Collection and Analysis of Arabidopsis Phloem Exudates Using the EDTA-facilitated Method
Summary
Knowledge of the composition of the phloem sap as well as the mechanism of its loading and long-distance transport is essential for the understanding of long-distance signaling in plant development and stress/pathogen response and of assimilate transport. This manuscript describes the collection of phloem exudates using the EDTA-facilitated method.
Abstract
The plant phloem is essential for the long-distance transport of (photo-) assimilates as well as of signals conveying biotic or abiotic stress. It contains sugars, amino acids, proteins, RNA, lipids and other metabolites. While there is a large interest in understanding the composition and function of the phloem, the role of many of these molecules and thus, their importance in plant development and stress response has yet to be determined. One barrier to phloem analysis lies in the fact that the phloem seals itself upon wounding. As a result, the number of plants from which phloem sap can be obtained is limited. One method that allows collection of phloem exudates from several plant species without added equipment is the EDTA-facilitated phloem exudate collection described here. While it is easy to use, it does lead to the wounding of cells and care has to be taken to remove contents of damaged cells. In addition, several controls to prove purity of the exudate are necessary. Because it is an exudation rather than a direct collection of the phloem sap (not possible in many species) only relative quantification of its contents can occur. The advantage of this method over others is that it can be used in many herbaceous or woody plant species (Perilla, Arabidopsis, poplar, etc.) and requires minimal equipment and training. It leads to reasonably large amounts of exudates that can be used for subsequent analysis of proteins, sugars, lipids, RNA, viruses and metabolites. It is simple enough that it can be used in both a research as well as in a teaching laboratory.
Introduction
Plants cannot move to escape adverse conditions. Consequently, they had to develop mechanisms to detect environmental stresses, elicit and transmit a related signal throughout the plant, and adjust development accordingly. Two transport systems exist for the distribution of water, nutrients and other (signaling) compounds. The first is the xylem; which typically transports water and minerals taken up by the roots throughout the plant. The second is the phloem. The view of the phloem has changed from a simple assimilate transport system to a conduit for RNA, protein, viruses, lipids and other small molecules. It plays an important role in assimilate and nutrient transport, response to biotic and abiotic stress, as well as in plant growth and development. It is now called the "information superhighway" of the plant18.
The phloem is comprised of several cell types: phloem parenchyma, as well as specialized companion cells and sieve elements. The sieve element is the site of long-distance movement. To allow for unobstructed longitudinal flow, sieve elements are missing most organelles as well as nuclei and are thought to contain at best a limited translational machinery21, 33. It is believed that companion cells synthesize proteins and other compounds, which travel in the phloem stream. These compounds are then transported into the sieve element via plasmodesmata and can function as long distance signals 4,16.
Several groups of compounds can be found in phloem exudates:
- Sugars are often the products of photosynthesis and are transported as "energy-carrying" molecules to other parts of the plant for storage or as building blocks. However, they can also function as antifreeze or as signaling compounds.
- Proteins can also be found in phloem exudates. They include metabolic enzymes but also have signaling function. One example of a protein which serves as a developmental signal is the Flowering locus T protein, which signals the induction of flowering as well as seasonal leaf abscission in plants.6
- Nucleic acids are present in phloem exudates in the form of mRNA, small and micro RNAs, and viral RNAs. They appear to be largely involved in signaling 27, 28, 39. At the same time, rRNA and tRNAs for almost all amino acids have been observed in the phloem sap of cucurbits42. They appear to be selectively transferred into the sieve tube system. The tRNAs show modifications necessary for the ability to transfer amino acids to the translational apparatus. Yet, rather than participating in translation, they appear to inhibit this process. Alternatively, they could serve as a source of cytokinin or signal the metabolic status of the plant42.
- Fatty acids, oxylipins, and other lipids have also been found in phloem exudates 3, 13, 14, 23. Jasmonic acid, an oxylipin moves through the phloem as part of the response to pathogen infection22, 29, 31, 32. While the role of most phloem lipids is still unclear, some of them likely have signaling function4.
The challenge in working with plant phloem lies in its capacity to seal itself upon wounding. There are four major methods used to collect phloem exudates, yet they work only in select species:
1) In cucurbits it is possible to obtain reasonable amounts of phloem exudate through cuts of the petiole. Once the initial drop with contamination of injured cells is removed, it is possible to obtain reasonably large amounts of pure phloem sap 1, 15. However, with increasing collection time, this exudate thickens, making it increasingly unsuitable for liquid chromatography approaches (unpublished). Recent publications suggest, that depending on the species, this phloem sap is derived from either the fascicular (FP) or the extrafascicular phloem (EP) and that, while it does contain "mobile" phloem sap from the sieve elements (FP), it is also prone to contamination from other cell types, including the xylem40, 41.
