Investigating the Pathogenesis of MYH7 Mutation Gly823Glu in Familial Hypertrophic Cardiomyopathy using a Mouse Model
Summary
Based on the familial hereditary cardiomyopathy family found in our clinical work, we created a C57BL/6N mouse model with a point mutation (G823E) at the mouse MYH7 locus through CRISPR/Cas9-mediated genome engineering to verify this mutation.
Abstract
Familial hypertrophic cardiomyopathy (HCM, OMIM: 613690) is the most common cardiomyopathy in China. However, the underlying genetic etiology of HCM remains elusive.
We previously identified a myosin heavy chain 7 (MYH7) gene heterozygous variant, NM_000257.4: c.G2468A (p.G823E), in a large Chinese Han family with HCM. In this family, variant G823E cosegregates with an autosomal dominant disorder. This variant is located in the lever arm domain of the neck region of the MYH7 protein and is highly conserved among homologous myosins and species. To verify the pathogenicity of the G823E variant, we produced a C57BL/6N mouse model with a point mutation (G823E) at the mouse MYH7 locus with CRISPR/Cas9-mediated genome engineering. We designed gRNA targeting vectors and donor oligonucleotides (with targeting sequences flanked by 134 bp of homology). The p.G823E (GGG to GAG) site in the donor oligonucleotide was introduced into exon 23 of MYH7 by homology-directed repair. A silenced p.R819 (AGG to CGA) was also inserted to prevent gRNA binding and re-cleavage of the sequence after homology-directed repair. Echocardiography revealed left ventricular posterior wall (LVPW) hypertrophy with systole in MYH7 G823E/- mice at 2 months of age. These results were likewise validated by histological analysis (Figure 3).
These results demonstrate that the G823E variant plays an important role in the pathogenesis of HCM. Our findings enrich the spectrum of MYH7 variants linked to familial HCM and may provide guidance for genetic counseling and prenatal diagnosis in this Chinese family.
Introduction
Hypertrophic cardiomyopathy (HCM, OMIM: 613690) is the most common cardiomyopathy in China, with an estimated incidence of 0.2%, affecting 150,000 people1,2.
The pathological anatomical feature that characterizes HCM is asymmetric ventricular hypertrophy, which often involves the ventricular outflow tract and/or interventricular septum3. The clinical manifestation is exertional dyspnea, fatigue, and chest pain. The individual phenotype of HCM has variability ranging from clinically insidious to severe heart failure. Patients with HCM require medical treatment, heart transplantation, life support equipment, and multidisciplinary follow-up4.
In the past century, PCR technology has changed the way we study DNA5. A DNA sequencing method for clinical diagnosis was discovered by Sanger and colleagues6. The Sanger technique was subsequently applied to the Human Genome Project, but this approach was costly and time-consuming7. The advent of whole-genome sequencing (WGS) brought insights into human genetic disease to new heights, but it remained prohibitive in terms of cost. Whole-exome sequencing (WES) technology has long been used to detect germline variants8 and has been successful in identifying somatic driver mutations in the exome of various cancers9. The detection of DNA exons or coding regions by WES can be used to reveal pathogenic variants in most Mendelian diseases. Today, with the decreasing cost of sequencing, WGS is expected to become an important tool in genomics research and can be widely used in the detection of pathogenic variants in the genome.
WES technology has also been used in inherited cardiomyopathy to identify pathogenic variants to further elucidate the etiology. Emerging evidence has implicated that genes coding sarcomere structural protein gene mutations, such as MYH710, MYH611, MYBPC312, MYL213, MYL314, TNNT215, TNNI316, TNNC117, and TPM118 are responsible for the genetic etiology of HCM. Awareness of pathogenic variants in rare disease-causing genes (e.g., obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF (OBSCN, OMIM: 608616)19, acting alpha 2 (ACTN2, OMIM: 102573)20, and cysteine and glycine rich protein 3 (CSRP3, OMIM: 600824)21) has also been associated with HCM. Current genetic studies have identified multiple distinct pathogenic variants in the sarcomeric protein gene in approximately 40%-60% of HCM patients, and genetic testing in HCM patients revealed that most pathogenic variants occur in the myosin heavy chain (MYH7) and myosin-binding protein C (MYBPC3). However,the genetic basis for HCM remains elusive. Exploring the pathogenicity of these variations that underlie the human HCM patients remains a major challenge22.
