Standardized Identification of Compound Structure in Tibetan Medicine Using Ion Trap Mass Spectrometry and Multiple-Stage Fragmentation Analysis
Summary
Here, we describe a general protocol and design that could be applied to identify trace amounts and minor constituents in the complex natural product formulations (matrixes) in Tibetan medicine.
Abstract
Tibetan medicines are complex and contain numerous unknown compounds, making in-depth research on their molecular structures crucial. Liquid chromatography-electrospray ionization time-of-flight mass spectrometry (LC-ESI-TOF-MS) is commonly used to extract Tibetan medicine; however, many unpredictable unknown compounds remain after using the spectrum database. The present article developed a universal method for identifying components in Tibetan medicine using ion trap mass spectrometry (IT-MS). The method includes standardized and programmed protocols for sample preparation, MS setting, LC prerun, method establishment, MS acquisition, multiple-stage MS operation, and manual data analysis. Two representative compounds in the Tibetan medicine Abelmoschus manihot seeds were identified using multiple-stage fragmentation, with a detailed analysis of typical compound structures. In addition, the article discusses aspects such as ion mode selection, mobile phase adjustment, scanning range optimization, collision energy control, collision mode switchover, fragmentation factors, and limitations of the method. The developed standardized analysis method is universal and can be applied to unknown compounds in Tibetan medicine.
Introduction
The qualitative analysis of trace components in traditional Chinese medicine (TCM) has become a crucial topic in research. Due to the high numbers of compounds in TCM, it is difficult to isolate them for nuclear magnetic resonance spectrometer (NMR) or X-ray diffractometer (XRD) analysis, making mass spectrometry (MS)-based methods that only require low sample volumes increasingly popular. Additionally, liquid chromatography (LC) coupled with MS has been widely used in TCM research in recent years for the improved separation of complex samples and qualitative analysis of chemical compounds1. One common method is liquid chromatography-electrospray ionization time-of-flight mass spectrometry (LC-ESI-TOF-MS), which is widely used in qualitative research on Tibetan medicine2. With this method, complex components are enriched and separated in an LC column, and the mass-to-charge ratio (m/z) of the adduct ions is observed using an MS detector. Searching tandem MS (MS/MS or MS2) databases is currently the fastest approach for confident compound annotations in small molecule analysis using quadrupole time-of-flight (Q-TOF) MS and Orbitrap MS3. However, the poor quality of databases and the presence of various isomers hinder the identification of unknown compounds. In addition, the information provided by the MS/MS database is limited4,5,6,7. It is significant to investigate the chemical compounds in each TCM using a general protocol that can be widely applied to other TCM.
IT-MS captures a wide range of ions by applying different radio frequency (RF) voltages to the ring electrodes8. IT-MS can perform time-series multiple-stage MS scans in diverse chronological orders, providing ingredient multiple-stage MS (MSn) fragmentation, where n is the number of product ion stages9. Linear IT-MS is considered the best for structure identification as it can be used for sequential MSn experiments10. Targeted ions can be isolated and accumulated in linear IT-MS1. The MSn (n ≥ 3) in IT-MS provides more fragment information than MS/MS in Q-TOF-MS. Since IT-MS cannot lock the target ion and its fragment ions, it is a powerful tool for the structure elucidation of unknown compounds, including isomers1. MSn technology has been widely applied to the structural analysis of unknown proteins, peptides, and polysaccharides11,12. The abundance level of fragment ions in MSn provides more molecular fragment information on targeted compounds in complex samples than MS/MS in Q-TOF-MS. Hence, applying MSn technology to structural identification in TCM is essential.
Tibetan medicine is a significant component of TCM13, and these medicines are primarily derived from animals, plants, and minerals found in the plateau area14. The Tibetan medicine Abelmoschus manihot seeds (AMS) is the seed of Abelmoschus manihot (linn.) medicus. AMS is a traditional herbal medicine used to treat conditions such as atopic dermatitis, rheumatism, and leprosy. It contains chalcone, which possesses antibacterial, antifungal, anticancer, antioxidative, and anti-inflammatory effects15. In the present study, MSn procedures were improved, and a detailed method was developed to identify compound structures in the Tibetan medicine AMS using IT-MS and MSn. Certain MS parameters, including the ion mode, scanning range, and collision mode, were optimized to overcome problems in identifying trace compounds. This study aims to promote the standardized structure identification of trace compounds in TCM.