2) A second approach is to obtain phloem sap through shallow cuts or punctures in stem or petiole. This method has been used successfully in lupine 17, 25, cucurbits36, and Brassica napus12. Here the contamination from injured cells is minimal and the exudates very pure. However, plants need to be healthy and well-watered since it is very challenging to selectively puncture just the sieve elements. If xylem vessels are nicked, all exudate is drawn into the xylem stream. This makes the method unsuitable for plants with very fragile or highly lignified petioles or stems.
3) Aphid stylectomy allows aphids to insert their stylet into the sieve elements then removing the aphid with a laser. Phloem sap is exuded through the remaining stylet2, 9, 10, 35, 38. In theory, any plant that can be infected by aphids can be used for this approach. However, most greenhouse or growth chamber managers will not support use of a pathogen. In addition, aphids introduce several proteins into the phloem with their saliva 19, 33. This leads to a limited transcriptional reprogramming 30 and has the potential to change the phloem composition26.
4) The method described here is the EDTA-facilitated exudation of phloem sap. This method employs EDTA to prevent sealing of the phloem20. EDTA chelates Ca2+ ions that would otherwise participate in processes that seal the phloem. While EDTA can lead to cell damage30, several groups have used this method and observed no adverse effect of EDTA concentrations from 10 mM to 20 mM on the cell ultrastructure or on phloem loading and transport5, 24. To reduce any damaging effect as well as interference of EDTA with chromatography and gel electrophoresis, plants are moved into water after 1 h and only the later part of the exudate is used14. Thus, rather than exudation into EDTA, phloem is exudated into water (EDTA-facilitated exudation). It is a straight-forward, low cost, and low-tech method for the collection of phloem exudates. Phloem sap obtained this way can be used to analyze proteins, small molecules, lipids, and RNAs and has been used successfully in many plants. While a limited amount of experiments has been performed in monocots11, the method appears to be more suitable for dicots (Perilla17, 20, Arabidopsis8, 13, 14, Poplar7). Collection of exudates has to occur in a humid environment to avoid loss of exudates through transpiration. Depending on the plant, incubation in EDTA for one to two hours is sufficient to prevent sealing of the phloem/ sieve elements. The collection can then occur into water. This has the benefit of preventing the negative effect EDTA has on cell structure and stability. It also eliminates the interference of EDTA with methods like HPLC or SDS-Page. It is shown as it applies to Arabidopsis. For larger plants, incubation in EDTA and exudate collections has to be scaled up and is performed in beakers rather than in 1.7 ml reaction tubes.
Protocol
1. Preparation of Plants
- Arabidopsis seeds can be germinated directly on soil or on MS plates. Grow the plants for four to six weeks in growth chambers with a 12-hr photoperiod (22 °C day, 15 °C night14). Water plants once to twice a week. Use bottom tray for watering; do not water plants from the top; allow the trays to dry before re-watering. Make sure plants are well hydrated at time of harvest.
2. Preparation of Solutions and Dishes
- K2-EDTA solution: Make a 100 mM K2-EDTA solution stock solution, which can be kept in the refrigerator. On the day of the collection, dilute the solution one to five (one part EDTA solution, four parts water) to obtain a 20 mM K2-EDTA solution.
- Fill a glass or Petri dish (diameter 7-15 cm) halfway with the 20 mM K2-EDTA solution. If collecting samples from different treatments (for example control and salt-stressed plants or different genotypes), prepare separate dishes for each treatment.
- Fill desired number of 1.7 ml reaction tubes with 1.4 ml of 20 mM K2-EDTA solution. Fill an equal number of reaction tubes with 1.4 ml autoclaved deionized or Millipore water.
- Put paper towels on the bench to set plants on; get one new razor blade and put on gloves.
3. Collection of Phloem Exudates (Depicted in Flowchart in Figure 1)
- Harvest rosette leaves from four to six week-old plants by cutting them with the razor blade at the base of the petiole close to the center of the rosette.
- Immediately place the leaves in the dishes containing 20 mM K2-EDTA. Make sure the cut end of the petiole is submerged in the solution (Figure 2).
- Once 15 leaves have been collected (ca. three to four plants), gently stack leaves on top of each other such that the cut petioles are aligned with each other. Recut the base of the petioles (ca. 1 mm) and immediately transfer the leaves into one of the reaction tubes with the 20 mM K2-EDTA solution. The leaves fit easiest if they are slightly rolled up. Avoid squeezing the leaves in since this might cause injury of cells and, thus, leads to the collection of leaf cell content.
