Quantitative Assessment of Human Neutrophil Migration Across a Cultured Bladder Epithelium
Summary
We developed an in vitro model that mimics an important component of the acute inflammatory response during infection of the bladder with uropathogenic Escherichia coli. The transuroepithelial neutrophil migration assay enables quantitative assessment of human neutrophil migration across bladder epithelia, cultured on permeable supports, in response to bacterial infection or chemoattractant substances.
Abstract
The recruitment of immune cells from the periphery to the site of inflammation is an essential step in the innate immune response at any mucosal surface. During infection of the urinary bladder, polymorphonuclear leukocytes (PMN; neutrophils) migrate from the bloodstream and traverse the bladder epithelium. Failure to resolve infection in the absence of a neutrophilic response demonstrates the importance of PMN in bladder defense. To facilitate colonization of the bladder epithelium, uropathogenic Escherichia coli (UPEC), the causative agent of the majority of urinary tract infections (UTIs), dampen the acute inflammatory response using a variety of partially defined mechanisms. To further investigate the interplay between host and bacterial pathogen, we developed an in vitro model of this aspect of the innate immune response to UPEC. In the transuroepithelial neutrophil migration assay, a variation on the Boyden chamber, cultured bladder epithelial cells are grown to confluence on the underside of a permeable support. PMN are isolated from human venous blood and are applied to the basolateral side of the bladder epithelial cell layers. PMN migration representing the physiologically relevant basolateral-to-apical direction in response to bacterial infection or chemoattractant molecules is enumerated using a hemocytometer. This model can be used to investigate interactions between UPEC and eukaryotic cells as well as to interrogate the molecular requirements for the traversal of bladder epithelia by PMN. The transuroepithelial neutrophil migration model will further our understanding of the initial inflammatory response to UPEC in the bladder.
Introduction
The movement of cells throughout the body, often across long distances, is required for growth and development, wound healing, and immune response. Cell migration is complex and requires the coordination of many different processes, including signaling cascades and the rearrangement of cytoskeletal components. Cells can move randomly (chemokinesis) as well as toward defined chemical gradients (chemotaxis). Many techniques have been developed to study cell migration in vitro. The oldest and most common technique, the Boyden chamber, consists of a vertical two-chamber system where a chemoattractant substance is placed in the bottom chamber and cells of interest are placed in the top chamber1. The movement of cells across the permeable filter, with pores of defined size, separating the two chambers is monitored. Additional techniques have been developed to investigate cell migration, including the Zigmond chamber2 and the Dunn chamber3. These collective approaches have yielded significant insight into the movement of many different cell types.
In addition to interrogating the basic principles of chemokinesis and chemotaxis, two-chamber assays have facilitated the investigation of cell migration through extracellular matrix components and both endothelial and epithelial cell layers. An advantage of two-chamber systems over other techniques is that the porous membrane can be coated with proteins such as collagen or fibrinogen, and cell migration across an extracellular matrix-like barrier can be assessed. Additionally, cultured cell lines can be grown and differentiated on the permeable supports. To investigate the movement of cells across an endothelial barrier, cultured endothelial cells are seeded and grown in the upper reservoir of the permeable supports. Motile cells, such as immune cells, are added to the upper reservoir and migration into the lower reservoir across the endothelial barrier in the physiologic apical-to-basolateral direction is observed. This model has been invaluable in understanding extravasation of immune cells out of the blood stream. In contrast to transendothelial migration, the movement of cells across an epithelial barrier typically occurs in the basolateral-to-apical direction. In order to model these events in vitro, researchers seed and grow cultured epithelial cells on the underside of the permeable supports. Motile cells are added to the upper reservoir and migration across the epithelial barrier, representing the basolateral-to-apical direction, is monitored. Such models of transepithelial migration have significantly contributed to our understanding of inflammatory responses at mucosal surfaces, particularly those of the lung and gut4,5.
