Fluorescent Labeling of COS-7 Expressing SNAP-tag Fusion Proteins for Live Cell Imaging
Summary
SNAP-tag and CLIP-tag protein labeling systems enable the specific, covalent attachment of molecules, including fluorescent dyes, to a protein of interest in live cells. Once cloned and expressed, the tagged protein can be used with a variety of substrates for numerous downstream applications without having to clone again.
Abstract
Protocol
Plating CHO-K1 Cells:
- Add 50-100μl of CHO-K1 cells that have been trypsinized according to standard protocols to 6ml of complete F-12K.
- Mix cell suspension by pipetting up and down several times, then aliquot 300 μl of trypsinized cell suspension to each well of a sterile 8-chamber Lab-Tek II Chambered Coverglass with #1.5 borosilicate coverglass chambers (Nalgene Nunc Int.; Product #155409; Lot: 100808-8-0).
- Incubate samples at 37°C, 5% CO2 overnight.
Transient Transfection of CHO-K1 Cells with pSNAP-ADRB2 and pSNAP-H2B:
- Check the cell culture chambers seeded on the previous day to look for cell density and health.
- Label the appropriate number of microfuge tubes.
- In a laminar flow hood, set up transfection complex mixtures according to the following sample chart, using either 0.3 μg pSNAP-ADBR2 Control Plasmid DNA or 0.3 μg of pSNAP-H2B Control Plasmid DNA, 1 μl of TransPass D2 and 1 μl of TransPass V per reaction.
plasmid number of reactions μl of serum-free medium μl of TransPass D2 μl of TransPass V μl of plasmid DNA μl of TransPass D2/reaction μl of TransPass V/reaction ng of DNA/ reaction DNA conc. (ng/μl) μl of plasmid DNA/ reaction SNAP-ADRb2 5 236.5 5 5 3.5 1 1 300 472.34 0.7 H2B-SNAP 5 237 5 5 3 1 1 300 515.89 0.6 Mock 5 240 5 5 0 1 1 0 0 0 Untransfected 5 250 0 0 0 0 0 0 0 0 - First, mix the serum free F-12K with the appropriate plasmid DNA, then add the TransPass D2 Transfection Reagent, then add the TransPass V Transfection Reagent. Mix the components well by pipetting up and down gently several times after the addition of each reagent.
- Incubate the reaction at room temperature for 30 minutes.
- During the incubation, wash cells once with 600 μl of complete DMEM and add 450 μl of complete DMEM to each sample.
- Add 50 μl of the transfection complex mixture to each sample, slowly and in a drop-wise fashion.
- Incubate the samples overnight at 37°C, 5% CO2.
SNAP-Cell TMR Star Labeling of CHO-K1 Cells Transiently Transfected with pSNAP-ADRB2 and pSNAP-H2B :
- Carefully remove the transfection complex media from each sample by vacuum suction. Wash cells twice with 600 μl of complete F-12K and replace media in each well with 600 μl of complete F-12K.
- Mix dye labeling media by adding 995 μl of complete F-12K and 51 μl 0.6 mM SNAP-Cell TMR-Star substrate. The final concentration of SNAP-Cell TMR-Star should be 3mM. Pipette up and down several times to mix after the addition of substrate.
- Carefully remove growth media from samples and add 200 μl of dye labeling media to each well. Incubate samples at 37°C, 5% CO2 for 30 minutes.
- While samples are incubating, prepare Hoechst 33342 solution for nuclear staining of samples by mixing 1 μl of Hoechst 33342, trihydrochloride, trihydrate (10 mg/ml solution in water, 16.2 mM solution; Invitrogen Molecular Probes) in 10 ml of complete F-12K.
- Remove the media from all samples and add 600 μl of the dilute Hoechst solution to each sample. Incubate samples at 37°C, 5% CO2 for 5 minutes.
- Wash samples three times with 600 μl of complete F-12K.
- Incubate the samples at 37°C, 5% CO2 for an additional 30 minutes to allow unincorporated fluorophore to diffuse out of the cells.
- After 30 minutes, change the media on the SNAP-Cell samples one last time and proceed to imaging.
- Image the cells using a Zeiss Apotome Axiovert 200M fluorescent microscope.
