Efficient and Consistent Generation of Retinal Pigment Epithelium/Choroid Flatmounts from Human Eyes for Histological Analysis
Summary
We describe a method to efficiently separate retinal pigment epithelium (RPE) from the retina in human eyes and generate whole RPE/choroid flatmounts for histological and morphometric analyses of the RPE.
Abstract
The retinal pigment epithelium (RPE) and retina are functionally and structurally connected tissues that work together to regulate light perception and vision. Proteins on the RPE apical surface are tightly associated with proteins on the photoreceptor outer segment surface, making it difficult to consistently separate the RPE from the photoreceptors/retina. We developed a method to efficiently separate the retina from the RPE of human eyes to generate complete RPE/choroid and retina flatmounts for separate cellular analysis of the photoreceptors and RPE cells. An intravitreal injection of a high-osmolarity solution of D-mannitol, a sugar not transported by the RPE, induced the separation of the RPE and retina across the entire posterior chamber without causing damage to the RPE cell junctions. No RPE patches were observed attached to the retina. Phalloidin labeling of actin showed RPE shape preservation and allowed morphometric analysis of the entire epithelium. An artificial intelligence (AI)-based software was developed to accurately recognize and segment the RPE cell borders and quantify 30 different shape metrics. This dissection method is highly reproducible and can be easily extended to other animal models.
Introduction
The retinal pigment epithelium (RPE) and the neural retina are strongly interconnected with each other because of the strong physiological dependence of the photoreceptors on the RPE. During dissection, the mechanical separation of the neural retina from the RPE causes tearing of the RPE cells, with the apical portions of the RPE remaining attached to the outer segments of the retinal photoreceptors. The extent of RPE-retinal adhesion is so great that the amount of pigment remaining on the retina after separation is used to quantify the strength of retinal adhesion1. Specifically, RPE tight junctions and the actin structure that connects them, which are located on the apical side, break off during mechanical separation. Therefore, staining RPE flatmounts for cell borders results in a patchy monolayer in which many cells have missing borders. This effect is exacerbated when the tissue is fixed with paraformaldehyde (PFA) before dissection, as the proteins become crosslinked.
Studies on intravitreal drug delivery have shown that injections of hyperosmotic solutions in the posterior chamber induce retinal detachment2,3. In these experiments, 50 µL of different solutions, ranging from 1,000 mOsm to 2,400 mOsm, injected in the mid-vitreous caused retinal detachment within minutes. Notably, even after long exposures to high-osmolarity solutions, the RPE tight junctions appeared intact in the transmission electron microscopic images of both rabbit and monkey eyes3. Following a similar strategy, we injected into the mid-vitreous a hyperosmotic solution of D-mannitol to induce an efficient retinal detachment before performing RPE dissection. As D-mannitol is not transported by the RPE4, a high intravitreal concentration is maintained, generating an osmotic gradient. The efficient separation of the RPE and retina across the entire posterior chamber guarantees the preservation of the RPE cellular junctions and allows for the study of RPE morphometry on the entire flatmount. In addition, we developed an artificial intelligence (AI)-based software that recognizes and segments fluorescently labeled RPE cell borders, quantifies 30 different shape metrics, and produces heatmaps of each metric for visualization5,6.
Protocol
Cadaver human globes were obtained from the Advanced Sight Network (Birmingham, AL). Work performed on cadaver tissue is exempted by the NIH Institutional Review Board from the research ethics committee.
1. Eye globe shipment
- After enucleation, ship fresh eye globes in a container filled with ice-cold DPBS 1x with Ca2+ and Mg2+.
NOTE: It is better to dissect the eye within 24 h after enucleation. The RPE morphology is not altered during this time window.
2. Silicone mold preparation
- Cut the bottom 20 mm of a 25 mm diameter round-bottom tube. Place it at the base of a square weighing boat (81 mm x 81 mm x 25 mm).
- Mix the two components of the Silicon Elastomer Kit at a 10:1 ratio, paying attention not to entrap air. Pour the mix into the weighing boat containing the spherical piece of the round-bottom tube.