- If more material is needed, gently cover the samples with a wet paper towel. Use a new set of dishes if for the collection of samples from different treatments or genotypes.
- Once all the samples are positioned in reaction tubes, collect the exudates either in the dark or in the light.
- For collection in the dark, line the floor of a large shallow dish with wet paper towels. Place the rack with the leaf-filled reaction tubes in the dish and either cover with wet paper towels or put into a black plastic back with slits for aeration. Place the entire setup in a cabinet or a dark room.
- For collection in the light, line the bottom of a clear plexiglass container with wet paper towels. Place the rack with the leaf-filled reaction tubes in the container and close it.
- After 1 hr, remove leaves gently from the container and the reaction tubes and wash thoroughly with distilled, deionized or Millipore water to remove all EDTA. This step serves three purposes: (1) it removes the EDTA to minimized damage to the cells, (2) it eliminates the interference of EDTA with SDS-PAGE and chromatography, and (3) removes any compounds that have accumulated from cut/damaged cells. Immediately transfer into the prepared reaction tubes containing autoclaved water and return to the humidified collection containers for the collection of exudates. At this point, add RNase inhibitor if the exudate needs to be used for RNA extraction. Optional: Proteinase inhibitor can be added if protein analysis is desired.
- After the intended collection time, pull out and discard the leaves and freeze the phloem exudates in liquid N2 for further use. If extended storage is needed, lyophilize the samples and store at -80 °C.
- In the case of Arabidopsis, metabolites can already be detected after collection of exudates for 1 hr. However, collection for 5-8 hr is advisable if analysis of less abundant components is planned. For larger plants like Perilla, longer collection times (8 hr) are recommended.
Exudate collected from 15 leaves is typically enough for one lane on an SDS-PAGE gel (proteins), for mRNA extraction, GC-MS (sugars and metabolites). For lipid analysis the pooling of 2-3 samples (exudate from 30-45 leaves) is recommended and leads to clearer results.
4. Subsequent Analysis (For Video We Only Show 4.4)
- For protein analysis, resuspend sample in 15 μl water and 30 μl Lämmli buffer, heat to 95 °C for 5 min, quickly spin down any precipitates and load entire sample onto a SDS-PAGE gel (45 μl loading pockets). Samples should be stained with Colloidal Coomassie or other sensitive stains since protein abundance is very low.
- For sugar and small metabolite analysis, add derivatizing agents to the dried sample as described in the literature14.
- mRNA can be analyzed using RT-PCR, microarray or RNA-Seq analysis.
- For lipid analysis immediately pool two to three samples and partition against Chloroform:Methanol (1:1) in the fume hood as follows: add an equal amount of Chloroform: Methanol to each sample, vortex for a few seconds, and centrifuge for 4 min at 400 x g. Collect the bottom (organic) phase in a glass tube with teflon cap and repeat partition with the top phase three more times. Dry the combined organic phases under a stream of N2 and submit to further analysis (Thin-layer chromatography: TLC14, 37; Liquid chromatography-Mass spectrometry: LC-MS14).
Representative Results
It has become clear over the past several years that the complexity of the phloem sap may be the answer to the question of how plants send varying developmental and stress signals for optimal response. Using EDTA-facilitated phloem exudation may provide the opportunity to analyze phloem exudates from plants that are not suitable for other approaches but may be of economic or physiological interest.
This method allows for the harvest of enough phloem exudates to identify phloem-localized proteins and detect changes in their level in response to development or stress (Figure 3). As the figure shows, proteins are in very low abundance, yet, their level is high enough for subsequent proteomics experiments using LC-MS/MS or for Western blot analysis. Findings in cucurbits suggest that the cutting of the stem leads to a disruption of the water potential balance and a subsequent influx of water and possible contaminants from the apoplast41. Yet collection for times ranging from one to eight hours does not affect the phloem protein profile/composition in Arabidopsis (only the absolute amount varies Guelette et al.), the amount of protein collected and the pattern of proteins found varies between species (Arabidopsis: A.t.; Perilla, lane I and NI) and treatments (Perilla grown at different day lengths, lane I and NI). As a result proteinaceous signals can be visualized, identified, and followed.
mRNA and micro RNAs can also be identified in phloem exudates by this method. However, treatment with RNAse inhibitor is necessary to prevent degradation of the mRNA during the extended exudation time. Since the sieve elements do not contain functional chloroplasts, Rubisco small or large subunit (RbcS or RbcL, respectively) mRNAs can be used as negative controls, while known phloem-localized mRNA like Ubiquitin-conjugating enzyme may be used as a positive control (Figure 4).