Protocol
1. Sample preparation
- Accurately weigh 1 g of the AMS sample, and place it in a conical flask with 30 mL of 80% methanol. Transfer the mixture to an ultrasound bath sonicator for 30 min of extraction at 25 °C. Centrifuge the sample at 14,000 x g for 5 min.
NOTE: The frequency of the ultrasound bath sonicator is 40 KHz. - Prepare an injection syringe and a microporous membrane filter (0.22 μm, organic only). Filter the supernatant into a 2 mL sample bottle.
2. MS setting
- Turn on the switch of the vacuum pump. Open the main valve of the argon cylinder and the partial pressure valve, and adjust the pressure to approximately 0.3 MPa. Open the nitrogen valve.
NOTE: Wait for at least for 8 h to ensure a sufficient vacuum degree for the experimental conditions. Check that the gas pressure of argon and nitrogen is high enough before analysis. - Launch the MS control software. Click on Heated SEI Source in the software panel, and enter the MS parameters, including the heater temperature (350 °C), sheath gas flow rate (35 arb), aux gas flow rate (15 arb), spray voltage (3.8 KV for positive mode, −2.5 KV for negative mode), and capillary temperature (275 °C). Click on the Apply button to activate the ion source.
3. LC prerun, method establishment, and MS acquisition
- Prepare mobile phase A and mobile phase B using 0.1% formic acid aqueous solution and pure acetonitrile, respectively. Degas them in an ultrasound bath sonicator for at least 15 min. Connect the solutions to the A and B fluid passages, respectively (Figure 1A). Prepare a methanol-water (1:9 v/v) solution, and then fill it into the cleanout fluid bottles of the pump and injector by hand.
NOTE: The frequency of the ultrasound bath sonicator is 40 KHz. - Launch the LC-MS control software.
- Click on the Direct Control button to open the LC control panel. Open the purge valve in the counterclockwise direction on the pump module (Figure 1B).
- Click on the More Option button to open the pump setting, and set the purge parameters at 5 mLmin−1 for 3 min. Click on the Purge button to start the bubble removal. Subsequently, close the purge valve.
- Click on the Prime Syringe, Wash Buffer Loop, and Wash Needle Externally buttons to rinse the syringe for three cycles, the loop for one cycle, and the needle for one cycle, respectively. Place the sample bottle in the sampler (Figure 1C).
- Click on the Instrument Setup button to open the method-editing window. Click on the Neu button to create a new LC-MS instrument method.
- Establish a total run time for the LC method. Next, enter values to set the pressure limit, total flow rate, flow gradient, sample temperature, column temperature, and ready temperature delta in the method-editing window.
NOTE: The default total flow rate of the mobile phase is constant at 0.3 mL/min with 50% A and 50% B and without column temperature in the absence of a chromatographic column. The default values of sample temperature and ready temperature delta are 15 °C and 0.1 °C, respectively. Other settings depend on the type of liquid chromatography column used. - Select the General MS or MSn experiment type for the MS method. Enter values to configure the acquisition time, polarity, mass range, divert value number, and divert value duration. Click on the Save button to configure the settings as an instrument method.
NOTE: The default settings without a chromatography column are as follows: acquisition time, 2 min; polarity, positive or negative; mass range, 100 to 1,200; divert value number, 2; and divert value duration, 1.99 min.
4. Operating multiple-stage mass spectrometry
- Click on the Sequence Setup button to open the sequence table.
- In the table, enter the following information: sample type, file name, path, sample ID, instrument method, position, and injection volume.
- Click on the Save button to record the sequence table, and then click on the Start Analysis button to implement the settings and initiate the MS acquisition.
NOTE: The default sample type is selected as unknown. The instrument method is the method saved in step 3.6. The sample bottle is placed in its unique location in the sample room. For example, RA1 is the first location in the first row of the red area in the sample room. The default injection volume is usually 2 μL, which depends on the concentration of the sample.
- Double-click on the raw file in explorer to load the MS data into the data processing software. In the base peak chromatogram (BPI), select the area with the maximum area under the curve (AUC) by clicking and dragging the mouse. The corresponding MS spectra will be displayed in the same window.