- Cure the silicone at room temperature overnight. Remove the weighing boat and the round-bottom tube from the cured silicone mold.
3. RPE dissection
- Clean the sclera of the fresh eye globe of muscles and connective tissue. Secure the eye to the silicone mold using 27 G needles threaded through the sclera. Fill the cavity of the mold with DPBS 1x with Ca2+ and Mg2+.
NOTE: Pay attention not to breach the eye chamber. The needles should only pass through the sclera. - Use a 1 mL syringe and a 21 G needle to inject ~400 µL of 1,700 mOsm D-mannitol solution into the vitreous. Insert the needle through the pars plana to avoid puncturing the anterior chamber of the eye. Leave the eye at room temperature for ~45 min.
- Cut open the anterior chamber at the level of the pars plana using a pair of fine scissors and forceps. Fill the posterior eye chamber with DPBS 1x with Ca2+ and Mg2+. Under the stereomicroscope, localize the macula (the yellow spot on the retina).
- If a surgical vitreous cutter is available, remove the vitreous and replace it with DPBS 1x with Ca2+ and Mg2+. Alternatively, try to lift the vitreous with forceps and cut it with fine scissors.
- Paying attention to preserving the macular region, cut the eye into quadrants: nasal, temporal, superior, and inferior. Remove the needles if they are in the way.
- Transfer the butterflied posterior chamber of the eye into a 100 mm Petri dish containing DPBS 1x with Ca2+ and Mg2+. Before removing the retina, mark the petal that contains the macula by making a little cut (V-shaped) in the ciliary margin. Lift and cut all the vitreous that lays on the retina.
- Gently lift the retina from multiple sides with a curved spatula or a pair of forceps to check if the retina has detached from the RPE and to let some fluid circulate between the two layers.
NOTE: The retina will still be attached at the very periphery (ciliary margins) and at the optic nerve. - Cut the retina from the ciliary margins in all the petals, ensuring not to scratch the RPE. Place the tissue in 4% PFA and incubate for ~1 h. Wash 3x with DPBS 1x with Ca2+ and Mg2+. Transfer the tissue into a container filled with DPBS 1x with Ca2+ and Mg2+ and store it at 4 °C.
NOTE: At this point, the retina is only attached at the optic nerve. This is a pause point in this experiment. - Transfer the sample to a 100 mm Petri dish containing DPBS 1x with Ca2+ and Mg2+. Punch out the optic nerve head with a 1.5 mm biopsy punch and collect the retina. Store the neural retina in DPBS 1x with Ca2+ and Mg2+ at 4 °C.
NOTE: Before punching out the optic nerve head, make sure to cut the optic nerve as much as possible on the scleral side. This will increase the precision of the punch. Punching out the optic nerve after the flatmount is fixed in 4% PFA reduces the damage to the RPE cells located around the optic nerve. - Remove the sclera from the RPE/choroid by gently lifting the RPE/choroid layer from the periphery and cutting the choroidal vessels and connective tissue that are between the sclera and the RPE with a pair of Vannas spring scissors. Once the RPE/choroid is completely separated from the sclera, collect the RPE/choroid layer. Transfer the tissue into a container filled with DPBS 1x with Ca2+ and Mg2+ and store it at 4°C.
NOTE: At this time, the experiment can be paused.
4. Staining
- Transfer the RPE/choroid to one well of a 6-well plate. Block and permeabilize the sample in DPBS 1x with Ca2+ and Mg2+ with 1% bovine serum albumin (BSA), 0.5% Tween 20, and 0.5% Triton X-100 for 1 h at room temperature.
- Incubate the sample with phalloidin conjugated with a 647 fluorophore at a 1:250 dilution in DPBS 1x with Ca2+ and Mg2+ with 1% BSA, 0.5% Tween 20, and 0.5% Triton X-100 for 1 h at room temperature. Wash 3x in DPBS 1x with Ca2+ and Mg2+.
- Transfer the RPE/choroid sample to a 50 mm x 75 mm glass slide and flatten it. Cut each "petal" into two to make the sample flatter. Pay attention to the macula. Draw a contour of the flatmount with a hydrophobic pen.
