8.0), 1 mM EDTA | |
| Annealing buffer | 1 M NaCl, 50 mM Tris-HCl pH 7.5, 100 mM MgCl2, 0,2 mM EDTA, 1 mM DTT | |
| 5x Gel Shift Binding Buffer | 20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, 50 mM Tris-HCl (pH 7.5) and 0.25 mg/ml poly(dI-dC). | |
| Loading buffer | 250 mM Tris-HCl (pH 7.5), 0.2% bromophenol blue and 40% glycerol | |
| TBE 5x | Dissolve 54 g of Tris Base and 27.5 g of boric acid in 1 L deionized water, which includes 20 ml 0.5 M EDTA, pH 8 | |
| 4% native gel | TBE 5x buffer | 6.0 ml |
| 29:1 acrylamide/bisacrylamide (30% w/v) | 10 ml | |
| 40% acrylamide (w/v) | 0.75 ml | |
| 100% glycerol | 1.5 ml | |
| Distilled water | 41.5 ml | |
| Ammonium persulfate 25% | 250 ml | |
| TEMED | 75 ml | |
| 4% Stacking Gel (6 ml) for SDS-PAGE | ddH2O | 4.1 ml |
| 30% Acrylamide | 1.0 ml | |
| 1.0 M Tris | 0.75 ml | |
| 10% SDS | 60 μl | |
| 10% APS | 60 μl | |
| TEMED | 6 μl | |
| 12% Separating Gel (10 ml) for SDS-PAGE | ddH2O | 3.3 ml |
| 30% Acrylamide | 4.0 ml | |
| 1.5 M Tris | 2.5 ml | |
| 10% SDS | 100 μl | |
| 10% APS | 100 μl | |
| TEMED | 4 μl |
Table 2. Composition of buffers.
Discussion
Studies on human placental function and development have been greatly improved by protocols aimed at optimizing isolation of various placental cell populations. One of the best studied placental cell population remains the villous cytotrophoblasts, the study of which has greatly benefited from optimized protocols permitting efficient and reliable isolation. This has further allowed a number of experiments, such as transfection and promoter studies. Using a previously described protocol 3, pure populations of primary villous cytotrophoblasts can be obtained. CK-7 is an intermediate filament protein, which is expressed in cytotrophoblasts. Monoclonal antibodies against this protein are used to determine the purity of this trophoblast population after their isolation from the placenta 10. Preparations are defined to be pure when 96% of cells are positive for CK-7. Cells can subsequently be used directly for transfection. Transfection protocols involving lipid-based approaches are broadly used to transfer nucleic acids into primary cells 11,12. However, toxicity of lipid formulation in some cells becomes a disadvantage. Microporation is an electroporation technology using a pipette tip as an electroporation space, which generates minimal heat, metal ion dissolution, pH variation and oxide formation; all of which can be deleterious to cells. Based on the data presented herein, microporation represents a more efficient approach for siRNA delivery than the lipid-based transfection protocol, as judged by Western blot analyses. Using an EMSA approach, a probe designed against a selected region of the Syncytin-2 promoter that is known to bind important factors for its activity 7 was tested. The probe was incubated with nuclear extract from isolated cytotrophoblasts. As presented in Figure 3, specific signals were obtained.
The different protocols described herein have been optimized in recent years. However, they all rely on high quality cytotrophoblast preparations, which depend on a certain number of critical steps. First and foremost, following birth, placentas need to be kept in a buffer solution and cells must be isolated from them no more than 5 hours after being immersed in the solution. Trypsin and DNase treatments are also of great importance and should be carefully undertaken to maximize the quality of cytotrophoblast preparation. Extensive scrubbing of the placenta villi and reduction of the time needed for their isolation are both additional improvements, which can strongly upgrade the efficiency of cell number isolation. The yield of this protocol further depends on optimal centrifugation during the density gradient step, which should be closely monitored. Failure to properly balance the tubes will also lead to distortion of the gradient and severely reduce cell numbers.
The transfection protocol described herein has been tested for over several years. Although conditions needed for microporation are generally straightforward, optimization of transfection for the various primary cells are nonetheless required. This also involves the optimization of the amount of transfected nucleic acid (siRNA or plasmid DNA). Thus, for the protocol described here, optimization of various siRNAs concentrations is recommended to identify an efficiently repressing siRNA. Addition of an appropriate suspension volume to the cell pellet before microporation and complete suspension of the cells are additional factors, which affect transfection efficiency.
