In Situ Nucleosome Assembly for Single-Molecule Correlative Force and Fluorescence Microscopy
Summary
This article presents a detailed experimental procedure for reconstituting nucleosome-containing DNA tethers for single-molecule correlative force and fluorescence microscopy. It further describes several downstream experiments that can be conducted to visualize the binding behavior of chromatin-interacting proteins and analyze changes in the physical properties of nucleosomes.
Abstract
Nucleosomes constitute the primary unit of eukaryotic chromatin and have been the focus of numerous informative single-molecule investigations regarding their biophysical properties and interactions with chromatin-binding proteins. Nucleosome reconstitution on DNA for these studies typically involves a salt dialysis procedure that provides precise control over the placement and number of nucleosomes formed along a DNA tether. However, this protocol is time-consuming and requires a substantial amount of DNA and histone octamers as inputs. To offer an alternative strategy, an in situ nucleosome reconstitution method for single-molecule force and fluorescence microscopy that utilizes the histone chaperone Nap1 is described. This method enables users to assemble nucleosomes on any DNA template without the need for strong nucleosome positioning sequences, adjust nucleosome density on demand, and use fewer reagents. In situ nucleosome formation occurs within seconds, offering a simpler experimental workflow and a convenient transition into single-molecule measurements. Examples of two downstream assays for probing nucleosome mechanics and visualizing the behavior of individual proteins on chromatin are further described.
Introduction
The primary packaging unit of eukaryotic chromatin is the nucleosome, in which ~147 base pairs (bps) of DNA are wrapped around an octamer of core histones proteins1,2. In addition to genome packaging, the nucleosome architecture serves as another rich layer of biophysical regulation that can be harnessed by chromatin-binding proteins when performing their various functions3,4. Experimentally accessing and measuring the physical characteristics of nucleosomes has been technically challenging since these units perform at minuscule scales (e.g., nanometer lengths, piconewton forces), and thus, their probing requires sufficient sensitivity and precision to meaningfully inform function. Moreover, chromatin-binding proteins often engage with their substrates transiently, and most ensemble approaches lack appropriate temporal resolution to inform the kinetics of these interactions5. Fortunately, the advent of single-molecule techniques has made it possible to visualize and manipulate individual proteins and their interactions in real-time, revealing mechanistic information about these molecular events occurring at the nanoscale6. In particular, single-molecule correlative force and fluorescence microscopy (smCFFM) harnesses both high-resolution fluorescence detection and force manipulation tools to simultaneously resolve the mechanics, composition, and coordination of chromatin and chromatin-protein complexes7,8.
In smCFFM that specifically employs "optical tweezers" as the force manipulation method, a tightly focused laser is used to trap a dielectric object, typically a micron-sized plastic bead, in three dimensions9. A biopolymer such as a piece of DNA can be conjugated to the bead (via, e.g., streptavidin-biotin, digoxigenin-anti-digoxigenin antibody linkage), and the user can both apply a calibrated force and measure the force/displacement generated by the system10. The added fluorescence module enables simultaneous multicolor fluorescence imaging to track protein composition and behavior.
The mainstream method for reconstituting nucleosome templates for smCFFM involves assembling nucleosomes on custom DNA templates by salt dialysis11,12,13. In this procedure, purified histone octamers are incubated with DNA templates in a high-salt buffer (~1 M sodium chloride), and as the salt is slowly dialyzed away, nucleosomes spontaneously assemble on the DNA in a sequence-dependent positional manner14,15. As such, it is customary that strong nucleosome positioning sequences (e.g., Widom 60116, X. laevis 5S rDNA17) are incorporated within the DNA templates to generate custom nucleosome arrays. Although this well-established method offers precise control over the placement and number of nucleosomes across DNA, it suffers from several disadvantages: (1) incorporation of strong nucleosome positioning DNA sequences may generate artificially stable nucleosomes that bias the activity of chromatin-binding proteins, (2) the process of salt dialysis for nucleosome formation requires high amounts of DNA and octamers, and (3) the procedure takes one to several days of work as well as a considerable effort to titrate the correct DNA to octamer ratio for optimal nucleosome array density.
In this manuscript, we describe an alternative method to form nucleosomes along DNA in situ within a single-molecule correlative force and fluorescence microscope using recombinant histone chaperone Nap1, which resembles the physiological pathway of nucleosome formation in vivo18,19,20,21,22. This method allows users to assemble nucleosomes on non-specific DNA sequences, easily adjust nucleosome density, and utilize significantly fewer amounts of reagents, all within minutes. Additionally, the formation of nucleosomal DNA tethers in situ enables simpler experimental workflow and the convenience of built-in single-molecule visualization and manipulation. Thus, when uniform and specific nucleosome positioning is not a necessity, this protocol offers a useful option for the investigation of nucleosome mechanics or protein behavior on chromatin, for which we also describe example assays.
