Differentiating Human Induced Pluripotent Stem Cells into Neurons on Micro-Electrode Arrays
Abstract
Source: Frega, M., et al., Rapid Neuronal Differentiation of Induced Pluripotent Stem Cells for Measuring Network Activity on Micro-electrode Arrays. J. Vis. Exp. (2017)
The video demonstrates a method for generating excitatory cortical neurons from human-induced pluripotent stem cells (hiPSCs). Upon doxycycline induction, the hiPSCs overexpress neurogenin-2, a neural-specific transcription factor. By using specific media and co-culturing with astrocytes, this process facilitates the hiPSCs' differentiation into neurons with synaptic connections.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. Differentiation of rtTA/Ngn2-positive hiPSCs to Neurons on 6-well MEAs and Glass Coverslips
NOTE: In this protocol, the details are provided for differentiating rtTA/Ngn2-positive hiPSCs on two different substrates, i.e. 6-well MEAs (devices composed of 6 independent wells with 9 recording and 1 reference embedded microelectrodes per well) and glass coverslips in the wells of a 24-well plate. The protocols, however, can easily be adapted for larger substrates (e.g., for the wells of 12- or 6-well plates), by scaling up the mentioned values according to the surface area.
- Prepare the MEAs or glass coverslips (day 0 and day 1)
- The day before the start of the differentiation, sterilize the MEAs according to the manufacturer's recommendation.
- Dilute the adhesion protein Poly-L-Ornithine (PLO) in sterile ultrapure water to a final concentration 50 µg/mL. Coat the active electrode area of 6-well MEAs by placing a 100 µL drop of the diluted PLO in each well. Place the coverslips in the wells of the 24-well plate using sterile tweezers. Add 800 µL of the diluted PLO in each well. Prevent the coverslips from floating by pushing them down with the 1000 µL pipette tip.
- Incubate the 6-well MEAs and 24-well plate O/N in a humidified 37 °C incubator with an atmosphere of 5% CO2. The next day, aspirate the diluted PLO. Wash the glass surfaces of the 6-well MEAs and the coverslips twice with sterile ultrapure water.
- Dilute laminin in cold DMEM/F12 to a final concentration of 20 µg/mL (for the 6-well MEAs) and 10 µg/mL (for the glass coverslips). Immediately coat the active electrode area of the 6-well MEAs by placing a 100 µL drop in each well. Similarly, add 400 µL of the diluted laminin in each well of the 24-well plate to coat the coverslips. Prevent the coverslips from floating by pushing them down with the 1,000 µL pipette tip.
- Incubate the 6-well MEAs and 24 well plate in a humidified 37 °C incubator with an atmosphere of 5% CO2 for at least 2 h.
- Plate the hiPSCs (day 1)
NOTE: The volumes that are mentioned in steps 3.2.1 – 3.2.4 assume that the rtTA/Ngn2-positive hiPSCs are cultured in a 6-well plate and that the cells of one well are harvested. The volumes that are required for plating the cells on the 6-well MEAs and/or the coverslips depends on the number of 6-well MEAs and/or the number of coverslips that are used in the experiment; the numbers specified in steps 3.2.6 – 3.2.8 allow scaling to different experiment sizes.- Warm DMEM/F12, CDS and E8 medium with 1% (v/v) penicillin/streptomycin to R/T. Add doxycycline to a final concentration of 4 µg/mL and ROCK inhibitor to the E8 medium.
- Aspirate the spent medium of the rtTA/Ngn2-positive hiPSCs and add 1 mL CDS to the hiPSCs. Incubate 3 – 5 min in a humidified 37 °C incubator with an atmosphere of 5% CO2. Check under the microscope whether the cells are detaching from one another.
- Add 2 mL DMEM/F12 in the well, gently suspend the cells with a 1,000 µL pipette and transfer the cells to a 15 mL tube. Add 7 mL DMEM/F12 to the cell suspension. Spin the cells at 200 x g for 5 min.
- Aspirate the supernatant and add 2 mL of the prepared E8 medium. Dissociate the hiPSCs by putting the tip of a 1,000 µL pipette against the side of the 15 mL tube and resuspending the cells gently. Check under the microscope whether the cells are dissociated.
- Determine the number of cells (cells/mL) using a hemocytometer chamber.