- Select a targeted ion for the next MS/MS analysis.
- Reopen the method-editing window. In the MSn Setting table, set the m/z of the targeted ion to one decimal place in the Parent Mass column.
- Select Collision mode, and enter the collision energy (CE) value. Set the MS/MS scan range. Click on the Save button to record the MS method, and enter a new file name in the sequence table. Click on the Start button to initiate the MS/MS acquisition.
NOTE: The MS/MS scan range was 40%-130% of the targeted parent ion. The default CE value in collision-induced dissociation (CID) mode is 35%.
- Double-click on the raw file in explorer to load the MS/MS raw file into the data processing software.
- Identify the strongest fragment ion in the MS/MS spectrum, and enter its m/z value into the MSn method list. In the MSn Setting table, set the MS3 parameters, including collision mode, CE value, and scan range.
- Click on the Save button to record the MS method, and enter a new file name in the sequence table. Click on the Start button to initiate the MS3 acquisition.
- Double-click on the raw file in explorer to load the MS3 raw file into the data processing software. Repeat step 4.4 to obtain the MS4 spectrum.
- Complete the MSn experiment when no stable fragment ions are observed in the spectrum.
5. Manual MSn data analysis
- Double-click on the raw files to open all the mass spectra from MS to MSn. Manually calculate the m/z difference values between the ion and the corresponding fragment ions.
NOTE: For example, the m/z difference value between the ion (m/z 617.25) and the corresponding fragment ions (m/z 571.28) was 45.97 in MS/MS, the m/z difference value between the ion (m/z 571.28) and corresponding fragment ions (m/z 525.38) was 45.90 in MS3, and the m/z difference values between the ion (m/z 525.38) and the corresponding fragment ions (m/z 344.93 and 273.16) were 180.45 and 252.22 in MS4, respectively. - Manually draw the "core" structure according to MS4 results (the last level of MSn). Manually derive the original structure using functional groups or molecular segments based on the m/z difference value. Manually draw the molecular cleavage paths according to each molecular structure in MSn. Examples of manual molecular derivation are detailed in the representative results section.
Representative Results
Cellobiose was used as a model to verify the feasibility of MSn in positive ion mode. As shown in Figure 2A, the ESI-MS (positive ion mode) of cellobiose [C12H22O11]+ produced the protonated molecule [M+H]+ at m/z 365. The product ion scan (CID-MS/MS) of [M+H]+ at m/z 365 resulted in the second fragment ion at m/z 305 (Figure 2B), which was further analyzed using MS3 and MS4 analyses (Figure 2C,D). MS3 analysis resulted in the third fragment ion at m/z 254, and the MS4 analysis resulted in the fourth fragment ion at m/z 185. The MS/MS analysis (Figure 2E) revealed that the lost fragment ion at m/z 60 indicated a sequence of ion fragmentation at m/z 365, namely ring-opening hydrolysis (marked in blue), C-C bond cleavage (marked in red), and dehydration (marked in green). Similarly, the MS3 analysis revealed that the lost fragment ion at m/z 60 indicated the C-C bond cleavage (marked in red) of an ion at m/z 305. The MS4 analysis showed that the lost fragment ion at m/z 60 implied hydrolysis (marked in blue) and dehydration (marked in green), resulting in the cleavage of the ion with m/z 245 into an ion with m/z 185. The step fracture in the MSn analysis indicated that this method was feasible for investigating the structure of carbohydrates.
The preliminary qualitative analysis of AMS using LC-Q-TOF-MS revealed the presence of numerous unknown compounds. One of these, an ion at m/z 617, was selected for MSn analysis in negative mode. The product ion scan (CID-MS/MS) of the [M-H]− at m/z 617 in AMS produced a second fragment ion at m/z 571. The MS3 analysis of this fragment ion produced a third fragment ion at m/z 525, and the MS4 analysis produced fourth fragment ions at m/z 345 and 273 (Figure 3A–D). The MS3 of m/z 571 afforded a fragment ion at m/z 525 by the loss of the CH2OH portion as methanol (−32 Da) and the OH portion (−18 Da) as water. These MS4 results were used for the manual identification of the "core" structure of the compound, and its original structure was determined by comparing the m/z values of the ion and its fragment ions. The molecular structure of the compound at m/z 617 and its cleavage paths in MSn are shown in Figure 3E. Another unknown compound at m/z 365 was analyzed in positive mode using MSn. The product ion scan (CID-MS/MS) of the [M+H]+ ion at m/z 365 in AMS produced second fragment ions at m/z 299, m/z 329, and m/z 347. The MS3 analysis of these fragment ions produced a third fragment ion at m/z 231 (Figure 4A–C). The molecular structure and cleavage mechanism of the compound at m/z 365 are shown in Figure 4E.