- To quench the lipofuscin autofluorescence, add 500 µL of the autofluorescence quencher solution diluted to 1:20 in 70% ethanol and incubate at room temperature for 2 min.
- Wash thoroughly (at least 3x) in DPBS 1x with Ca2+ and Mg2+. Remove the DPBS and add the mounting medium. Place a coverglass on the flatmount and seal with nail polish.
- Image the flatmount with a fluorescence microscope (preferably using a 10x or 20x objective).
5. REShAPE analysis
NOTE: As the REShAPE AI-based algorithm was trained on 10x and 20x images, it is, therefore, highly recommended to use a 10x or 20x objective when imaging. If not, the images will need to be rescaled accordingly.
- If the images are acquired with more than one fluorescent channel, isolate the channel used to acquire the cell borders. Export the images as 16-bit greyscale TIF files.
NOTE: Images acquired with the file extension .czi do not need to be exported as TIF files, but the channel containing the cell borders still needs to be isolated. - Install the software on Windows x64 or Linux (Centos 7) platforms.
NOTE: The software and the instructions for installation can be found at https://github.com/nih-nei/REShAPE. - Open the software (Figure 1).
- In the Directories tab, select the input and output folders. In the input directory, select the folder that contains the images. In the output directory, let the software automatically change the path to "input directory/Processed". Alternatively, change the output directory manually.
NOTE: The software will iterate through all the images contained in the input folder. - In the NN Segmentation Options tab, choose the following settings for image segmentation:
- Batch Size: Type a value of 20 in the text box as a good starting point, but lower it if the system runs out of graphics processing units (GPUs).
NOTE: The original image is split into smaller tiles for processing. The batch size specifies how many tiles can be processed at once. - Use GPU?: If the system does not have enough GPU resources, check the box No.
- Internal Scale: Using the dropdown menu, change the internal scaling from None in cases where the images were not acquired with a 10x or 20x objective. For example, downscale 40x images to half the size using the 1/2 button. The rescaling options available in the dropdown menu are as follows : 5x, 2x, None, 1/2, 1/5.
NOTE: The machine learning was trained with 10x and 20x images. It may not recognize cell borders from images taken at other magnifications. In this case, binary images will appear completely black. - Arti Filter: Use the artifact filter tab to remove very bright particles (debris or small parts of the retina or connective tissue around the optic nerve) that may be present in the image and may interfere with cell border segmentation. No filter is used by default. The artificial filter options in the dropdown menu are as follows: Regular, Strong, and Weak.
- Batch Size: Type a value of 20 in the text box as a good starting point, but lower it if the system runs out of graphics processing units (GPUs).
- In the Tiled Image Option tab, insert a value in the text box to specify the amount of overlap between the image tiles by adjusting the Pad Tiles By parameter. An overlap of 100 pixels usually works well.
- In the Output Graphic Options tab, tune the settings for heatmap generation:
- Reconstruct Tiled Images?: if the system does not have enough resources for the analysis of large images, check the box No to allow the software to complete the analysis by saving individual tiles without reconstructing the entire image.
- Create Color Images: To generate heatmaps, select Alle from the dropdown menu.
- For Coloring, choose any of the different color palettes available in the dropdown menu: Thermal, Green, Mpl-magma, Phase, Fire, Jet, Cyan Hot.
- For Image Format, save the images as TIF or PNG by selecting one of the options in the tab.
- With respect to the Use Manual Limits? feature, check the box No to let the software use the minimum and maximum values detected in each image. Check the box Yes to manually adjust the range of values for each shape metric heatmap, and click on the Set Limits button to choose ranges for the individual parameters by inserting values in the text boxes. After changing the values of interest, click on Save. Click on Load Defaults to reset all the limits.
NOTE: Use manual limits if heatmaps from multiple images need to be compared, such as when comparing the effects of different compounds on cell shape. This way, the same range of values is used. The set of values to be entered manually varies depending on the type of sample. It is recommended to run a few iterations to choose the optimal range.