NOTE: A 6-well plate well at 80 – 90% confluency will typically yield 3.0 – 4.0 x 106 cells in total. - Aspirate the diluted laminin. For the 6-well MEAs, dilute the cells to obtain a cell suspension of 7.5 x 105 cells/mL. Plate the cells by adding a drop of 100 µL of the cell suspension on the active electrode area in each well of the 6-well MEAs. For the coverslips, dilute the cells to obtain a cell suspension of 4.0 x 104 cells/mL. Plate the cells by adding 500 µL of the cell suspension to the wells of the 24-well plate.
NOTE: The final cell density on the MEAs is higher than on the coverslips (Figure 1A and B). We found that this high cell density was required to properly record the network activity. In the protocol, the numbers are provided that were optimal for the assays. - Place the 6-well MEAs and 24-well plate in a humidified 37 °C incubator with an atmosphere of 5% CO2 for 2 h (MEAs) or O/N (24-well plate).
- After 2 h, carefully add 500 µL of the prepared E8 medium to each well of the 6-well MEAs. Place the 6-well MEAs O/N in a humidified 37 °C incubator with an atmosphere of 5% CO2.
- Change the medium (day 2)
- The next day, prepare DMEM/F12 with 1% (v/v) N-2 supplement, 1% (v/v) non-essential amino acids, and 1% (v/v) penicillin/streptomycin. Add human recombinant neurotrophin-3 (NT-3) to a final concentration of 10 ng/mL, human recombinant brain-derived neurotrophic factor (BDNF) to a final concentration of 10 ng/mL, and doxycycline to a final concentration of 4 µg/mL. Warm the medium to 37 °C.
- Add laminin to a final concentration of 0.2 µg/mL to the medium. Filter the resulting medium. Aspirate the spent medium from the wells of the 6-well MEAs and the 24-well plate and replace it with the prepared medium. Incubate the 6-well MEAs and 24-well plate O/N in a humidified 37 °C incubator with an atmosphere of 5% CO2.
- Add rat astrocytes (day 3)
NOTE: The volumes that are mentioned in this protocol assume that the rat astrocytes are cultured in T75 culture flasks. It is critical that the rat astrocytes that are added to the cultures are of good quality. We use two criteria to check if the rat astrocytes are of good quality. First, the rat astrocyte culture should be able to grow confluent within ten days after the isolation from the rat embryonic brains. Second, after splitting the rat astrocyte culture, the rat astrocytes should be able to form a confluent, tessellated monolayer (Figure 1C). If the rat astrocyte culture does not fulfill these two criteria, we advise not to use this culture for differentiation experiments.- Warm 0.05% trypsin-EDTA to RT. Warm the DPBS and DMEM/F12 with 1% (v/v) penicillin/streptomycin to 37 °C.
- Aspirate the spent medium of the rat astrocyte culture. Wash the culture by adding 5 mL DPBS and swish it around gently.
- Aspirate the DPBS and add 5 mL 0.05% trypsin-EDTA. Swish the trypsin-EDTA around gently. Incubate in a humidified 37 °C incubator with an atmosphere of 5% CO2 for 5 – 10 min.
- Check under the microscope whether the cells are detached. Detach the last cells by hitting the flask a few times.
- Add 5 mL of DMEM/F12 to the flask. Triturate the cells gently inside the flask with a 10 mL pipette. Collect the cell suspension in a 15 mL tube. Spin the tube at 200 x g for 8 min.
- Aspirate the supernatant and resuspend the cells in 1 mL of DMEM/F12. Determine the number of cells (cells/mL) using a hemocytometer chamber.
- Add 7.5 x 104 astrocytes per well of the 6-well MEAs. Add 2.0 x 104 astrocytes per well of the 24-well plate. Incubate the MEAs and the 24-well plate O/N in a humidified 37 °C incubator with an atmosphere of 5% CO2.
- Change the medium (day 4)
- Prepare neurobasal medium with 2% (v/v) B-27 supplement, 1% (v/v) L-alanyl-L-glutamine and 1% (v/v) penicillin/streptomycin. Add NT-3 to a final concentration of 10 ng/mL, BDNF to a final concentration of 10 ng/mL, and doxycycline to a final concentration of 4 µg/mL. In addition, add cytosine β-D-arabinofuranoside to a concentration of 2 µM.
NOTE: Cytosine β-D-arabinofuranoside is added to the medium to inhibit astrocyte proliferation and to kill the remaining hiPSCs that are not differentiating into neurons. - Filter the medium and warm to 37 °C. Aspirate the spent medium from the wells of the 6-well MEAs and the 24-well plate and replace it with the prepared medium. Maintain the 6-well MEAs and the 24-well plate in a humidified 37 °C incubator with an atmosphere of 5% CO2.