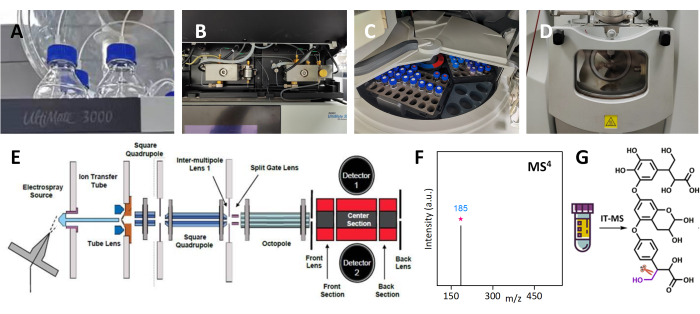
Figure 1: Identifying unknown compound structures in Tibetan medicine using IT-MS and multiple-stage mass spectrometry analysis. (A) The mobile phase forliquid chromatography. (B) The liquid chromatography pump. (C) The sample room. (D) The ion source for MS. (E) The internal structure of the ion trap module in MS. (F) The MS4 spectrum. (G) The molecular structure information from the MS4 results. Please click here to view a larger version of this figure.

Figure 2: Multiple-stage fragmentation of cellobiose via IT-MS in positive ion mode. (A) Original mass spectrum of cellobiose. (B) Fragment ions in the MS/MS spectrum. (C) Fragment ions in the MS3 spectrum. (D) Fragment ions in the MS4 spectrum. (E) The cleavage mechanism and molecular structure of cellobiose. Please click here to view a larger version of this figure.
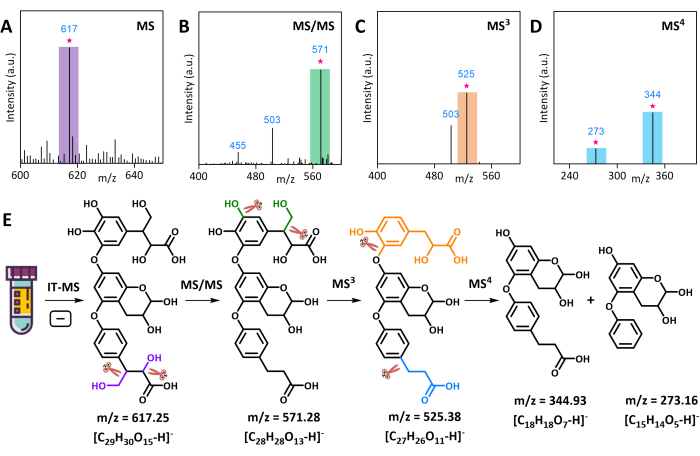
Figure 3: Multiple-stage fragmentation and structural analysis of the unknown AMS compound ion at m/z 617 via IT-MS in negative ion mode. (A) Partial mass spectrum of AMS. (B) Fragment ions in the MS/MS spectrum. (C) Fragment ions in the MS3 spectrum. (D) Fragment ions in the MS4 spectrum. (E) The cleavage mechanism and molecular structure of the AMS compound ion at m/z 617. Please click here to view a larger version of this figure.

Figure 4: Multiple-stage fragmentation structural analysis of the unknown AMS compound ion at m/z 365 via IT-MS in positive ion mode. (A) Partial mass spectrum of AMS. (B) Fragment ions in the MS/MS spectrum. (C) Fragment ions in the MS3 spectrum. (D) The cleavage mechanism and molecular structure of the AMS compound ion at m/z 365. Please click here to view a larger version of this figure.