- In the Cell Size Restrictions for Analysis tab, select a cell size threshold for the analysis:
- In Lower Cell Size, insert in the text box the size of the smallest cell to be included in the analysis.
- In Upper Cell Size, insert in the text box the size of the largest cell to be included in the analysis.
NOTE: The unit of cell size changes from pixels to micrometers squared depending on the option chosen in the Automated Unit Conversion tab.
- In the Automated Unit Conversion tab, choose the preferred unit for the analysis:
- In Convert pixels to real units?, check the No box to run the analysis in pixel units. Check the Yes box to run the analysis in micrometers.
- In Length of scale bar (Pixels), enter the pixel value in the text box.
- In Length of scale bar (Microns), enter in the text box the corresponding distance in micrometers.
- To start the analysis, press Go For It.
NOTE: The software can also measure cell viability when the cells are stained with 4',6-diamidino-2-phenylindole (DAPI) and propidium iodide (CZI options tab), but this does not apply to RPE flatmounts.
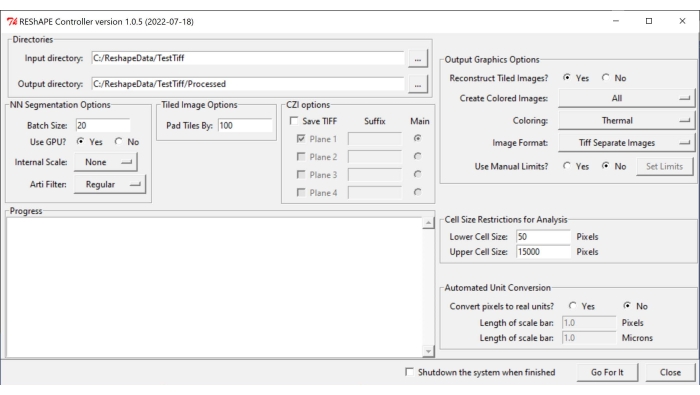
Figure 1: REShAPE graphical user interface. The GUI has different tabs for selecting the working directories (Directories tab), modifying the segmentation options (NN Segmentation Options and Tiled Image Options tabs), specifying the parameters for analysis (Cell Size Restrictions for Analysis and Automated Unit Conversion tabs), and for heatmap generation (Output Graphics Options tab). Abbreviation: GUI = graphical user interface. Please click here to view a larger version of this figure.
Representative Results
This protocol results in a single-plane image of a flatmount, where the cell location and 30 shape metrics are measured for every correctly identified RPE cell (Figure 2). A folder named "Processed" is automatically generated inside the input directory. This folder contains four subdirectories, named "Analysis," "Color Coded," "Combined Files," and "Segmented Images," and some temporary files generated during the analysis. The "Combined Files" folder contains a spreadsheet with all the shape measurements and a spreadsheet with the frequencies of the cell neighbor counts of all the files combined. The "Analysis" folder contains a spreadsheet with all the shape measurements and a spreadsheet with the frequencies of the cell neighbor counts for each image separately. The "Segmented Images" directory contains the final binary masks of the RPE cell borders; it can be used to evaluate the quality of the segmentation. The "Color Coded" directory contains heatmaps for each shape measurement to visualize the shape patterns in each image. The shape metric definitions and abbreviations can be found in Table 1.
Sometimes RPE flatmounts can contain residual pieces of retina that were not cleanly removed, especially around the optic nerve. Phalloidin staining of the sample results in a strong signal coming from the retina, and this can cause problems for RPE cell border segmentation. Some tiles will appear completely black, while the surrounding tiles will show normal segmentation. Other bright objects that may be present in the image will also cause the generation of black tiles (Figure 3). In these cases, choosing one of the filtering options (Weak, Regular, Strong) available in the Arti Filter dropdown menu will prevent the formation of black tiles.