Similarly, sugars or small metabolites can be analyzed using either GC-MS or LC-MS. Here it should be mentioned that the phloem exudates contain a large number of functional enzymes, including almost the entire glycolytic pathway12. Hence, using several time points may reveal metabolic processes during the exudate collection. An example is shown in Figure 5: In all exudates, sucrose is the most abundant metabolite; this is most obvious after a collection for 1 hour. However, after five hours the sucrose to fructose ratio is slightly reduced. If the same exudate is collected for one hour and left on the bench for the next four hours, the sucrose to fructose ratio is reduced to a much larger extent, suggesting that active enzymes in the phloem exudates lead to a degradation of sucrose at room temperature (for more peak interpretations see 14). This is consistent with findings that phloem loading and transport of molecules from companion cells into the sieve elements may continue during exudation until the system loses its vitality34.
Lipids in the phloem exudates and, by extension, long-distance lipid signaling are a fairly recent field of interest in plant science. Due to their hydrophobic nature lipids are only present in low concentrations and may be bound to other molecules for solubilization. Yet, the EDTA-facilitated exudation allows for the collection of sufficient material to visualize (TLC; Figure 6) and identify (LC-MS; Figure 7) phloem lipids from several plant species. As the figure shows it is possible to identify and separate several lipid species. LC-MS allows to identify different lipid species and to monitor changes within the lipid profiles of different genotypes or treatments. This allows for the study of the role of lipids during plant development and stress response.
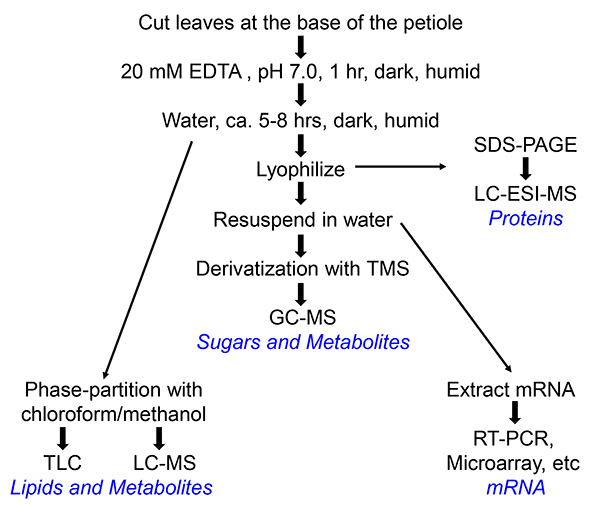
Figure 1. Flow chart of the collection of phloem exudates of Arabidopsis or Perilla. Different paths lead to different end products, which are indicated in blue. For preparation of RNA, RNAse inhibitor (100 U/ml, Roche) is added to the water into which the phloem exudate is collected.
Click here to view larger image.

Figure 2. Setup of materials for phloem exudate collection. The setup displays all materials needed for protocol steps 3.1-3.4.
Click here to view larger image.
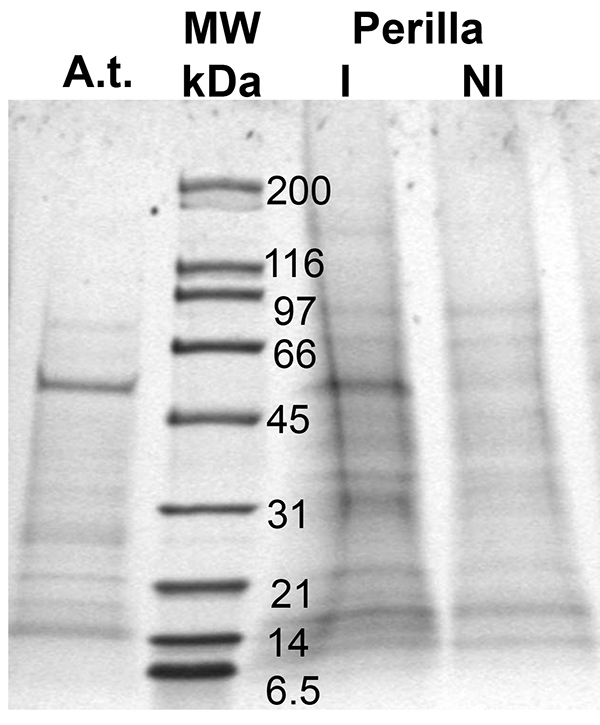
Figure 3. Proteins found in phloem exudates of two different plant species, Arabidopsis (A.t.) and Perilla (I and NI; different day lengths); MW: molecular weight marker (lane 2). Proteins were separated using a 10-20% gradient SDS-PAGE.