Protocol
The details of the reagents and the equipment used in the study are listed in the Table of Materials.
1. Preparation of biotinylated DNA
- Prepare a 120 µL volume reaction containing 20 µg of λ DNA, 33 µM of biotin-dCTP, 33 µM of biotin-dUTP, 33 µM of biotin-dATP, 33 µM of dGTP, 10 units of Klenow enzyme, and 1x NEB Buffer 2.
NOTE: This procedure specifically utilizes linear double-stranded (ds) methylation-free bacteriophage λ genomic DNA (48,502 bps), which contains 12-nucleotide (nt) 5' single-stranded (ss) DNA overhang23. However, the protocol can be adapted to make use of any length or sequence of linear dsDNA provided it contains at least several nts of ss 5' overhangs, the length and nt sequence dictating the number of biotinylated nts installed on each end for downstream tethering between optical traps. Additionally, the reaction can be scaled down. For example, 5 µg of DNA can be used in a 30 µL volume reaction. Additionally, the DNA substrates generated in this way are not torsionally constrained. For protocols to create torsionally constrained DNA, please refer to the previously published reports24,25. - Incubate the reaction at room temperature for 15 min.
- Add 10 mM of EDTA and incubate at 75 °C for 20 min.
- Perform ethanol precipitation to obtain the biotinylated DNA product.
- Add 0.1x volume of 3 M sodium acetate and 3x volume of ice cold 100% ethanol. Keep at -20 °C for at least 1 h up to overnight. Centrifuge at 13,500 x g for 30 min at 4 °C.
- Carefully remove the supernatant without disturbing the pellet and add 1 mL of 75% ethanol. Centrifuge at 13,500 x g for 1 min at 4 °C.
- Repeat the supernatant removal, ethanol addition, and centrifugation steps.
- Carefully remove all the supernatant and let the pellet air dry for 10 min. Dissolve the pellet in 100-200 µL of TE buffer or ddH2O (can incubate at 37 °C to facilitate dissolving). Measure the concentration using a Nanodrop spectrophotometer.
2. Preparation of channels in a flow cell
NOTE: This procedure is based on a commercially available microfluidic chip (see Table of Materials) but can be adapted to any smCFFM instrument with a multi-channel flow cell.
- Passivate channels (ch.) 4 and 5 (or any ch. that will contain proteins) (see schematic of channels in Figure 1A). Add 500 µL of 0.1% BSA to each channel and flow at 0.8 bar (~0.45 µL/s for tubing with a 0.010 inch inner diameter) for 8 min. Remove solution.
- Add 500 µL of 0.05% Pluronic-F127 to each channel and flow at 0.8 bar for 8 min. Remove the solution and rinse out the syringes with 1x PBS. Add 2 mL of 1x PBS to each channel and flow at 0.8 barfor 27 min.
- Vortex the stock solution of streptavidin-coated beads (this procedure uses 3.13 µm size beads) and add 2.15 µL of the bead solution to 1 mL of 1x PBS. Add to ch. 1.
NOTE: The size and concentration of the streptavidin-coated beads can be adjusted according to user preference. - Add 30-80 ng of biotinylated λ DNA to 1 mL of 1x PBS. Add to ch. 2.
NOTE: The concentration of biotinylated DNA can be adjusted to user preference. Greater amounts of DNA will increase the frequency of forming DNA tethers but also raise the frequency of tethering multiple molecules of DNA at once, which can add to the difficulty of the experiment.
3. Nap1-mediated in situ nucleosome formation
- Add 500 µL of protein-free image buffer to ch. 3.
NOTE: The buffer composition in ch. 3 is flexible and should be appropriate for both nucleosomes and the specific protein(s) of interest. Optionally, users may choose to add oxygen scavenging systems for reduced photobleaching of fluorophores and/or competitor DNA to remove histone octamers that are not properly wrapped into nucleosomes. - Add 2 nM S. cerevisiae Nap1 and 2-5 nM LD655-labeled H4 histone octamer (HO) to 500 µL of 1x HR buffer18. Add to ch. 4.
NOTE: Histone octamers and Nap1 can be commercially purchased or prepared in-house (see Table of Materials). The number of nucleosomes to be formed along each tethered DNA depends on the octamer concentration as well as the time spent in this channel. The user's desired and approximate nucleosome density along each DNA molecule can be modulated by adjusting either or both of these parameters. Additionally, the user may choose to label histone octamers with their preferred fluorophore. - Optional: Add chromatin-binding protein or other protein of interest to ch. 5 in an appropriate buffer.