In contrast, immune cell trafficking through the epithelium of the urinary tract has received much less attention. To further our understanding of innate immune responses in the urinary tract during infection with uropathogenic Escherichia coli (UPEC), we developed an in vitro assay, the transuroepithelial neutrophil migration assay, that enables investigation of polymorphonuclear leukocyte (PMN; neutrophil) movement across a bladder epithelial barrier 6-8. As with other two-chamber models of transepithelial migration, cultured human bladder epithelial cells are grown on the underside of a permeable support and form confluent epithelial layers. Human neutrophils, isolated from venous blood, are applied to the basolateral side of the epithelial layer, and migration across the epithelium in the physiologically relevant basolateral-to-apical direction is quantified in response to infection with different strains of E. coli or the presence of chemoattractant molecules. Much research had focused on neutrophil movement, both chemokinesis and chemotaxis, in the absence of additional cell types. The transuroepithelial neutrophil migration assay is advantageous as it takes into account complex interactions between bladder epithelial cells and immune cells during infection. This tractable in vitro model has the potential to permit the detailed investigation of immune responses at uroepithelial surfaces.
Protocol
1. Culturing 5637 Bladder Epithelial Cells
Perform the following steps in a laminar flow tissue-culture hood prepared with UV irradiation and wiped down with 70% ethanol.
- Prepare RPMI-1640 medium containing 10% fetal bovine serum (FBS) [termed RPMI+], filter sterilize using a 0.22 µm pore size filter, and warm to 37 °C in a water bath.
- Thaw a cryovial of 5637 cells (American Type Culture Collection HTB-9; derived from bladder carcinoma) in a 37 °C water bath. Quickly transfer the thawed cells to a 75 cm2 tissue culture flask containing 20 ml RPMI+.
- Incubate the flask at 37 °C in a humidified atmosphere with 5% CO2 until the cells are approximately 95% confluent (~6 x 106 cells per flask), about 4 days.
- To subculture the 5637 cells:
- Remove the medium from the flask. Wash the cells with 10 ml Dulbecco's Phosphate-Buffered Saline (DPBS) at room temperature.
- Add 6 ml warm (37 °C) 0.05% trypsin, 0.02% EDTA solution to the flask, and incubate at 37 °C with 5% CO2 for 15 min.
- Transfer the cells to a 15 ml conical tube, and pellet by centrifugation at 300 x g for 5 min. Remove the trypsin/EDTA solution.
- Resuspend the cells in 6 ml RPMI+ (106 cells/ml), and transfer 1 ml (106 cells) to a new 75 cm2 flask containing 20 ml RPMI+.
- Repeat step 1.4 to subculture cells every 4 days or when the cells reach 95% confluence.
2. Seeding and Growing 5637 Cells on Permeable Supports
Perform the following steps in a tissue culture hood using aseptic technique. For optimal results, use 5637 cells that have undergone fewer than 10 subcultures.
- Using sterile forceps, invert the permeable supports in a sterile 25 mm deep tissue culture dish.
- Trypsinize a 75 cm2 flask of 5637 cells at 95% confluence according to steps 1.4.1-1.4.3.
- Using a 1,000 µl pipette, gently resuspend the cells in ~2 ml RPMI+ to a concentration of 3 x 106 cells/ml, determined by cell counting using a hemocytometer.
- Apply 50 µl of the cell suspension (1.5 x 105 cells) to each permeable support without touching the membrane. Place the lid on the dish, and carefully place the dish at 37 °C with 5% CO2 for no more than 16 hr.
- Using sterile forceps, right the permeable supports into a 24-well plate containing 0.6 ml RPMI+ per well. Add 0.1 ml RPMI+ to the upper reservoir of each permeable support. Incubate at 37 °C with 5% CO2.
- Replace the medium every 2 days. First, aspirate medium from the upper reservoir followed by the lower reservoir, and then apply fresh RPMI+ in the reverse order, to the lower reservoir (0.6 ml) and then the upper reservoir (0.1 ml).
- Seven days after seeding the permeable supports with 5637 cells, assess confluence of the cells. Fill the upper reservoir with RPMI+ (~0.35 ml). The cells are sufficiently confluent when the medium does not equilibrate between the upper and lower reservoirs.
3. Preparation of the Bacterial Inoculum
- Using aseptic technique, add 20 ml of appropriate culture broth to a 250 ml flask with a cap. Use Luria-Bertani (LB) broth for E. coli cultures, and add antibiotics to the broth where appropriate.
- Using a sterile inoculation loop, inoculate the broth with bacteria from a glycerol stock or a streak plate. For UPEC strains, incubate the bacterial culture at 37 °C for approximately 16 hr, without shaking (to promote production of type 1 pili).
- Transfer the bacterial culture to a centrifuge tube. Pellet the bacteria by centrifugation at 8,000 x g for 10 min.