REShAPE takes 8-bit or 16-bit greyscale images as input but not RGB images. Using RGB images for the REShAPE analysis will produce entirely black binary images. If this occurs, converting the RGB images to greyscale will produce correctly segmented binary images (Figure 4). On some occasions where the RPE borders are not recognized correctly, for example, if the staining is not optimal or if the sample is damaged by a scratch (Figure 5A), large clumps of cells may be identified as a single very large cell (Figure 5B). In this case, large objects can be excluded from the analysis by reducing the cell size threshold (Figure 5C). This can be achieved by inserting a lower value in the Upper Cell Size text box. However, this will result in a change in the range of the heatmap. If a researcher chooses to do so, it is also possible to maintain the original heatmap range (Figure 5D) by checking the box Yes in the Use Manual Limits? feature. Subsequently, the researcher must left-click on the Set Limits button and insert the desired values in the text boxes to specify the manual limits.

Figure 2: Complete morphometric analysis of an entire human RPE monolayer. (A) A low magnification view of an entire human RPE/choroid flatmount (magenta: phalloidin). (B) A zoomed-in view of phalloidin-stained RPE cells. (C) REShAPE-generated segmentation of the RPE cell borders for an entire human RPE/choroid flatmount and (D) the corresponding zoomed-in view. (E) A software-generated heatmap illustrating the cell area of the individual RPE cells in the entire human flatmount. The thermal scale on the top-left corner shows the range of values used. (F) The corresponding zoomed-in view showing individual RPE cells colored by area. Scale bars = (B,D,F) 50 µm, (A,C,E) 5 mm. Abbreviation: RPE = retinal pigment epithelium. Please click here to view a larger version of this figure.

Figure 3: Filtering of bright artifacts. (A) A human RPE flatmount stained for cell borders (magenta: phalloidin) can present bright areas (green rectangles) that interfere with segmentation. (B) The RPE cell border segmentation of the entire flatmount contains three completely black tiles (green arrows) corresponding to bright regions of fluorescence. (C,E) Two of the black tiles correspond to areas containing bright dots, which are possibly some debris. (D) One of the black tiles was generated by a piece of neural retina around the optic nerve that was not correctly removed. The pieces of neural retina are considerably brighter than the RPE layer and hinder cell segmentation. Please click here to view a larger version of this figure.
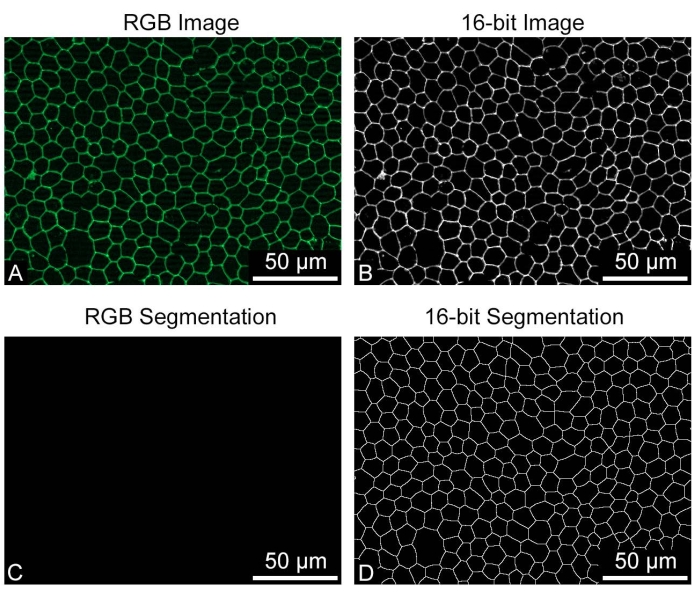
Figure 4: Input image requisite. The RPE cells stained for cell borders were saved as (A) RGB or as (B) greyscale 16-bit images for REShAPE analysis. (C) The output of the RBG image analysis is a black binary image, (D) while the analysis of the greyscale image produces a correctly segmented binary of the cell borders. REShAPE can only analyze 8-bit or16-bit greyscale images. Scale bars = 50 µm. Please click here to view a larger version of this figure.