Click here to view larger image.
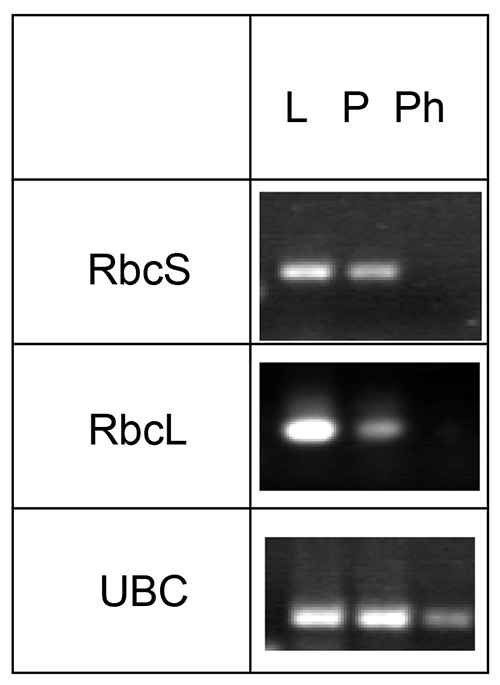
Figure 4. Analysis of the presence of mRNA for Rubisco small and large subunit (RbcS and RbcL, respectively) and ubiquitin-conjugating enzyme (UBC). mRNA was collected from Arabidopsis leaves (L), petioles (P), and phloem exudates (Ph), reverse transcribed and specific transcripts visualized using PCR. The figure was modified from Guelette et al.14.
Click here to view larger image.
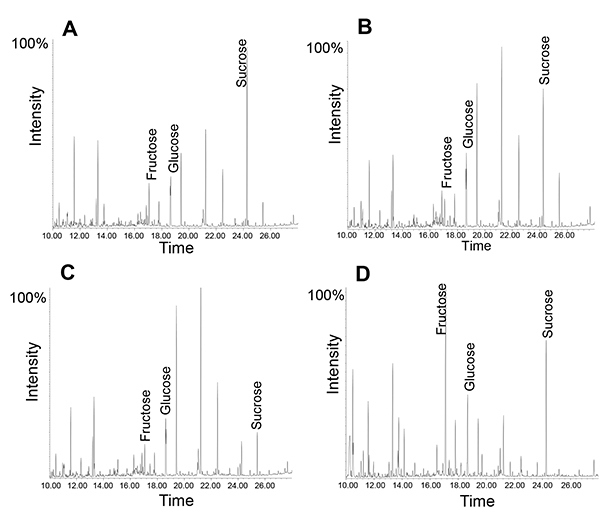
Figure 5. GC-MS profile of phloem exudates collected for different amount of times. Note how the relative amount of sucrose changes from 1 hour collection (A) to 5 hours of collection time (B). Exudate profile after collection for 1 hour followed by "incubation" at room temperature (RT) for four hours (C). Leaf metabolite profile (D).
Click here to view larger image.
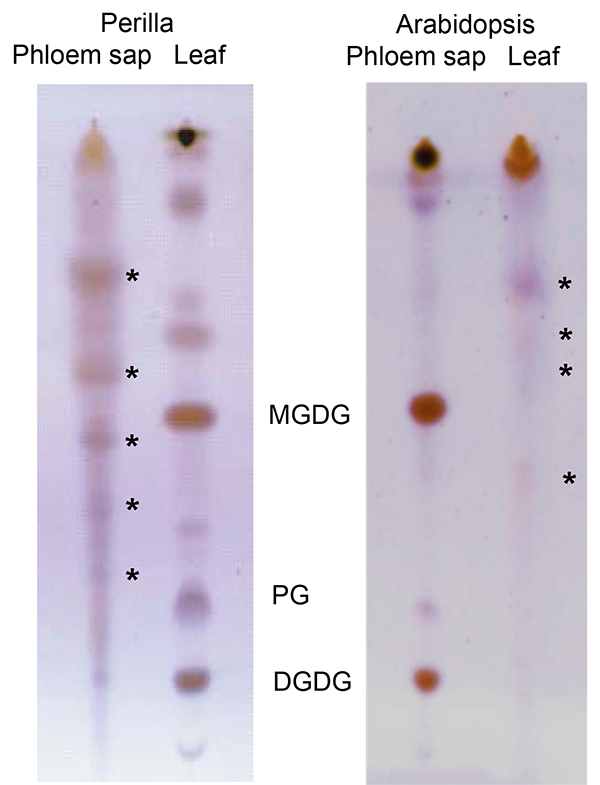
Figure 6. Thin-layer chromatogram comparing lipids from Perilla (left) and Arabidopsis (right) leaves and phloem exudates. Asterisks indicate lipids specific for phloem exudates. DGDG: digalactosyldiacylglycerol; PG: phosphatidylglycerol; MGDG: monogalactosyldiacyglycerol; The part of the figure displaying Arabidopsis lipids has been modified from Guelette et al.14.