- Decant the supernatant, and resuspend the bacteria in PBS to OD600 = 1.000, equivalent to ~109 CFU/ml.
- To generate a heat-killed bacterial stimulus, incubate an aliquot (e.g. ~1.5 x 108 CFU in 150 µl) of the resuspended bacteria at 55 °C for 30 min. Plate an aliquot of the heat-killed suspension to confirm bacterial death.
- Immediately before use, dilute the resuspended bacteria (live or heat-killed) 10-fold to 108 CFU/ml in warm serum-free RPMI [termed RPMI-] in a microfuge tube.
4. Isolation of Human Neutrophils from Peripheral Blood
The collection of blood from adult volunteers requires advance review and approval from an institutional review board. Wear appropriate personal protective equipment and properly dispose of hazardous materials to avoid exposure to human blood.
- Approximately 25 ml of venous blood from a healthy adult volunteer should be drawn into 3 sterile sodium heparin-containing blood collection tubes by staff trained in phlebotomy.
- Tightly wrap a rubber tourniquet around the upper arm, above the elbow.
- Disinfect the entry site, the antecubital fossa, using a sterile 70% alcohol wipe.
- Attach a plastic tube holder to a winged butterfly needle system.
- Insert the needle into an antecubital vein at a 30° angle or less, bevel facing up. The needle has punctured the vein when a spurt of blood appears in the plastic tubing.
- Insert a blood collection tube into the plastic tube holder. When the first collection tube is full, replace with the second collection tube. Repeat with the third tube.
- When the third collection tube is approximately half full, remove the tourniquet.
- Cover the puncture site with a sterile cotton ball and slowly withdraw the needle from the vein. Slide the protective shield over the needle and place in a biohazard sharps container.
- Apply pressure to the puncture site. Cover the puncture site and cotton ball with an adhesive bandage.
- Gently invert the blood collection tubes to disperse the heparin.
Perform the following steps in a tissue culture hood using aseptic technique.
- Using a 10 ml pipette, gently transfer the blood to a fresh, sterile 50 ml conical tube. Add an equivalent volume of 3% (w/v) dextran in 0.9% NaCl and mix by inversion. Incubate the tube upright at room temperature for 20 min.
- Without disrupting the lower layer, carefully aspirate the upper layer and transfer it to a new 50 ml conical tube. Pellet the cells by centrifugation at 300 x g for 10 min. Discard the supernatant.
- Resuspend the cell pellet in a volume of 0.9% NaCl equivalent to the starting volume of blood.
- Layer 10 ml of density centrifugation solution under the cell suspension, preserving the interface between the two phases. Centrifuge at 400 x g for 30 min with no brake. Discard the supernatant.
- To lyse remaining red blood cells, resuspend the cell pellet in 10 ml cold 0.2% NaCl. Incubate for 30 sec, and then promptly add 10 ml cold 1.6% NaCl to restore isotonicity.
- Pellet the cells by centrifugation at 300 x g for 6 min. Discard the supernatant.
- Repeat steps 4.6-4.7 until the cell pellet appears to be free of red blood cells, typically 3 rounds of lysis.
- Using a 1,000 µl pipette, resuspend the cell pellet, primarily PMN, in warm (37 °C) RPMI- to a concentration of 107 cells/ml, determined by cell counting using a hemocytometer. Keep the cells at 37 °C until use. Typically, PMN viability and purity are >99% as assessed by trypan blue exclusion and visualization of nuclear morphology after staining, respectively.
5. Transuroepithelial Neutrophil Migration Assay
Perform the following steps in a tissue culture hood using aseptic technique. Use only permeable supports bearing confluent 5637 cell layers, as determined in step 2.7. The 24-well plates containing RPMI- can be prepared in advance and kept at 37 °C with 5% CO2 until use.
- Aliquot 1 ml warm RPMI- per well in a 24-well plate; prepare 3 wells per permeable support.
- Aspirate the medium from the upper and lower reservoirs of the permeable supports.
- Using sterile forceps, transfer the permeable supports to the 24-well plate prepared in step 5.1. Wash the permeable supports three times by transferring the supports from well to well.
- If a bacterial inoculum is to be used, invert the permeable supports in a sterile 25 mm deep tissue culture dish. If a chemoattractant (e.g., N-Formyl-Met-Leu-Phe (fMLF) or IL-8) is to be used, proceed to step 5.6.