Discussion
IT-MS and its MSn technology offer a new approach to identifying the structure of trace TCM compounds. Unlike Q-TOF-MS, which could not deeply identify the fragment ions, IT-MS with MSn technology excels due to its ability to isolate and accumulate ions. This article outlines a method for identifying trace compounds in Tibetan medicine using the IT-MS and MSn technique. The method utilizes the n value in MSn to determine the amount of fragment ion information provided. The crucial steps in this method include selecting the appropriate scan range and adjusting the CE value, which lead to the identification of valuable fragments.
In general, the MSn analysis of saccharides is best performed in positive ion mode16, while phenolic acids and alkaloids are best analyzed in negative ion mode. The response of the compound in the ESI source can be improved by adjusting the mobile phase with additives such as formic acid, acetic acid, and ammonium acetate17. An atmospheric-pressure chemical ionization source can be considered for compounds with weak polarity. Choosing an appropriate scan range can increase the intensity of the fragment ions, which is beneficial for the next stage of MSn because of the inevitable energy decay in each MSn. The m/z of the fragment ion should be located in the central region of the scanning range to obtain the best corresponding intensity. If an ion has double or multiple charges, fragment ions with higher m/z values can be obtained by decreasing the charge number during fragmentation. In this case, the end m/z of the scanning range should be set to be larger. The CID mode is suitable for most compounds in MSn analysis18. If the intensity of the fragment ion is insufficient, the CE value can be increased by 5% at a time. When there are multiple, complex fragment ions in MSn, a lower CE value is needed to control the ion dissociation. The pulsed-Q collision-induced dissociation mode, which is suitable for small molecules, provides more detailed information about low-molecular weight fragment ions than CID mode19. The electron transfer dissociation (ETD) model is dominant in peptide fracture and protein identification but is rarely used to identify the TCM components20. The ETD mode can be used to investigate unknown compounds containing disulfide bonds21.
Although the MSn method has many advantages for structural identification compared to other MS techniques, there are still some limitations. First, none of the collision modes are suitable for all TCM compounds. A reasonable choice of collision mode and manual adjustment of the collision energy can improve the fragment ions. In addition, with the MSn method, it is difficult to distinguish the position of functional groups in large molecules with complex isomers. Identifying the functional group sites is a challenging task that requires experienced researchers. Manual post-analysis and long MSn data processing time are also significant barriers that discourage researchers from utilizing this technology. Q-TOF-MS is popular among researchers due to its high measurement accuracy, resolution, and ease of use with databases. However, IT-MS is a good solution for unidentified ions and trace ions due to its ability to isolate and accumulate ions and perform multiple stages of analysis. The integration of Q-TOF and IT-MS could provide a optimal solution for the full qualitative analysis of TCM samples. MSn technology is widely used in fields such as food, environmental science, and medicine, and its popularity and use in various fields are expected to increase with the improvement of IT-MS instrumentation.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Xinglin Talent Program of Chengdu University of TCM (No. 030058191), the Nature Science Foundation of Sichuan (2022NSFSC1470), and the National Natural Science Foundation of China (82204765).
Materials
| Acetonitrile | Thermo Scientific | CAS 75-05-8 | LC-MS grade |
| Formic Acid | Knowles | CAS 64-18-6 | HPLC grade |
| Linear ion trap mass spectrometer | Thermo Scientific | LTQ XL | |
| liquid chromatograph | Thermo Scientific | U3000 | |
| LTQ Tune | Thermo Scientific | version 2.8.0 | MS control software |
| Methanol | Thermo Scientific | CAS 67-56-1 | LC-MS grade |
| Pure water | Thermo Scientific | CAS 7732-18-5 | LC-MS grade |
| Xcalibur | Thermo Scientific | version 2.0 | LC-IT-MS operational software |
Referenzen
- Chen, X. -. F., Wu, H. -. T., Tan, G. -. G., Zhu, Z. -. Y., Chai, Y. -. F. Liquid chromatography coupled with time-of-flight and ion trap mass spectrometry for qualitative analysis of herbal medicines. Journal of Pharmaceutical Analysis. 1 (4), 235-245 (2011).