- Prepare neurobasal medium with 2% (v/v) B-27 supplement, 1% (v/v) L-alanyl-L-glutamine and 1% (v/v) penicillin/streptomycin. Add NT-3 to a final concentration of 10 ng/mL, BDNF to a final concentration of 10 ng/mL, and doxycycline to a final concentration of 4 µg/mL. In addition, add cytosine β-D-arabinofuranoside to a concentration of 2 µM.
- Refresh the medium (day 6 – 28)
NOTE: Starting from day 6, refresh half of the medium every two days. From day 10 onwards, the medium is supplemented with FBS to support the astrocyte viability.- Prepare neurobasal medium with 2% (v/v) B-27 supplement, 1% (v/v) L-alanyl-L-glutamine, and 1% (v/v) penicillin/streptomycin. Add NT-3 to a final concentration of 10 ng/mL, BDNF to a final concentration of 10 ng/mL, and doxycycline to a final concentration of 4 µg/mL. From day 10 onwards, also supplement the medium with 2.5% (v/v) FBS. Filter the resulting medium and warm it to 37 °C.
- Remove half of the spent medium from the wells of the 6-well MEAs and the 24-well plate using a 1000 µL pipette and replace it with the prepared medium. Maintain the 6-well MEAs and the 24-well plate in a humidified 37 °C incubator with an atmosphere of 5% CO2.
Representative Results
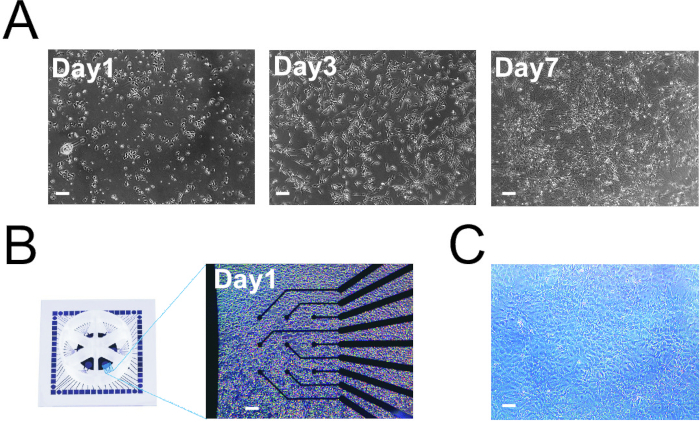
Figure 1: hiPSC Differentiation into Neurons. A. Three time points of hiPSCs differentiation into neurons on coverslips. B. Plating of hiPSCs on MEAs. C. Astrocytes at 100% confluency in T75 flask (note that the cells form a tessellated monolayer). Scale bars: 150 µm.
Divulgaciones
The authors have nothing to disclose.
Materials
| B-27 supplement | Gibco | 0080085SA | |
| Ca2+/Mg2+-free HBSS | Gibco | 14175-095 | |
| 0.05 % Trypsin-EDTA | Gibco | 25300-054 | |
| 2.5 % Trypsin | Gibco | 15090-046 | |
| High-glucose DMEM | Gibco | 11965-092 | |
| FBS | Sigma-Aldrich | F2442-500ML | |
| Penicillin/Streptomycin | Sigma-Aldrich | P4333 | |
| 70 µm cell strainer | BD Falcon | 352350 | |
| DPBS | Gibco | 14190-094 | |
| Basement membrane matrix | Gibco | A1413201 | |
| DMEM/F12 | Gibco | 11320-074 | |
| Cell detachment solution | Sigma-Aldrich | A6964 | |
| E8 medium | Gibco | A1517001 | |
| ROCK inhibitor | Gibco | A2644501 | Alternatively, ROCK inhibitors like thiazovivin can be used. |
| 6-well MEAs | Multi Channel Systems | 60-6wellMEA200/30iR-Ti-tcr | |
| Glass coverslips | VWR | 631-0899 | |
| Poly-L-ornithine | Sigma-Aldrich | P3655-10MG | |
| Laminin | Sigma-Aldrich | L2020-1MG | |
| Doxycyclin | Sigma-Aldrich | D9891-5G | |
| N-2 supplement | Gibco | 17502-048 | |
| Non-essential amino acids | Sigma-Aldrich | M7145 | |
| NT-3, human recombinant | Promokine | C66425 | |
| BDNF, human recombinant | Promokine | C66212 | |
| Trypsin-EDTA | Gibco | 25300-054 | |
| Neurobasal medium | Gibco | 21103-049 | |
| Cytosine β-D-arabinofuranoside | Sigma-Aldrich | C1768-100MG |