Click here to view larger image.
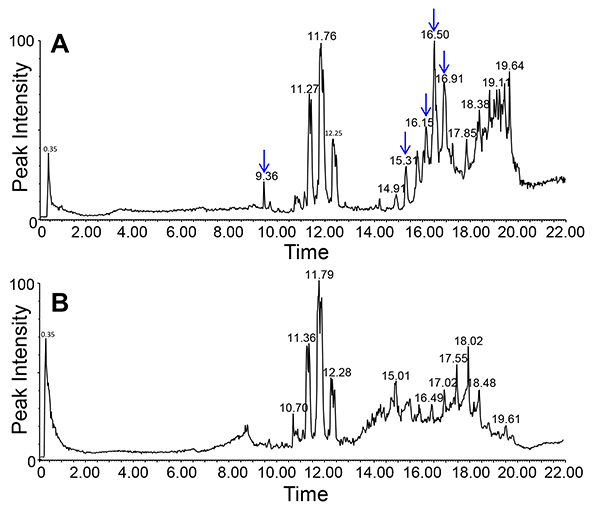
Figure 7. Lipid profile of phloem exudates of Arabidopsis (chloroform phase) using LC-MS/ negative ion mode. Graphs in the top (mutant, A) and bottom (wild-type, B) show the differences in the lipid profiles between two different genotypes (indicated by arrows).
Click here to view larger image.
Discussion
The EDTA-facilitated collection of phloem exudates is very simple, needs minimal equipment, and is applicable to most plants. Overall, this method extends the ability to analyze phloem exudates from more species. Though the exudate is diluted, it is easy to collect many different samples or from many plants to scale up to a large amount of material. This then allows for the detection of novel compounds that are in very low abundance and would otherwise be overlooked.
This method can be used for multiple plant species and works as described for the ones listed in this manuscript. If new species are explored, the two aspects that might need modification are the number of leaves used and the time of exudation. In most cases, a five to eight hour exudation should be suitable. To confirm the use of this method for a new plant species, exudate needs to be collected and one or more of the subsequent analysis as described in protocol part 4 needs to be performed. Depending on the intensity of the signal observed, the collection volume may need to be scaled up. In general, lipid and protein analysis will require more material than sugar and metabolite analysis since the former are less abundant in phloem exudates.
Because the EDTA-facilitated exudate collection involves cut surfaces as well as a potentially damaging effect of EDTA, several points have to be considered: (1) after the one hour incubation with EDTA, the petioles have to be washed thoroughly. This removes any compounds obtained from wounded cells as well as the EDTA itself, which could damage cells or interfere with the extraction. (2) Positive and negative controls need to be included to ensure data obtained are derived from phloem sap and not from injured cells (see critical steps below). (3) Phloem sap does contain several enzymes, which could be active during exudate collection. Hence, in some cases it may be useful to collect for different amounts of time. (4) Since the method involved exudation into water, an absolute quantification of the compounds is not possible.
Critical steps: The key to the successful use of this method is the use of caution while handling the sample to not injure any cells as well as the use of appropriate controls (see ref. 14). One suitable control is the collection of phloem exudates without prior treatment with EDTA. In this case the phloem will seal itself and no exudate can be collected. Any contaminating compounds coming from the wounded cells will be detected here. A second control would be the analysis of sugars in the exudate. In Arabidopsis, sucrose should be the predominant metabolite within the phloem sap, with fructose to sucrose ratio of between 1:4 and 1:8 (ref 8, 14).
For RNA extraction the use of an RNAse inhibitor is necessary. In addition, a positive control of previously described phloem-localized mRNA (UBC9: Ubiquitin conjugating enzyme 9, At4g27960; 8, 14) and a negative control of for example a chloroplast mRNA (Rubisco LSU) are advisable.