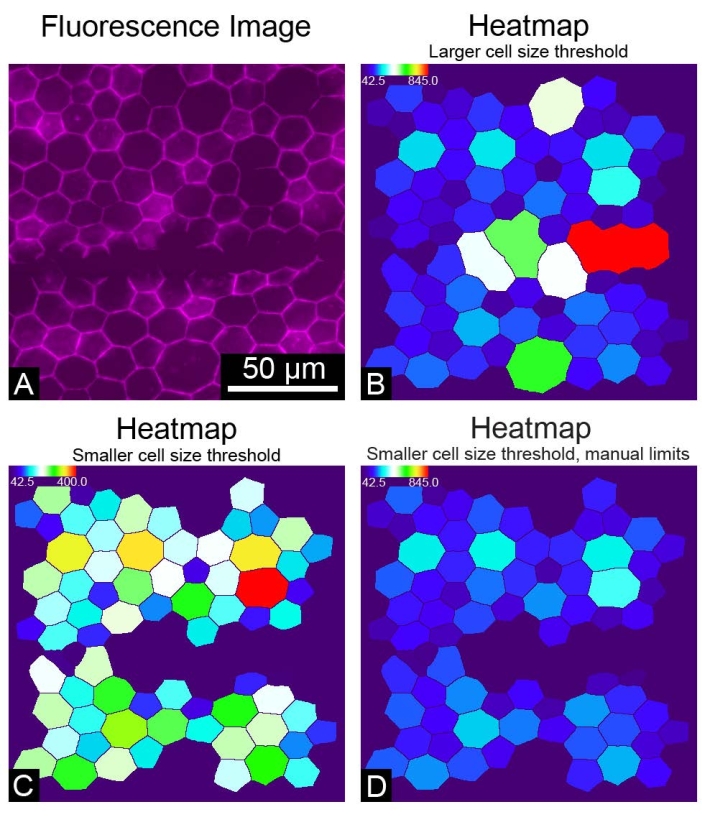
Figure 5: Suboptimal results. (A) An image of a portion of the RPE monolayer where the cells stained with phalloidin were accidentally scratched. (B) A heatmap of RPE cells colored by the dimension of cell area. A large upper cell size threshold includes large objects in the analysis. (C) A cell area heatmap in which a smaller upper cell size threshold was chosen to exclude large objects from the analysis. (D) A cell area heatmap in which a smaller upper cell size threshold was chosen and manual limits were set to maintain the heatmap range used originally. Please click here to view a larger version of this figure.
Table 1: REShAPE parameters. The table reports the definition of each parameter and the abbreviations used in the raw spreadsheets ("_Data.csv" files) and for the heatmaps. Please click here to download this Table.
Discussion
The consistent and efficient separation of human RPE and retinas can be achieved using this protocol. This method allows for the study of regional differences in RPE shape across entire human retinas5. A crucial step in the protocol is the physical separation of the RPE and retina. If the two tissues are not completely detached in some areas, one should gently lift the retina, ensuring not to break the tissues. The REShAPE analysis of large flatmounts may require the use of systems with considerable RAM resources. In this case, the reassembly of the entire image can be disabled to allow the software to successfully finish the analysis despite a lack of processing resources.
The main limitation of using REShAPE to segment human RPE flatmounts is that the AI algorithm was mostly trained on images of induced pluripotent stem cell-derived RPE. As a consequence, the segmentation of human RPE flatmounts is less accurate. RPE cells from aged donors contain a large amount of lipofuscin7, and the broad spectrum of its autofluorescence interferes with cell border segmentation. In the future, more images of RPE flatmounts will be used to improve cell border segmentation in this kind of sample. Despite this limitation, REShAPE was specifically trained to recognize and segment RPE cell borders and performs better than other existing methods, such as Voronoi8 and CellProfiler9 segmentation of RPE cells.
Moreover, compared to manual segmentation10, REShAPE provides the advantage of analyzing large images quickly (~130,000 pixels x 130,000 pixels were tested). In conclusion, this dissection method is highly reproducible and can be easily extended to other animal models. In addition, the software can be used to study RPE shape in eye flatmounts or in cell culture models to examine the effect of certain treatments. Finally, REShAPE's versatility makes it broadly applicable for the analysis of other types of epithelial cells.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank the National Eye Institute (NEI) histology core for the use of the Zeiss Axio Scan.Z1. We also thank the donors, their families, the Advancing Sight Network, and the Lions Eye Institute for their generosity. This work was supported by NEI IRP funds (grant number ZIA EY000533-04).