- Flow all channels at 1.0 bar (~0.55 µL/s in the current experimental setup) for 30 s to flush the flow cell channels with samples.
- Set the flow to 0.2 bar (~0.16 µL/s in the current experimental setup). Move the optical traps (OTs) to ch. 1 and catch a suitable pair of streptavidin-coated beads.
- Move the OTs to ch. 3, conduct a force calibration for the bead pair, turn on the red confocal laser (or another laser line of choice), and calibrate the imaging area.
- Set the flow to 0.2 bar and move OTs to ch. 2. Catch biotinylated DNA.
- Move the OTs to ch. 3 and stop the flow. Increase the distance between the OTs to stretch the DNA and monitor the associated force-distance (FD) curve to confirm the presence of a single DNA tether (as opposed to multiple tethers). The curve should follow the worm-like-chain model for dsDNA at the given contour length26.
- Adjust the distance between the OTs so that the DNA tension reads approximately 1 pN.
NOTE: 1 pN of tension is low enough to allow nucleosomes to remain wrapped by DNA27 but is high enough to place the entire DNA molecule within the line scanning imaging axis. - Turn on line scanning to start collecting a kymograph with the red laser on.
NOTE: Kymographs or 2D scans of tethered DNA can be obtained, as per user preference. - Move the OTs to ch. 4 with the flow off. Facilitated by Nap1, HOs are loaded onto DNA and wrapped to form nucleosomes in real-time. Nucleosome loading is visualized as fluorescent trajectories appearing on the DNA within kymographs. Concurrently, nucleosome formation on the DNA can be monitored by the force increase as more HOs are loaded when the positions of OTs are held constant.
NOTE: Fluorescence excitation laser can be turned off during the nucleosome formation steps to avoid photobleaching of fluorophores. Additionally, non-specific binding of histone octamers to DNA wherein the octamers are not wrapped into nucleosomes could occur using this protocol. These binding events will not generate a "rip" when the DNA is stretched, as described in the Unwrapping Nucleosomes (step 4) protocol below, and can sometimes be washed off by a gentle buffer flow, particularly if the buffer contains free competitor DNA. - When the approximate desired number of nucleosomes is formed, start collecting data.
4. Unwrapping nucleosomes
- Complete step 2 and step 3.
- After forming a DNA tether containing nucleosomes, move the OTs to ch. 3.
- Prepare for nucleosome unwrapping experiments by reducing the distance between OTs until the force is approximately zero. Zero the force.
- Begin moving the OT 1 away from the OT 2 at a constant speed of 0.1 µm/s. On a kymograph, the bead in OT 1 will visually move farther from the bead in OT 2. On the FD curve, the force will rise as the distance increases. As nucleosomes unwrap, there will appear "rips" or transitions in the force (~8-37 pN) that signify the unraveling of the inner wrap of individual nucleosomes27,28,29.
NOTE: The force at which unwrapping occurs depends on the pulling rate, which is expected for non-equilibrium processes. Sometimes, multiple nucleosomes unwrap at once, producing a larger distance change in the transition on the FD curve. Additionally, the unraveling of the outer wrap of the nucleosome is not readily visible in this experimental setting (see the Discussion section).
5. Visualizing protein binding to chromatin
- Complete step 2 and step 3.
- Add 10 nM of Cy3-labeled linker histone H1.4 to 500 µL of image buffer in ch. 5.
NOTE: Any protein of interest can be added to ch. 5 at a concentration of choice (typically below 100 nM for single-molecule visualization). - After forming a DNA tether containing nucleosomes, turn on the green laser in addition to the red laser and move OTs to ch. 5 for real-time imaging of H1 binding to chromatin.
- Continue collecting kymographs or scans until sufficient statistics have been achieved (at least ~10-20 kymographs depending on the frequency of events and experimental difficulty).
Representative Results
Using the setup described in step 3 (Figure 1A), nucleosome formation along the DNA tether was visualized as the appearance of red fluorescent foci on a 2D scan (Figures 1B, left) or trajectories over time on a kymograph (Figure 1B, right). Properly wrapped nucleosomes yielded fluorescence trajectories that were stationary over time within the diffraction limit of confocal detection (~300 nm). Notably, multiple nucleosomes formed near each other under the diffraction limit cannot be spatially resolved and appear as brighter foci or trajectories. Nevertheless, the photobleaching steps of individual nucleosomal trajectories were analyzed and frequently showed 1-step (Figure 1C) or 2-step (Figure 1D) photobleaching events, which indicates that foci mostly contained mononucleosomes given the approximately 70% labeling rate of the H4 protein in this sample. Moreover, based on the average photon count for a single LD655 fluorophore in this experimental condition, the number of labeled H4 copies in each nucleosome trajectory was estimated and showed that the nucleosome loci predominately (>75%) contained 1 or 2 labeled H2A copies (Figure 1E), again consistent with the notion that they were primarily loaded as spatially separated mononucleosomes. The other brighter foci contained more than 3 copies of labeled H4, likely representing multiple nucleosomes within the diffraction limit.