- For experiments with live or killed bacterial stimuli, inoculate the apical side of each permeable support with 60 µl RPMI- (mock infection) or with the bacterial inoculum (6 x 106 CFU) prepared in step 3.5. Place the lid on the dish and incubate at 37 °C with 5% CO2 for 1 hr.
- Add 0.6 ml RPMI- per well to a 24-well low-attachment plate; prepare 1 well per permeable support. Prepare 3 wells containing 0.5 ml RPMI- to enumerate PMN input.
- If a chemoattractant is being used in place of bacteria, add the chemoattractant to 0.6 ml RPMI- in the 24-well plate prepared in step 5.6.
- Right the permeable supports into the 24-well low-attachment plate.
- Add 0.1 ml PMN (106 PMN), prepared in step 4.9, to the upper reservoir (basolateral side) of each permeable support. Add 100 µl PMN directly to wells containing 500 µl RPMI-. Incubate at 37 °C with 5% CO2 for 1 hr.
- Using sterile forceps, gently scrape the membrane of the permeable support against the edge of the well to remove additional PMN from the apical side and then dispose of the support.
- Collect PMN by gently scraping the bottom of each well with the 1,000 µl pipette tip, and transfer PMN suspensions to microfuge tubes.
- Enumerate PMN using a hemocytometer.
- To calculate the total number of PMN in the lower reservoir, multiply the number of PMN in 1 mm2 (100 nl) by 6,000. Data can be reported as a percentage of input PMN, or as PMN numbers normalized to 106 input PMN.
Representative Results
The transuroepithelial neutrophil migration assay enables the quantitative assessment of human PMN migration across cultured bladder epithelial cell layers in response to various stimuli (Figure 1B). While the protocol is straightforward, there are a number of variables that can influence PMN migration and consequently affect the reproducibility of this assay. Measures should be taken while preparing the permeable supports and the PMN to reduce variability between technical and biological replicates. For example, only permeable supports containing sufficiently confluent 5637 cell layers should be used in an experiment. Confluence of the 5637 cells is assessed using a functional assay that measures impermeability to liquid. If medium added to the upper reservoir equilibrates across the permeable support, then the 5637 cells are not sufficiently confluent to conduct the experiment. If the volume in the upper reservoir is maintained, then the permeable support can be used to assess PMN migration. We have measured transepithelial electrical resistance in this system, which rises modestly upon confluence of the cells; if this method is chosen, care should be taken not to contaminate the otherwise sterile setup. Confluence of the 5637 cells 7 days after seeding can be influenced by multiple factors, including the passage number of the cells and the number of cells seeded on the permeable support. In addition, the amount of time that the 5637 cells are incubated on the permeable support in the inverted position during seeding should not exceed 16 hr (Figure 1A). For optimal reproducibility, the protocol should be followed precisely. Finally, permeable supports containing confluent 5637 cell layers should be used within 1-2 days, and the membranes of the supports should never be touched during either growth of the 5637 cells or during the transuroepithelial neutrophil migration assay.
In addition to the 5637 cells, variability can also be introduced during PMN preparation. Using the protocol detailed above to isolate PMN, 1 ml of human blood typically yields about 106 PMN, although this number varies from individual to individual. Once the typical yield of an individual donor's blood is known, the isolation protocol can be scaled up or down accordingly. PMN from unhealthy or ill individuals should be avoided, and different PMN donors should be used for biological replicates to ensure that results observed are reproducible. PMN should be handled gently and aseptically to avoid activation during isolation. Lastly, the timing of the experimental procedures is crucial, as PMN do not survive for extended periods of time once removed from the body. We utilize PMN within 1 hr of completing the isolation procedure. Given these considerations, at least 3 technical replicates should be included in each biological replicate.
The number of PMN in the lower reservoir after 1 hr is shown in (Figure 2) normalized to 106 input PMN. Alternatively, PMN numbers can be compared to an internal control after normalization to input PMN, which may reduce variation between biological replicates. Adherence to the protocol outlined above with attention to detail enables the enumeration of PMN migration in response to stimuli including bacteria (Figure 2A) and chemoattractant substances (Figure 2B).