- Ou, C., et al. Systematically investigating the pharmacological mechanism of Dazhu Hongjingtian in the prevention and treatment of acute mountain sickness by integrating UPLC/Q-TOF-MS/MS analysis and network pharmacology. Journal of Pharmaceutical and Biomedical Analysis. 179, 113028 (2020).
- Kind, T., et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrometry Reviews. 37 (4), 513-532 (2018).
- Phetsanthad, A., Vu, N. Q., Li, L. Multi-faceted mass spectrometric investigation of neuropeptides in Callinectes sapidus. Journal of Visualized Experiments. (183), e63322 (2022).
- Seetaloo, N., Phillips, J. J. Millisecond hydrogen/deuterium-exchange mass spectrometry for the study of alpha-synuclein structural dynamics under physiological conditions. Journal of Visualized Experiments. (184), e64050 (2022).
- Karas, B. F., et al. Dose uptake of platinum-and ruthenium-based compound exposure in zebrafish by inductively coupled plasma mass spectrometry with broader applications. Journal of Visualized Experiments. (182), e6358 (2022).
- Chang, H. -. L., et al. Uracil-DNA glycosylase assay by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis. Journal of Visualized Experiments. (182), e63089 (2022).
- Wang, S., et al. Structural characterization and identification of major constituents in Jitai tablets by high-performance liquid chromatography/diode-array detection coupled with electrospray ionization tandem mass spectrometry. Molecules. 17 (9), 10470-10493 (2012).
- Pang, B., Zhu, Y., Lu, L., Gu, F., Chen, H. The applications and features of liquid chromatography-mass spectrometry in the analysis of traditional Chinese medicine. Evidence-Based Complementary and Alternative Medicine. 2016, 3837270 (2016).
- Ichou, F., et al. Comparison of the activation time effects and the internal energy distributions for the CID, PQD and HCD excitation modes. Journal of Mass Spectrometry. 49 (6), 498-508 (2014).
- Fu, X., et al. Suppression of oligomer formation in glucose dehydration by CO2 and tetrahydrofuran. Green Chemistry. 19 (14), 3334-3343 (2017).
- Fu, X., et al. Solvent effects on degradative condensation side reactions of fructose in its initial conversion to 5-Hydroxymethylfurfural. ChemSusChem. 13 (3), 501-512 (2020).
- Yang, S., Wang, Z., Zhao, H., Ren, X. Modern research of Tibetan medicine. World Journal of Traditional Chinese Medicine. 5 (2), 131-138 (2019).
- Shang, X., et al. Ethno-veterinary survey of medicinal plants in Ruoergai region, Sichuan province, China. Journal of Ethnopharmacology. 142 (2), 390-400 (2012).
- Su, J., et al. Chalcone derivatives from Abelmoschus manihot seeds restrain NLRP3 inflammasome assembly by inhibiting ASC oligomerization. Frontiers in Pharmacology. 13, 932198 (2022).
- Fu, X., et al. Mapping out the reaction network of humin formation at the initial stage of fructose dehydration in water. Green Energy & Environment. , (2022).
- Hua, Y., Jenke, D. Increasing the sensitivity of an LC-MS method for screening material extracts for organic extractables via mobile phase optimization. Journal of Chromatographic Science. 50 (3), 213-227 (2012).
- Kumar, S., Singh, A., Bajpai, V., Kumar, B. Identification characterization and distribution of monoterpene indole alkaloids in Rauwolfia species by Orbitrap Velos Pro mass spectrometer. Journal of Pharmaceutical and Biomedical Analysis. 118, 183-194 (2016).
- Bayat, P., Lesage, D., Cole, R. B. Tutorial: Ion activation in tandem mass spectrometry using ultra-high resolution instrumentation. Mass Spectrometry Reviews. 39 (5-6), 680-702 (2020).
- Wu, S. -. L., et al. Mass spectrometric determination of disulfide linkages in recombinant therapeutic proteins using online LC−MS with electron-transfer dissociation. Analytical Chemistry. 81 (1), 112-122 (2009).
- Echterbille, J., Quinton, L., Gilles, N., De Pauw, E. Ion mobility mass spectrometry as a potential tool to assign disulfide bonds arrangements in peptides with multiple disulfide bridges. Analytical Chemistry. 85 (9), 4405-4413 (2013).