Nucleosome formation was also monitored by an increase in the tension along the tether when the positions of the OTs remained constant as more DNA was wrapped around histone octamers (Figure 1B, bottom). In the absence of Nap1, histone octamers can still bind DNA but remain mostly unwrapped, as the associated tension when the OT positions were held remained constant (Figure 1F). It is possible that non-specific binding of histone octamers to DNA occurs even in the presence of Nap1. In another experiment, the same protocol to form nucleosomes containing Cy3-labeled H2A was performed, which yielded similar results as those performed with LD655-labeled H4 (Figure 1G), indicating the assembly of intact octamers into nucleosomes.
Using the setup described in step 4 (Figure 2A), nucleosome unwrapping was visualized over time on a kymograph as the tether distance increased (Figure 2B). It was previously reported that histone octamers often remain bound to the DNA after being unwrapped30,31, and therefore, fluorescent trajectories often persist even at high tensions. Nucleosome unwrapping was also monitored on the associated FD curve (Figure 2C). Quantifiable features of the FD curve include the force at which nucleosome(s) unwrap (see "Transition F" in Figure 2C inset) as well as the length of DNA that was released from the histone octamer (see "ΔL" in Figure 2C inset), which exists in multiples of ~24 nm, consistent with independent unwrapping of the inner turns of DNA of individual nucleosomes27,30. These parameters can be used to estimate the number of nucleosomes formed on each tether or for additional downstream analysis of nucleosome mechanics. As such, the number of inner-turn ruptures obtained from mechanical pulling experiments was compared to the estimated number of labeled H4 copies at each nucleosome trajectory based on the monomer intensity, which showed that the numbers are largely consistent (examples shown in Figures 2D–G). However, it was noted that they do not perfectly match due to the ~70% labeled efficiency of the H4 protein in the sample and any residual Nap1-independent histone binding to DNA, as shown in Figure 1F. Finally, as expected, stretching Nap1-bound bare DNA tethers (without histones) do not generate force-induced transitions (Figure 2H), indicating that Nap1 alone does not significantly distort DNA.
Employing the setup described in step 5 (Figure 3A), the binding of linker histone H1 to chromatin was visualized as the appearance of green fluorescent foci along the chromatin tether (Figure 3B). Notably, H1-bound nucleosomes generated dual-color trajectories (see white arrows in Figure 3B) that appeared stationary over time. In addition, diffusing H1 trajectories across nucleosome-free DNA regions were observed (see green stars in Figure 3B).
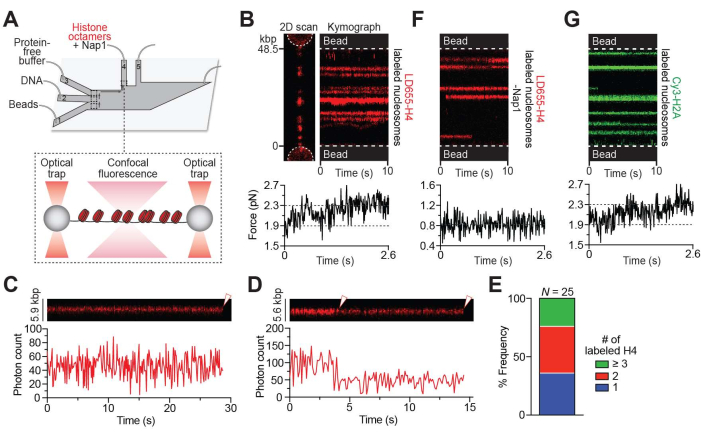
Figure 1: Nap1-mediated in situ nucleosome formation. (A) Schematic of the experimental setup described in the Nap1-mediated in situ nucleosome formation protocol (step 3). A pair of beads were optically trapped in ch. 1, and a single λ DNA molecule was tethered between the beads through biotin-streptavidin linkage in ch. 2. The tether was then moved to ch. 4, containing LD655-labeled histone octamers (red) and Nap1 for real-time formation of nucleosomes across the DNA. (B) Representative 2D scan (left) and kymograph (right) of an LD655-H4-labeled nucleosome-containing DNA tether. The associated force reading (bottom) shows an increase in force immediately as the tether is moved to ch. 4 when the OT positions are held constant, indicating nucleosome wrapping. Brighter trajectories signify multiple adjacent nucleosomes located within the diffraction limit of ~300 nm. (C) Example kymograph showing a 1-step photobleaching event (white arrow) of a nucleosome containing LD655-H4. (D) Example kymograph showing a 2-step photobleaching event (white arrows) of a nucleosome containing LD655-H4. (E) Relative frequency of the number of LD655-labeled H4 per trajectory estimated from their fluorescence intensity. (F) Representative kymograph of a tether bound with histone octamers in the absence of Nap1. The associated force reading (bottom) remains unchanged even as histone octamers bind the DNA. (G) Representative kymograph of a Cy3-H2A-labeled nucleosome-containing DNA tether. The associated force reading (bottom) shows an increase in force immediately as the tether is moved to ch. 4 (containing octamers and Nap1), as nucleosomes are formed when the OT positions are held constant. Please click here to view a larger version of this figure.