Because the exudate contains active enzymes, a time course is advised to make sure changes in the phloem profile are not simply due to variations in the collection time. In some cases, it may be beneficial to use proteinase inhibitors. However, the investigators need to keep in mind, that the collected exudate will be concentrated and that any added compounds will be largely concentrated. A second critical step within the protocol is the recutting of the petiole under EDTA. This step serves a dual purpose: First, it removes any sealed sieve plates while preventing the formation of new plugs thus allowing for free flow of the phloem sap. Second, it removes the cavitation bubble in the xylem generated while cutting and thus allows for xylem transport. The size of the second cut depends on the plants used. In plants with larger petioles it should be several millimeters (up to 1 cm) above the first cut, in plants like Arabidopsis, 1-2 mm are sufficient.
The use of this method can lead to the discovery of novel compounds in phloem exudates that could have signaling, developmental or biomedical implications. It has prompted a more in-depth look at long-distance lipid signaling, which was a little-studied field within plant science due to the lack of access to the phloem.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Science Foundation NSF-IOS grant#1144391 to SHB.
Materials
| K2-EDTA | Sigma-Aldrich | ED2P-500G | |
| Shallow Glass or plastic Petri Dish (7-15 cm) | PYREX or Corning | Any clean, shallow dish will work | |
| Chloroform | EMD | CX1054-1 | Only open containers in fume hood |
| Methanol | J.T.Baker | 9070-03 | |
| Screw cap tubes | VWR International | 53283-800 | |
| Screw cap tubes | Sun Sri | 13-425 | |
| Eppendorf tubes | Denville | C2170 |
Referencias
- Balachandran, S., Xiang, Y., Schobert, C., Thompson, G. A., Lucas, W. J. Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc. Natl. Acad. Sci. USA. 94, 14150-14158 (1997).
- Barnes, A., Bale, J., Constantinidou, C., Aston, P., Jones, A., Pritchard, J. Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J. Exp. Bot. 55, 1473-1481 (2004).
- Behmer, S. T., Grebenok, R. J., Douglas, A. E. Plant sterols and host plant suitability for a phloem feeding insect. Functional Ecology. , (2010).
- Benning, U. F., Tamot, B., Guelette, B. S., Hoffmann-Benning, S. New aspects of phloem-mediated long-distance lipid signaling in plants. Front.Plant.Sci. 3, 53> (2012).
- Chen, S., Petersen, B. L., Olsen, C. E., Schulz, A., Halkier, B. A. Long-distance phloem transport of glucosinolates in Arabidopsis. PlantPhysiol. 127, 194-201 (2001).
- Corbesier, L., et al. FT protein movement contributes to long distance signalling in floral induction of Arabidopsis. Science. 316, 1030-1033 (2007).
- Dafoe, N. J., Gowen, B. E., &Constabel, C. P. Thaumatin-like proteins are differentially expressed and localized in phloem tissues of hybrid poplar.BMC. Plant Biology. 10, (2010).
- Deeken, R., Ache, P., Kajahn, I., Klinkenberg, J., Bringmann, G., Hedrich, R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. The Plant Journal. 55, 746-759 (2008).
- Doering-Saad, C., Newbury, H. J., Bale, J. S., Pritchard, J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 53, 631-637 (2002).
- Fisher, D. B., Wu, Y., Ku, M. S. B. Turnover of soluble-proteins in the wheat sieve tube. Plant Physiol. 100, 1433-1441 (1992).
- Gaupels, F., Buhtz, A., Knauer, T., Deshmukh, S., Waller, F., van Bel, A. J. E., Kogel, K. -. H., Kehr, J. Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J Exp Bot. 59, 3297-3306 (2008).
- Giavalisco, P., Kapitza, K., Kolasa, A., Buhtz, A., Kehr, J. Towards the proteome of Brassica napus phloem sap. Proteomics. 6, 896-909 (2006).
- Guelette, B. S., Chamberlin, B., Benning, U. F., Hoffmann-Benning, S., Benning, C., Ohlroggeeds, J. Indications of lipids/lipid signaling in the phloem exudates of Arabidopsis thaliana and Perilla ocymoides. , 92-95 (2007).
- Guelette, B. S., Benning, U. F., Hoffmann-Benning, S. Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 63, 3603-3616 (2012).
- Haebel, S., Kehr, J. Matrix-assisted laser desorption/ionization time of flight mass spectrometry peptide mass fingerprints and post source decay: a tool for the identification and analysis of phloem proteins from Cucurbita maxima Duch. separated by two-dimensional polyacrylamide gel electrophoresis. Planta. 214, 3328 (2001).