EMSA experiments have also necessitated optimization. Given the heterogeneity associated with placenta donors and the varying cytotrophoblast isolation efficiencies and purities, EMSA experiments need to be repeated up to 5 times to confirm the results. In addition, having high cell numbers is important to ensure sufficient protein levels in nuclear extract preparations and subsequent detection of protein/ DNA complexes. Furthermore, highly radioactive oligonucleotide probes should be prepared, and should be prepared fresh from recently purchased radioactive ATP.
A series of limitations must be underscored in the presented protocol. It first needs to be stressed that flow cytometry analyses cannot assess the potential presence of syncytiotrophoblast fragments in cell preparations. Other protocols have been proposed to solve this issue, such as sorting using flow cytometry or the use of antibody-bound magnetic beads 13,14. It should be mentioned though that these fragments do not normally adhere to cell culture wells and are often removed following the cell washing step. Another limitation to the presented protocol is that primary villous cytotrophoblasts have a very limited survival time in cell culture. Hence, regardless of the selected method, stable transfection of villous cytotrophoblasts will remain unattainable. EMSA analyses, as described in this protocol, also have certain limitations. Although specificity of the signals can easily be assessed by competition experiments with excess unlabeled oligonucleotides, bound factors can only be identified using antibodies against specific transcription factors resulting in supershifted signals. EMSA experiments furthermore remain limited as it studies in vitro DNA-protein interactions. Experiments with more representative settings, such as ChIP assay, and more recent in vivo protocols are needed to further confirm the EMSA result.
The protocols described herein for cytotrophoblast isolation and transfection have important applications for studies in which transient silencing or overexpression of specific genes is needed to understand their role in function, proliferation or differentiation of a specific trophoblast cell type. In addition, upon transfection, tagged versions of expressed proteins will be suitable for intracellular tracking and analyses by standard techniques, such as confocal microscopy and flow cytometry. The EMSA protocol can be used to study intricate details pertaining to promoter regulation and the implicated transcription factors. Furthermore, this experimental approach will allow researchers to identify factors involved in the development of certain diseases related to placentation deficiency and the altered regulation of genes expressed in villous cytotrophoblasts.
In conclusion, human primary cytotrophoblasts can be easily isolated from placentas and are amenable to standard protocols, such as siRNA transfection and EMSA. Although different transfection procedures, such as lipofection 15 and nucleofection 16 are available, microporation seems a very efficient method for siRNA transfection of isolated villous cytotrophoblasts, offering an approach with limited cell death and relatively low cost. In the context of villous cytotrophoblasts, this approach should permit the silencing of different genes and the assessment of their implication in various functions or characteristics of the cell type. In addition, transfection of expression vectors is also feasible using this protocol and will yield complementary data to the siRNA-based approach. With respect to EMSA, this technique provides a more rapid and simpler method of evaluating protein-DNA complexes when compared to alternative approaches, such as DNase I footprinting, methylation interference assay, chromatin immunoprecipitation and chromatin conformation analyses. Hence, this technique remains valuable in standard promoter analyses or testing of any other enhancer/suppressor-containing DNA regions. Our demonstration of its reliable use for villous cytotrophoblasts is important, as it adds information on the regulation of genes, with potential implications at the functional level. Despite their limited time in culture, primary villous cytotrophoblasts are suitable for standard studies and should be adaptable to more recent as well as informative methods.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from the National Sciences and Engineering Research Council of Canada (NSERC) (#298527) (BB). CT was supported by an institutional FARE scholarship. AV was supported by a NSERC Graham Bell Ph.D. scholarship. BB held a Canada Research Chair in Human Retrovirology (Tier 2). Thanks to Beatrix Beisner for help in revising the text.