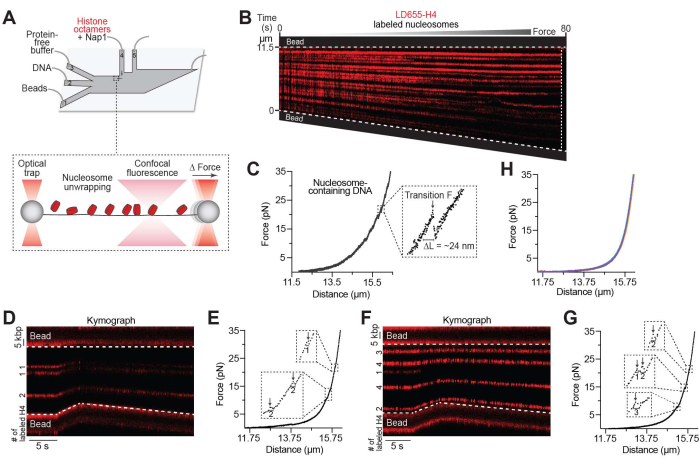
Figure 2: Nucleosome unwrapping assay. (A) Schematic of the experimental setup described in the unwrapping nucleosomes protocol (step 4). The DNA tether containing nucleosomes was moved to ch. 3 containing a protein-free image buffer where the inter-bead distance was increased and the nucleosomes were unwrapped. (B) Representative kymograph of a nucleosome-containing DNA tether pulled to high forces by gradually increasing the inter-bead distance. The decrease in fluorescence intensity toward the bottom bead was likely due to the DNA tilting slightly out of line scanning focus. (C) The associated FD curve showing force-induced transitions of inner turn unwrapping occurring between 8-37 pN. The inset shows a zoom-in view of an example transition for which the distance unwrapped (ΔL = ~24 nm for the inner turn of DNA of individual nucleosomes) and the transition force (Transition F) can be recorded and further analyzed. For this tether, 26 nucleosomes were unwrapped, and the tether ruptured at ~65 pN, as denoted by the white dotted line in the kymograph. (D,E) Example LD655-H4 nucleosome-containing tether pulled to high forces where the number of H4 per trajectory is estimated via fluorescence intensity (indicated in the kymograph, D) and the number of wrapped nucleosomes is estimated via the size of force-induced distance change (indicated in the associated force-distance curve, E). (F,G) Another example as in (D,E). (H) DNA tethers (n = 5) pulled to high forces after being incubated in a channel containing Nap1 only. Please click here to view a larger version of this figure.
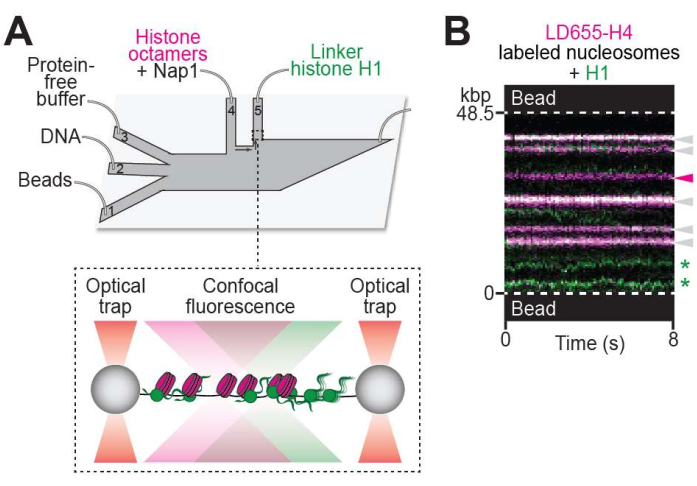
Figure 3: Visualizing protein-chromatin interaction. (A) Schematic of the experimental setup described in the visualizing protein binding to chromatin protocol (step 5). The DNA tether containing nucleosomes was moved to ch. 5, containing 10 nM of Cy3-labeled linker histone H1 (green). (B) Representative two-color kymograph of H1 binding to a nucleosome-containing DNA tether. H1-nucleosome colocalization events are denoted with white arrows. Unbound nucleosome trajectory is denoted with the magenta arrow. H1 molecules diffusing on bare DNA are denoted with green stars. Please click here to view a larger version of this figure.