- Hayashi, H., Fukuda, A., Suzui, N., Fujimaki, S. Proteins in the sieve element-companion cell complexes: their detection, localization and possible functions. Aust. J. Plant Physiol. 27, 489-496 (2000).
- Hoffmann-Benning, S., Gage, D. A., McIntosh, L., Kende, H., Zeevaart, J. A. D. Comparison of peptides in the phloem sap of flowering and non-flowering Perilla and lupine plants using microbore HPLC followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Planta. , 216-2140 (2002).
- Jorgensen, R. A., Atkinson, R. G., Forster, R. L., Lucas, W. J. An RNA-Based Information Superhighway in Plants. Science. 6, 1486-1487 (1998).
- Kehr, J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 57, 767-774 (2006).
- King, R. W., Zeevaart, J. A. Enhancement of phloem exudation from cut petioles by chelating-agents. Plant Physiol. 53, 96-103 (1974).
- Lin, M. -. K., Lee, Y. -. J., Lough, T. J., Phinney, B., Lucas, W. J. Analysis of the pumpkin phloem proteome provides functional insights into angiosperm sieve tube function. Mol. Cell. Proteomics. 8, 343-356 (2009).
- Lough, T. J., Lucas, W. J. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu.Rev. Plant Biol. 57, 203-232 (2006).
- Madey, E., Nowack, L. M., Thompson, J. E. Isolation and characterization of lipid in phloem sap of canola. Planta. 214, 625-634 (2002).
- Maeda, H., Song, W., Sage, T. L., DellaPenna, D. Tocopherols playa crucial role in low-temperature adaptation and phloem loading in Arabidopsis. The Plant Cell. 18, 2710-2732 (2006).
- Marentes, E., Grusak, M. A. Mass determination of low-molecular-weight proteins in phloem sap using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Exp. Bot. 49, 903-911 (1998).
- Prado, E., Tjallingii, W. Behavioral evidence for local reduction of aphid-induced resistance. Journal ofInsect Science. 7, 48 (2007).
- Ruiz-Medrano, R., Xoconostle-Cázares, B., Lucas, W. J. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development. 126, 4405-4419 (1999).
- Ryabov, E. V., Robinson, D. J., Taliansky, M. E. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. USA. 96, 1212-1217 (1999).
- Schilmiller, A. L., Howe, G. A. Systemic signaling in the wound response. Curr.Opin. Plant Biol. 8, 369-377 (2005).
- Thompson, G. A., Goggin, F. L. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 57, 755-766 (2006).
- Thorpe, M. R., Ferrieri, A. P., Herth, M. M., Ferrieri, R. A. (11)C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta. (11), 226-541 (2007).
- Truman, W., Bennett, M. H., Kubigsteltig, I., Turnbull, C., Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA. 104, 1075-1080 (2007).
- van Bel, A. J. E., Knoblauch, M. Sieve element and companion cell: the story of the comatose patient and the hyperactive nurse. Aust. J. Plant Physiol. 27, 477-487 (2000).
- van Bel, A. J. E., Hess, P. H. Hexoses as phloem transport sugars: the end of a dogma. J. Exp. Bot. 59, 261-272 .
- Walz, C., Juenger, M., Schad, M., Kehr, J. Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J. 31, 189-197 (2002).
- Walz, C., Giavalisco, P., Schad, M., Juenger, M., Klose, J., Kehr, J. Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry. 65, 1795-1804 (2004).
- Wang, Z., Benning, C. Arabidopsis thaliana Polar Glycerolipid Profiling by Thin Layer Chromatography (TLC) Coupled with Gas-Liquid Chromatography (GLC). J. Vis. Exp. (49), e2518 (2011).
- Will, T., van Bel, A. J. E. Physical and chemical interactions between aphids and plants. J. Exp. Bot. 57, 729-737 (2006).
- Yoo, B. C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y. M., Lough, T. J., Lucas, W. J. A systemic small RNA signaling system in plants. Plant Cell. 16, 1979-2000 (2004).
- Zhang, B., Tolstikov, V., Turnbull, C., Hicks, L. M., Fiehn, O. Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. PNAS USA. 107, 13532-13537 (2010).
- Zhang, C., Yu, X., Ayre, B. G., Turgeon, R. The Origin and Composition of Cucurbit “Phloem” Exudate. Plant Physiology. 158, 1873-1882 (2012).
- Zhang, S., Sun, L., Kragler, F. The Phloem-Delivered RNA Pool Contains Small Noncoding RNAs and Interferes with Translation. Plant Physiol. 150, 378-387 (2009).