Materials
| HBSS without Ca2+, Mg2+ | Sigma | #H2387 | |
| HBSS (10X) | Sigma | #14060-057 | |
| DMEM High Glucose without Hepes | Gibco | #12100-061 | |
| Hepes (1 M) | Gibco | #15630-080 | |
| Penicillin-Streptomycin-Neomycin (100X) | Gibco | #15640-055 | |
| Amphotericin B | Sigma | #A2411 | |
| CaCl2 | Sigma | #C4901 | |
| MgSO4.7H2O | Sigma | #M | |
| Fetal bovine serum | Gibco | #16170-078 | |
| Percoll | Sigma | #P-1644 | For density gradient |
| Syncytin-2 siRNA | Ambion Life technologies | #AM16708 | |
| Scrambled siRNA | Qiagen | # SI03650318 | |
| DNase I | Sigma-Aldrich | #D5025 | |
| Trypsine, type I | Sigma | #T8003 | |
| DharmaFECT Lipotransfection reagents | GEhealthcare | # T-2001-01 | |
| Trypsin/EDTA | Life technologies | #25300-062 | |
| Protease Inhibitor Cocktail | Roche Diagnostic | #11873580001 | |
| Pierce BCA Protein Assay Kit | Thermo Scientific | #23225 | |
| BSA | Sigma | #A7906 | |
| TWEEN 20 | Sigma | #P9416 | |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling | #7074 | |
| BM Chemiluminescence Western Blotting Substrate (POD) | Roche Diagnostic | #11500708001 | |
| DPBS | Life technologies | #14287-080 | |
| T4 Polynucleotide Kinase | NEB | #M0201S | |
| ATP, [γ-32P] | Perkin Elmer | #BLU002A100UC | |
| Acrylmide | Sigma | #A9099 | |
| TEMED | Life technologies | #17919 | |
| Ammonium Persulfate | Sigma | #A3678 | |
| Anti-human cytokeratin-7 antibody clone LP5K, FITC conjugated | Millipore, | CBL194F | Dilution1:200 |
| FcR blocking reagent | Miltenyi Biotec | 130- 059-901 | Dilution 1:10 |
| Flow Cytometer BD Acuri system | Becton Dickinson | ||
| Microporator MP-100 apparatus | Digital Bio | ||
| Resuspension Buffer R (Neon Transfection System 100 µL Kit) | Life technologies | MPK10096 | |
| PVDF membrane | Millipore | IPVH00010 | Activate with methanol |
| Anti-human GAPDH antibody | Santa Cruz Biotechnology | sc-137179 | 1:500 |
| HorseRadish Peroxidase (HRP)-conjugated goat anti-rabbit antibody or anti-mouse antibody | Cell Signalling | #7074 | 1:10,000 |
| HorseRadish Peroxidase (HRP)-conjugated goat anti-mouse antibody | Cell Signalling | #7076 | 1:10,000 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagent | Thermo Scientific | #78833 | |
| G-25 column | GE Healthcare | #27-5325-01 | |
| Chemiluminsescence and fluorescence imaging device | Montréal Biotech | Fusion FX5 | |
| 4 % native gel | Home made | ||
| PBS | Home made | 1X | |
| Personal Molecular Imager (PMI) System | BioRad | ||
Referencias
- Huppertz, B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension. 51, 970-975 (2008).
- Huppertz, B. IFPA Award in Placentology Lecture: Biology of the placental syncytiotrophoblast–myths and facts. Placenta. 31, S75-S81 (2010).
- Le Bellego, F., Vaillancourt, C., Lafond, J. Isolation and culture of term human cytotrophoblast cells and in vitro methods for studying human cytotrophoblast cells’ calcium uptake. Methods Mol. Biol. 550, 73-87 (2009).
- Morrish, D. W., et al. In vitro cultured human term cytotrophoblast: a model for normal primary epithelial cells demonstrating a spontaneous differentiation programme that requires EGF for extensive development of syncytium. Placenta. 18, 577-585 (1997).
- Forbes, K., Desforges, M., Garside, R., Aplin, J. D., Westwood, M. Methods for siRNA-mediated reduction of mRNA and protein expression in human placental explants, isolated primary cells and cell lines. Placenta. 30, 124-129 (2009).
- Vargas, A., et al. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 392, 301-318 (2009).
- Toufaily, C., Lokossou, A. G., Vargas, A., Rassart, E., Barbeau, B. A CRE/AP-1-Like Motif Is Essential for Induced Syncytin-2 Expression and Fusion in Human Trophoblast-Like Model. PLoS One. 10, e0121468 (2015).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254 (1976).
- Smith, P. K., et al. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76-85 (1985).
- Blaschitz, A., Weiss, U., Dohr, G., Desoye, G. Antibody reaction patterns in first trimester placenta: implications for trophoblast isolation and purity screening. Placenta. 21, 733-741 (2000).
- Desforges, M., et al. The SNAT4 isoform of the system A amino acid transporter is functional in human placental microvillous plasma membrane. J. Physiol. 587, 61-72 (2009).
- Felgner, P. L., et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 84, 7413-7417 (1987).
- Guilbert, L. J., et al. Preparation and functional characterization of villous cytotrophoblasts free of syncytial fragments. Placenta. 23, 175-183 (2002).
- Petroff, M. G., Phillips, T. A., Ka, H., Pace, J. L., Hunt, J. S. Isolation and culture of term human trophoblast cells. Methods Mol. Med. 121, 203-217 (2006).
- Ma, B., Zhang, S., Jiang, H., Zhao, B., Lv, H. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J. Control. Release. 123, 184-194 (2007).
- Freeley, M., Long, A. Advances in siRNA delivery to T-cells: potential clinical applications for inflammatory disease, cancer and infection. Biochem. J. 455, 133-147 (2013).