Discussion
The described protocol provides several advantages for nucleosome reconstitution including minimizing reagents and time as well as enabling chaperone-dependent nucleosome formation along any (potentially native) DNA sequence. Moreover, the in situ method allows simpler experimental workflow and a convenient transition into single-molecule assays of nucleosome mechanics and protein-chromatin interaction. On the other hand, limitations of this approach include the inability to direct nucleosome positioning along the DNA (even with the incorporation of strong nucleosome positioning sequences) and precisely control the number of nucleosomes formed per DNA tether (although this can be empirically tuned by standardizing the experimental workflow). In effect, those who choose this method trade in precise control of nucleosome positioning and density for greater workflow ease and low reagent requirement, the choice of which depends on each specific experiment. It is also noted that the outer turn of DNA is not resolved in the current protocol due to the long length of the tether (~48.5 kbps) and buffer conditions32. To obtain information about the outer turn of DNA as well as inter-nucleosome stacking, the user may choose to shorten the DNA length and load a higher density of nucleosomes onto the DNA and/or use the salt dialysis method to place nucleosomes on strong positioning sequences that are separated by regular linker lengths. Finally, non-specific binding of histone octamers to DNA is likely to occur; therefore, it is important to consider both fluorescence and force signals when interpreting the results.
One critical aspect of the current protocol is ensuring the user has achieved a high fluorescent labeling rate for histone octamers (if visualization is required) so that most, if not all, nucleosomes can be visually detected. Another critical aspect is confirming the user has chosen an appropriate buffer for the imaging channel that is compatible with nucleosomes and any other proteins of interest. User-specific modifications to the protocol may include fluorophore conjugation to a different histone (see reports33,34 for examples using this method to form nucleosomes with labeled H3 or H2B), choosing an alternative fluorophore, incorporating DNA tethers of different sequences and lengths, adding an oxygen-scavenging system to extend the lifetime of fluorophores, choosing a different bead size, and utilizing an alternative DNA-bead conjugation system (e.g., digoxigenin-anti-digoxigenin antibody linkage).
In summary, the described method provides smCFFM with a convenient and versatile approach to generate nucleosome-containing tethers that can be easily employed to study a wide array of chromatin-focused questions in biology. Applicable research areas include chromatin mechanics as regulated by histone variants and post-translational modifications, biophysical engagement and kinetics of chromatin-binding proteins, and nucleosome-directed higher-order assembly processes such as biomolecular condensation35,36,37,38.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
G. N. L. C. acknowledges support from the National Institute of Mental Health of the National Institutes of Health (NIH) under award number F31MH132306. S. L. is supported by the Robertson Foundation, the International Rett Syndrome Foundation, and the NIH (award number R01GM149862).
Materials
| 1x HR buffer | N/A | N/A | 30 mM tris acetate pH 7.5, 20 mM magnesium acetate, 50 mM potassium chloride, 0.1 mg/mL BSA |
| 1x PBS (Phosphate-buffered saline) | N/A | N/A | 137 mM sodium chloride, 2.7 mM potassium chloride, 10 mM sodium phosphate dibasic, 1.8 mM potassium phophate monobasic |
| Acetic acid, glacial | Millipore Sigma | AX0074-6 | Use to make tris acetate |
| Biotin-11-dUTP | Jena Bioscience | NU-803-BIOX-S | |
| Biotin-14-dATP | Jena Bioscience | NU-835-BIO14-S | |
| Biotin-14-dCTP | Jena Bioscience | NU-956-BIO14-S | |
| Bovine Serum Albumin | Millipore Sigma | A9418-50G | Dissolve in H2O and run through 0.22 um filter |
| Cy3 Maleimide Mono-Reactive Dye | Cytiva | PA23031 | Maleimide functionalized Cy3 fluorophore |
| dGTP | New England Biolabs | N0442S | |
| Eppendorf Centrifuge 5425 R | Fisher Scientific | 05-414-051 | Benchtop centrifuge with cooling |