Materials
| Biopsy punch 1.5 mm | Acuderm Inc. | P1525 | |
| Bovine albumin | MP Biomedicals | 160069 | |
| Coverglass 50 x 75 mm, #1.5 thickness | Brain Research Laboratories | 5075-1.5D | |
| Curved spatula | Katena | K3-6600 | |
| D-Mannitol | Sigma | M9546 | |
| DPBS 1x with Ca2+ and Mg2+ | Gibco | 14040-133 | |
| Fine Scissors | Fine Science Tools | 14558-11 | |
| Fluormount-G | Southern Biotech | 0100-01 | |
| Forceps – Dumont #5 | Fine Science Tools | 11252-23 | |
| Microscope slides 50 x 75 x 1.2 mm | Brain Research Laboratories | 5075 | |
| Needles 21 G x 1-1/2" hypodermic | Becton Dickinson (BD) | 305167 | |
| Needles 27 G x 1-1/4" hypodermic | Becton Dickinson (BD) | 305136 | |
| Paraformaldehyde 16% | Electron Microscopy Sciences | 15710 | |
| Petri dish 100 mm | Corning | 430167 | |
| Phalloidin-iFluor 647 | Abcam | ab176759 | |
| Razor blades | PAL (Personna) | 62-0177 | |
| Round bottom tubes 50 mL | Newegg | 9SIA4SR9M88854 | |
| Silicon Elastomer Kit | Dow Corning Corporation | 4019862 | |
| Square weighing boat (81 mm x 81 mm x 25 mm) | Sigma | W2876 | |
| Surgical Vitrectomy System | BD Visitrec | 585100 | optional |
| Syringe 1 mL | Becton Dickinson (BD) | 309659 | |
| Triton X-100 | Sigma | T9284 | |
| TrueBlack | Biotium | 23007 | autofluorescence quencher |
| Tween 20 | Affymetrix | 20605 | |
| Vannas Spring Scissors – 3 mm cutting edge | Fine Science Tools | 15000-10 |
Referenzen
- Endo, E. G., Yao, X. Y., Marmor, M. F. Pigment adherence as a measure of retinal adhesion: Dependence on temperature. Investigative Ophthalmology and Visual Science. 29 (9), 1390-1396 (1988).
- Marmor, M. F. Retinal detachment from hyperosmotic intravitreal injection. Investigative Ophthalmology and Visual Science. 18 (12), 1237-1244 (1979).
- Marmor, M. F., Martin, L. J., Tharpe, S. Osmotically induced retinal detachment in the rabbit and primate. Electron microscopy of the pigment epithelium. Investigative Ophthalmology and Visual Science. 19 (9), 1016-1029 (1980).
- Ban, Y., Rizzolo, L. A culture model of development reveals multiple properties of RPE tight junctions. Molecular Vision. 3, 18 (1997).
- Ortolan, D., et al. Single-cell-resolution map of human retinal pigment epithelium helps discover subpopulations with differential disease sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 119 (19), 2117553119 (2022).
- Schaub, N. J., et al. Deep learning predicts function of live retinal pigment epithelium from quantitative microscopy. Journal of Clinical Investigation. 130 (2), 1010-1023 (2020).
- Beatty, S., Koh, H. -. H., Phil, M., Henson, D., Boulton, M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Survey of Ophthalmology. 45 (2), 115-134 (2000).
- Liu, Z., Kocaoglu, O. P., Miller, D. T. 3D imaging of retinal pigment epithelial cells in the living human retina. Investigative Ophthalmology and Visual Science. 57 (9), 533-543 (2016).
- Bhatia, S. K., et al. Analysis of RPE morphometry in human eyes. Molecular Vision. 22, 898-916 (2016).
- vonder Emde, L., et al. Histologic cell shape descriptors for the retinal pigment epithelium in age-related macular degeneration: A comparison to unaffected eyes. Translational Vision Science & Technology. 11 (8), 19 (2022).