Representative Results
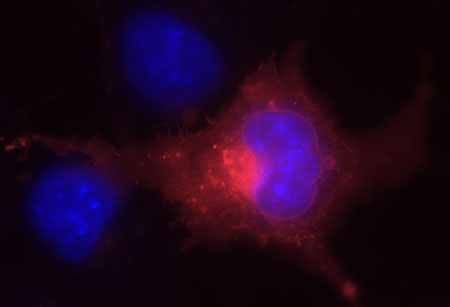
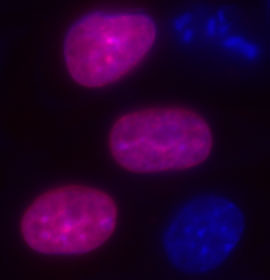
Figure 1. Here are some representative results of fluorescent imaging using standard fluorescent microscopy of live COS-7 cells that are expressing SNAP-tag fusions. This first image shows live COS-7 cells expressing pSNAP-H2B (nuclear histone) labeled with SNAP-Cell TMR-Star. The pSNAP-H2B construct was generated using the pSNAP-tag(m) vector. The cells were labeled with SNAP-Cell TMR-Star for 30 minutes and counterstained with Hoechst 33342 (blue) for nuclei. The pink fluorescence demonstrates the overlay of red fluorescence where histone protein is present in addition to the blue fluorescence indicating the nucleus. The expression of the H2B protein is clearly and easily identified.
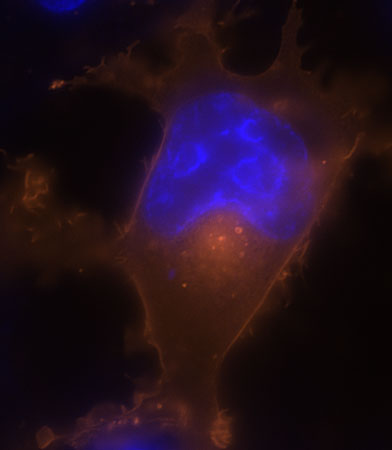
Figures 2 and 3. These next two images show live COS-7 cells transiently transfected with pSNAP-ADRβ2.
Cells were labeled with SNAP-Cell TMR-Star (red) and counterstained with Hoechst 33342 (blue). The pSNAP-ADRβ2 construct was generated using the pSNAP-tag(m) vector. The cells were labeled with SNAP-Cell TMR-Star for 30 minutes and counterstained with Hoechst 33342 (blue) for nuclei. The red fluorescence indicates the presence of cell surface receptor protein ADRB2 while blue fluorescence identifies the nucleus.
Discussion
There are several variables affecting the observed results such as focus, laser intensity, equipment capabilities, lens used, etc. The photostability of the fluorophore determines how quickly the fluorescence must be captured before fading.
This method of protein labeling is versatile and suitable for many applications. SNAP-tag and CLIP-tag technologies for the specific labeling of fusion proteins with synthetic probes enable various aspects of protein function, including a variety of dynamic processes in live cells and in cell lysates. SNAP-, CLIP-, MCP- and ACP-tag technologies provide simplicity and extraordinary versatility to the imaging of proteins in live and fixed cells and to the study of proteins de vitro. The creation of a single gene construct yields a tagged fusion protein capable of forming a covalent linkage to a variety of functional groups, including fluorophores, biotin, or beads. The protein of interest must be cloned and expressed only once and then the fusion can be used with a variety of fluorescent substrates for numerous downstream applications such as simultaneous protein labeling inside live cells, protein localization and translocation, pulse-chase experiments, receptor internalization studies, selective cell surface labeling, protein pull down assays, protein detection in SDS-PAGE, flow cytometry, high-throughput binding assays in microtiter plates, biosensor interaction experiments, and FRET-based binding assays.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
Kai Johnsson
Ivan Correa
Andreas Brecht
Katrin Müentener
Chris Noren
Luo Sun
Ming Xu
Theodore Davis
Salvatore Russello
Aihua Zhang
Inca Ghosh
Elissa Maunus
Nicholas Labarthe
James MacFarland
John Buswell
Brenda Desmond
James Ellard
Don Comb
Materials
| Material Name | Tipo | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| CHO-K1 Cells | ATCC | CCL-61 | ||
| Serum free F-12K | ||||
| Lab-Tek II Chambered Coverglass with #1.5 borosilicate coverglass chambers | Nalgene Nunc Int. | 155409 | Lot: 100808-8-0 | |
| DMEM | ||||
| Hoechst 33342, trihydrochloride, trihydrate (10 mg/ml solution in water, 16.2 mM solution) | Invitrogen Molecular Probes | H3570 | 470519 | |
| Zeiss Apotome Axiovert 200M fluorescent microscope | Zeiss | |||
| Transpass V | New England Biolabs | M2561S | 0030709 | |
| Transpass D2 | New England Biolabs | M2554S | 0060802 | |
| SNAP-Cell TMR Star | New England Biolabs | S9105S | 0010811 | |
| pSNAP-ADBR2 Control Plasmid | New England Biolabs | N9178S | 0010811 | |
| pSNAP-H2B Control Plasmid | New England Biolabs | N9179S | 0010811 |
Referencias
- Johnsson, K. Visualizing biochemical activities in living cells. Nat Chem Biol. 5, 63-65 (2009).
- Johnsson, K. SNAP-tag Technologies: Novel Tools to Study Protein Function. NEB Expressions. 3.3, 1-3 (2008).