| Ethyl alcohol, Pure | Millipore Sigma | 459844 | |
| Ethylenediaminetetraacetic acid (EDTA) | Millipore Sigma | E9884 | Dissolve in H2O to 0.5 M |
| Human histone octamer (H4, L50C; H2A, K119C) | N/A | N/A | Recombinant histone proteins and those harboring labeling mutations were purified in-house as described previously (see refs. 33, 34, 35, 36). Briefly, recombinant histones were expressed in BL21 (De3) pLySS cells (Promega). Inclusion bodies were isolated after sonication, and histones were extracted under denaturing conditions. Histones were dialyzed into buffer A (7 M urea, 10 mM tris hydrochloride pH 8.0, 100 mM sodium chloride, 1 mM EDTA, and 5 mM 2-mercaptoethanol), and the solution was added to a gravity column loaded with Q Sepharose Fast Flow (Cytiva). The flow through was then added to a gravity column loaded SP Sepharose Fast Flow (Cytiva), and the histones were eluted from the column by adding buffer A supplemented with 600 mM sodium chloride. Histones harboring labeling mutations (H4, L50C; H2A, K119C) were purified and then conjugated to the desired fluorophore via maleimide-functionalized dyes (Cytiva, Lumidyne) using a 20:1 dye-to-protein molar ratio (see refs. 33, 34). Histone octamers were assembled by adding an equal molar ratio of each wild-type or fluorophore-labeled histone under denaturing conditions, dialyzed into a high-salt buffer containing 2 M sodium chloride, and then purified by size exclusion chromatography as described previously (see refs. 36, 37). Alternatively, individual histone proteins and ready-made histone octamers can be purchased commercially (e.g., Epicypher). |
| Image buffer | N/A | N/A | 20 mM tris hydrochloride pH 8.0, 100 mM sodium chloride |
| Klenow Fragment (3' to 5' exo-) | New England Biolabs | M0212S | |
| Lambda DNA (dam-, dcm-) | Thermo Fisher Scientific | SD0021 | Methylation-free λ DNA |
| LD655-MAL | Lumidyne Technologies | 9 | Maleimide functionalized LD655 fluorophore |
| Linker histone H1.4 (A4C) | N/A | N/A | Purified recombinant protein made in-house (see ref. 38) |
| LUMICKS C-Trap Dymo | LUMICKS | N/A | Dual-trap configuration; standard materials for instrument provided by manufacturer |
| Magnesium acetate solution | Millipore Sigma | 63052-100ML | Use to make HR buffer |
| NEBuffer 2 | New England Biolabs | B7002S | Included with Klenow Fragment kit |
| Pluronic F-127 | Millipore Sigma | P2443-250G | Dissolve in H2O and run through 0.22 um filter |
| Potassium chloride | Millipore Sigma | P3911 | Use to make PBS and HR buffer |
| Potassium phosphate monobasic | Millipore Sigma | P0662 | Use to make PBS |
| S. cerevisiae Nap1 | N/A | N/A | Nap1 was purified in-house as previously described (see refs. 33, 34). Alternatively, Nap1 can be purchased commercially (e.g., Active Motif). |
| Sodium acetate | Millipore Sigma | 241245 | Dissolve in H2O to 3 M |
| Sodium chloride | Millipore Sigma | S9888 | Use to make PBS and image buffer |
| Sodium phosphate dibasic | Millipore Sigma | S9763 | Use to make PBS |
| SPHERO Biotin Coated Particles (3.0-3.4 µm) | Spherotech | TP-30-5 | |
| Thermo Scientific NanoDrop 2000/2000c Spectrophotometer | Fisher Scientific | ND2000 | NanoDrop Spectrophotometer |
| Tris Base | Fisher Scientific | BP152-500 | Dissolve in H2O and adjust to appropriate pH; use to make image buffer and tris acetate |
Referenzen
- Kornberg, R. D. Chromatin structure: A repeating unit of histones and DNA. Science. 184, 868-871 (1974).
- Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., Crystal Richmond, T. J. structure of the nucleosome core particle at 2.8 A resolution. Nature. 389, 251-260 (1997).
- Zhou, K., Gaullier, G., Luger, K. Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol. 26, 3-13 (2019).
- McGinty, R. K., Tan, S. Principles of nucleosome recognition by chromatin factors and enzymes. Curr Opin Struct Biol. 71, 16-26 (2021).
- Fierz, B., Poirier, M. G. Biophysics of chromatin dynamics. Annu Rev Biophys. 48, 321-345 (2019).
- Selvin, P. R., Ha, T. . Single-molecule techniques: A laboratory manual. , (2008).
- Chua, G. N. L., Liu, S. When force met fluorescence: Single-molecule manipulation and visualization of protein-DNA interactions. Annu Rev Biophys. , (2024).