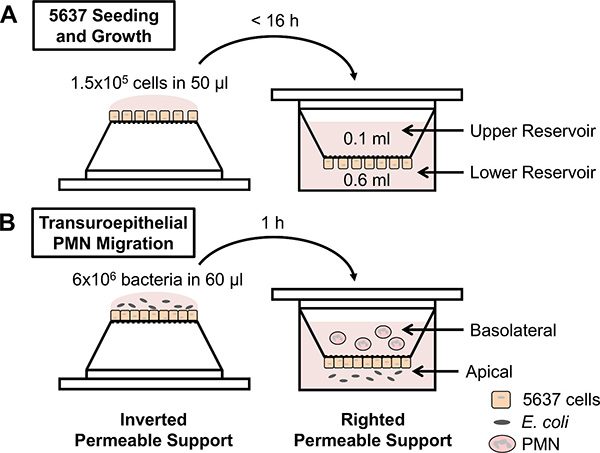
Figure 1. Schematic of experimental design. (A) 5637 bladder epithelial cells are seeded on inverted permeable supports, the supports are righted into a 24-well plate, and the cells are grown to confluence. (B) Permeable supports containing confluent 5637 cells are inverted and infected with E. coli on the apical side of the epithelial layers. Alternatively, chemoattractants can be placed in the lower reservoir. The permeable supports are righted into a low-attachment plate, and freshly isolated human PMN are applied to the upper reservoir (representing the basolateral side of the epithelial layers). PMN migrate across the epithelium and are enumerated from the lower reservoir using a hemocytometer.
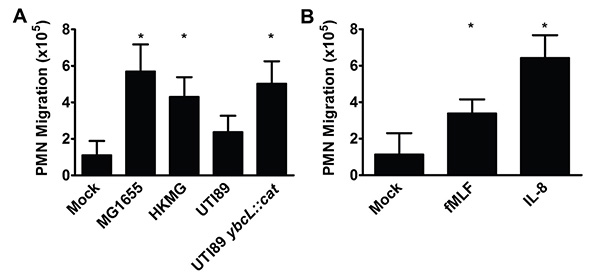
Figure 2. PMN migrate across bladder epithelia in response to various stimuli. (A) Infection with nonpathogenic E. coli strain MG1655, heat-killed MG1655 (HKMG) or UPEC mutant UTI89 ybcL::cat elicits significantly more PMN migration than mock infection or infection with wild-type UPEC strain UTI89, a cystitis isolate (*, p <0.001). (B) The addition of fMLF (100 nM) or IL-8 (100 ng/ml) to the lower reservoir results in significantly more PMN migration than mock treatment (*, p <0.001). Data represent the mean and standard deviation from at least 3 biological replicates. Statistically significant differences were determined using an unpaired Student's t test.
Discussion
Using a cultured bladder epithelial cell line and freshly isolated human PMN, we established an in vitro model of transuroepithelial neutrophil migration. This model has been instrumental in beginning to dissect the complexities of the innate immune response during urinary tract infection (UTI), an extremely common bacterial infection typically caused by UPEC9. During infection of the bladder, or cystitis, recruitment of PMN to the bladder lumen is essential for bacterial clearance10. To establish a foothold in the face of an inflammatory response, UPEC delay the arrival of PMN to the bladder, which prolongs the period during which UPEC can invade the bladder epithelium in the absence of immune pressure6,11 . This phenotype, suppression of PMN migration by UPEC at early time points, is observed in our in vitro model of transuroepithelial PMN migration. Infection with nonpathogenic E. coli MG1655 elicits significantly more PMN migration than infection with uropathogenic E. coli UTI89 (Figure 2A); co-infection with MG1655 and UTI89 yields the uropathogenic phenotype (i.e. low levels of PMN migration)6. Furthermore, we identified a UPEC protein, YbcL, that contributes to the suppressive phenotype, as deletion of ybcL resulted in significantly more PMN in the lower reservoir compared to wild-type UTI89 (Figure 2A)7. High levels of PMN migration were also elicited by heat-killed MG1655, fMLF and IL-8 (Figures 2A and B). In comparison to infection with a live bacterial stimulus, the use of chemoattractants may simplify both the experimental protocol and the interpretation of results. It is likely that other chemoattractants (e.g. bacterial products or chemokines) would also elicit PMN in this model. Thus far, the transuroepithelial neutrophil migration assay has facilitated investigations into suppression of the early inflammatory response by UPEC, revealing phenotypes that have been verified in in vivo models6-8, and will be an invaluable tool in future endeavors.
While this assay has the potential to address a number of questions, there are a few limitations. Given the number of variables inherent to this assay, care must be taken while preparing and conducting experiments to ensure reproducibility between replicates. Enumerating PMN by counting is prone to error, as counting can be time intensive and PMN are short-lived once removed from the body, especially in the presence of bacteria. Reducing the time between PMN sample collection and PMN enumeration will result in more accurate data collection. Researchers have described colorimetric assays that measure myeloperoxidase (MPO) enzyme activity as a surrogate for PMN12,13. Assays utilizing colorimetric substrates such as 3,3',5,5',-tetramethylbenzidine (TMB) or 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) are not sufficiently sensitive to detect the concentrations of PMN present in the lower reservoirs using the transuroepithelial neutrophil migration protocol detailed above (Lau and Hunstad, unpublished data). The parameters of the migration assay could be manipulated to increase the PMN density in the lower reservoir samples. Alternatively, an MPO assay with greater sensitivity, potentially utilizing a fluorescent substrate, could be established. Measurement of MPO activity represents an alternative to PMN counting and may enable more accurate enumeration of PMN in the lower reservoir samples. Additionally, such an assay may also allow enumeration of PMN adherent to the epithelial layers and remaining in the upper reservoir. A validated MPO-based protocol would represent a powerful tool that could expand the amount and type of data that could be collected from the transuroepithelial neutrophil migration assay.
Transepithelial neutrophil migration assays modeling the innate immune response in the gastrointestinal tract and lung are widespread and are responsible for our current understanding of the traversal of epithelial barriers by PMN4,5. In contrast, PMN movement across uroepithelial barriers has received far less attention. Säve and colleagues have reported a model that uses a polarized epithelium composed of differentiated UROtsa cells, an immortalized cell line derived from ureter tissue, grown to confluence on permeable supports14. Agace and colleagues have reported the use of undifferentiated bladder (J82) and kidney (A498) epithelial cells in a similar model15. In the transuroepithelial neutrophil migration model detailed herein, though the 5637 cell layers are not stratified and likely not formally polarized, tight junctions are formed, assessed by impermeability to macromolecular flux and the expression and localization of tight junction proteins (Lau and Hunstad, unpublished data). Of note, the epithelial layer also maintains such impermeability during the infection conditions we have described. The protocols reported by Säve and Agace model the inflammatory response after infection with UPEC isolates for 24 hr. In these models, UPEC elicit robust PMN migration. In contrast, the epithelial layers in our model are exposed to UPEC for a relatively short period of time, 1 hr, in order to examine the initial interactions between host and pathogen. Furthermore, the use of a bladder epithelial cell line and a cystitis-derived UPEC isolate enables potential translation of in vitro findings to the murine cystitis model 16. Lastly, the studies mentioned above utilized large permeable supports that require 6 well tissue culture dishes. The use of smaller permeable supports, as in our model, reduces reagent use and increases the number of inserts that can be manipulated per experiment. Although each of these model systems has advantages and disadvantages, the collective potential of these models to define events required for PMN migration into urinary tract tissues is substantial.
Although less well understood than migration across endothelial barriers, the passage of PMN across gastrointestinal and pulmonary epithelial barriers has received much study4,5. Transepithelial neutrophil migration models that employ permeable supports and cultured epithelial cells have revealed some of the signaling events and adhesion molecules involved in the movement of PMN through these epithelia. Preliminary studies using cultured urinary epithelial cells suggest the involvement of intercellular adhesion molecule-1 (ICAM-1) and the β-integrin CD11b/CD18 (Mac-1) in PMN migration across urinary tissues15. It is unclear which additional signaling pathways and adhesive molecules are involved in these complex processes in the urinary tract. Using the transuroepithelial neutrophil migration model described herein, perhaps augmented with microscopic examination14,15, these basic questions and many others can be interrogated. Additionally, in conjunction with bacterial genetics, this model can be used to further evaluate pathogen-specific phenotypes such as suppression of PMN migration by UPEC. It is unclear at which point in the multi-step process of PMN migration UPEC exerts its suppressive effect. Also, the mechanism by which YbcL influences PMN migration has yet to be elucidated. By first understanding the basic requirements for the passage of PMN across the bladder epithelium, we can then begin to probe how UPEC manipulates these processes to facilitate disease.