- Hashemi Shabestari, M., Meijering, A. E. C., Roos, W. H., Wuite, G. J. L., Peterman, E. J. G. Recent Advances in biological single-molecule applications of optical tweezers and fluorescence microscopy. Methods Enzymol. 582, 85-119 (2017).
- Ashkin, A., Dziedzic, J. M., Bjorkholm, J. E., Chu, S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 11, 288 (1986).
- Neuman, K. C., Block, S. M. Optical trapping. Rev Sci Instrum. 75, 2787-2809 (2004).
- Hansen, J. C., van Holde, K. E., Lohr, D. The mechanism of nucleosome assembly onto oligomers of the sea urchin 5 S DNA positioning sequence. J Biol Chem. 266, 4276-4282 (1991).
- Peterson, C. L. Salt gradient dialysis reconstitution of nucleosomes. CSH Protoc. , (2008).
- Leicher, R., Liu, S. Probing the interaction between chromatin and chromatin-associated complexes with optical tweezers. Methods Mol Biol. 2478, 313-327 (2022).
- Segal, E., et al. A genomic code for nucleosome positioning. Nature. 442, 772-778 (2006).
- Ioshikhes, I. P., Albert, I., Zanton, S. J., Pugh, B. F. Nucleosome positions predicted through comparative genomics. Nat Genet. 38, 1210-1215 (2006).
- Lowary, P. T., Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 276, 19-42 (1998).
- Chipev, C. C., Wolffe, A. P. Chromosomal organization of Xenopus laevis oocyte and somatic 5S rRNA genes in vivo. Mol Cell Biol. 12, 45-55 (1992).
- Crickard, J. B., Moevus, C. J., Kwon, Y., Sung, P., Greene, E. C. Rad54 drives ATP hydrolysis-dependent DNA sequence alignment during homologous recombination. Cell. 181, 1380-1394.e18 (2020).
- Vlijm, R., et al. Comparing the assembly and handedness dynamics of (H3.3-H4)2 tetrasomes to canonical tetrasomes. PLoS One. 10, e0141267 (2015).
- Mazurkiewicz, J., Kepert, J. F., Rippe, K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J Biol Chem. 281, 16462-16472 (2006).
- Nakagawa, T., Bulger, M., Muramatsu, M., Ito, T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem. 276, 27384-27391 (2001).
- Andrews, A. J., Chen, X., Zevin, A., Stargell, L. A., Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 37, 834-842 (2010).
- Wu, R., Taylor, E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 57, 491-511 (1971).
- King, G. A., Peterman, E. J., Wuite, G. J. Unravelling the structural plasticity of stretched DNA under torsional constraint. Nat Commun. 7, 11810 (2016).
- Paik, D. H., Roskens, V. A., Perkins, T. T. Torsionally constrained DNA for single-molecule assays: An efficient, ligation-free method. Nucleic Acids Res. 41, e179 (2013).
- Bustamante, C. J., Chemla, Y. R., Liu, S., Wang, M. D. Optical tweezers in single-molecule biophysics. Nat Rev Methods Primers. 1, 25 (2021).
- Diaz-Celis, C., et al. Assignment of structural transitions during mechanical unwrapping of nucleosomes and their disassembly products. Proc Natl Acad Sci USA. 119, e2206513119 (2022).
- Leicher, R., et al. Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin. Proc Natl Acad Sci USA. 117, 30465-30475 (2020).
- Chua, G. N. L., et al. Differential dynamics specify MeCP2 function at methylated DNA and nucleosomes. bioRxiv. , (2023).
- Brower-Toland, B. D., et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA. 99, 1960-1965 (2002).
- McCauley, M. J., et al. Human FACT subunits coordinate to catalyze both disassembly and reassembly of nucleosomes. Cell Rep. 41, 111858 (2022).
- Mihardja, S., Spakowitz, A. J., Zhang, Y., Bustamante, C. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci USA. 103, 15871-15876 (2006).
- Li, S., et al. Nucleosome-directed replication origin licensing independent of a consensus DNA sequence. Nat Commun. 13, 4947 (2022).
- Li, S., et al. Origin recognition complex harbors an intrinsic nucleosome remodeling activity. Proc Natl Acad Sci USA. 119, e2211568119 (2022).
- Harada, B. T., et al. Stepwise nucleosome translocation by RSC remodeling complexes. Elife. 5, e10051 (2016).
- Luger, K., Rechsteiner, T. J., Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 119, 1-16 (1999).
- Rogge, R. A., et al. Assembly of nucleosomal arrays from recombinant core histones and nucleosome positioning DNA. J Vis Exp. (79), e50354 (2013).
- Leicher, R., et al. Single-stranded nucleic acid binding and coacervation by linker histone H1. Nat Struct Mol Biol. 29, 463-471 (2022).
Tags

.