In summary, while numerous techniques exist to study the movement of cells, fewer approaches are available to interrogate cell migration across cellular barriers. Modifications to the Boyden chamber have been integral to investigating cell migration across endothelial and epithelial barriers. A tractable in vitro model of the acute inflammatory response in the urinary tract, such as the transuroepithelial neutrophil migration assay detailed herein, is a valuable tool for interrogating these complex processes. Lastly, modifications to this assay could facilitate investigations into other disease states of the urinary tract.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants R01-DK080752 and P50-DK064540. We thank J. Loughman for her efforts in establishing this assay.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Transwell Inserts (i.e. permeable supports) | Corning | 3472 | 6.5 mm, 3 μm pore, polyester membrane insert |
| BD Vacutainer Blood Collection Tubes | Becton, Dickinson and Company (BD) | 366480 | 16 mm x 100 mm x 10 ml green cap tubes containing sodium heparin |
| BD Safety-Lok Blood Collection and Infusion Set | Becton, Dickinson and Company (BD) | 367281 | 21 G x 0.75 in needle x 12 in tubing |
| BD Vacutainer one-use, nonstackable holder | Becton, Dickinson and Company (BD) | 364815 | |
| Dextran | Sigma-Aldrich | D4876 | From Leuconostoc mesenteroides |
| Ficoll-Paque PLUS density centrifugation solution | GE Healthcare | 17-1440-02 | |
| Ultra-Low Attachment Plates | Corning | 3473 | 24-well, clear flat bottom |
| N-Formyl-Met-Leu-Phe (fMLF) | Sigma-Aldrich | F3506 | Reconstituted in DMSO to 10 mM |
| Recombinant Human CXCL8/IL-8 | R&D Systems | 208-IL | Reconstituted in PBS to 100 μg/ml |
Referencias
- Boyden, S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 115, 453-466 (1962).
- Zigmond, S. H. Orientation chamber in chemotaxis. Methods Enzymol. 162, 65-72 (1988).
- Zicha, D., Dunn, G. A., Brown, A. F. A new direct-viewing chemotaxis chamber. J. Cell Sci. 99 (Pt. 4), 769-775 (1991).
- Chin, A. C., Parkos, C. A. Pathobiology of neutrophil transepithelial migration: implications in mediating epithelial injury. Annu. Rev. Pathol. 2, 111-143 (2007).
- Zemans, R. L., Colgan, S. P., Downey, G. P. Transepithelial migration of neutrophils: mechanisms and implications for acute lung injury. Am. J. Respir. Cell Mol. Biol. 40, 519-535 (2009).
- Loughman, J. A., Hunstad, D. A. Attenuation of human neutrophil migration and function by uropathogenic bacteria. Microbes Infect. 13, 555-565 (2011).
- Lau, M. E., Loughman, J. A., Hunstad, D. A. YbcL of uropathogenic Escherichia coli suppresses transepithelial neutrophil migration. Infect. Immun. 80, 4123-4132 (2012).
- Loughman, J. A., Hunstad, D. A. Induction of indoleamine 2,3-dioxygenase by uropathogenic bacteria attenuates innate responses to epithelial infection. J. Infect. Dis. 205, 1830-1839 (2012).
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 7, 653-660 (1038).
- Haraoka, M., et al. Neutrophil recruitment and resistance to urinary tract infection. J. Infect. Dis. 180, 1220-1229 (1999).
- Billips, B. K., et al. Modulation of host innate immune response in the bladder by uropathogenic Escherichia coli. Infect. Immun. 75, 5353-5360 (2007).
- Bozeman, P. M., Learn, D. B., Thomas, E. L. Assay of the human leukocyte enzymes myeloperoxidase and eosinophil peroxidase. J. Immunol. Methods. 126, 125-133 (1990).
- Lee, W. Y., Chin, A. C., Voss, S., Parkos, C. A. In vitro neutrophil transepithelial migration. Methods Mol. Biol. 341, 205-215 (2006).
- Säve, S., Mohlin, C., Vumma, R., Persson, K. Activation of adenosine A2A receptors inhibits neutrophil transuroepithelial migration. Infect. Immun. 79, 3431-3437 (2011).
- Agace, W. W., Patarroyo, M., Svensson, M., Carlemalm, E., Svanborg, C. Escherichia coli induces transuroepithelial neutrophil migration by an intercellular adhesion molecule-1-dependent mechanism. Infect. Immun. 63, 4054-4062 (1995).
- Hung, C. S., Dodson, K. W., Hultgren, S. J. A murine model of urinary tract infection. Nat. Protoc. 4, 1230-1243 (2009).