In this study, we report a pathogenic variant in MYH7 in a Chinese Han family with HCM by WES. In order to verify the pathogenicity of this variant, we established a C57BL/6N-Myh7em1(G823E) knockin mice using the CRISPR/Cas9 system. We also discuss plausible mechanisms of this variant.
Protocol
The histories of the families were obtained by interviewing the family members. The study was approved by the Ethics Committee of the Guangdong Provincial Hospital of Chinese Medicine (No. 2019074). Informed written consent was obtained from all the family members. All the animals are treated in accordance with the ethical guidelines of the Guangdong Provincial Hospital of Chinese Medicine (Guangzhou, China).
1. Study subjects
NOTE: The proband III-3 sought medical advice in the Department of Cardiovascular Surgery of the Guangdong Provincial Hospital of Chinese Medicine in July 2019.
- Obtain detailed family medical history of the proband. Notify and call all the family members of the proband. All the family members have undergone meticulous physical examinations.
- Perform a systematic review of all clinical data, including medical records, ECGs, echocardiograms, and cardiac catheterization reports.
- Reconfirm cardiac phenotypes in all the patients during intervention or surgery.
- Select a total of 174 population-based healthy controls from the local database. Collect approximately 4.0 mL of peripheral venous blood from each patient.
2. DNA extraction
NOTE: DNA is extracted with a commercial blood kit according to the manufacturer's instructions.
- Lyse blood cells with an anionic detergent in the presence of a DNA stabilizer following the manufacturer's protocol.
- Add 10 µL of RNase A solution to the cell lysate and mix by inverting the tube 25 times. Incubate at 37 °C for 15 min to 1 h.
- Remove proteins by salt precipitation. Use protein precipitation solution (0.25 mg of bovine serum albumin dissolved in 25 mL of distilled water) and 100% isopropanol to precipitate proteins. Then, use 200 µL of 70% ethanol and perform centrifugation at 12,000 x g/min at 4 °C for 15 min to wash the DNA pellet.
- Recover the genomic DNA by precipitation with 500 µL of 70% ethanol. Centrifuge at 12,000 x g for 30 s. Aspirate and then dissolve the pellet in hydration solution (1 mM EDTA, 10 mM Tris·Cl, pH 7.5).
- Use a spectrophotometer (e.g., NanoDrop 2000) to determine purity. Purified DNA typically has an A260/A280 ratio between 1.7 and 1.9 and is up to 200 kb in size.
- Store the DNA for a long term at 2-8, -20, or -80 °C.
3. Whole exome sequencing and variant analysis
NOTE: To systematically search for disease-causing gene mutations, exome sequencing in affected individuals (II-5, II-7, III-3, III-7, III-8, III-9, and IV-3) and unaffected individuals (III-2, III-5, IV-4) was performed.
- Use exome probe according to the manufacturer's instructions to perform the exome capture.
- Apply the qualified libraries to 2 × 150 bp paired-end sequencing on the HiSeq X-ten platform. Please see Supplemental File 1.
- Align FASTQ files to the human reference genome (hg19/GRCh37) with BWA v0.7.1318,19. Sort the aligned files (sam/bam format files) with samtools, and then flag duplicates using Picard. Please see Supplemental File 1.
- Use GATK, realign reads locally and recalibrate base qualities. Please see Supplemental File 1.
- Generate mapping statistics that include coverage and depth from recalibrated files by BEDTools and in-house perl/python scripts.
- Genotype variants (SNVs and indels) from recalibrated BAM files using the multi-sample processing mode of the HaplotypeCaller tool from GATK.
- Use VQSR (variant quality score recalibration) to reduce false positives of variant calling.
- Annotate SNVs and indels using ANNOVAR software21 against multiple databases, including the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org), the Exome Sequencing Project (ESP) (https://esp.gs.washington.edu), and 1,000 G (http://www.1000genomes.org). Interpret the pathogenicity of the sequence variants according to the ACMG guidelines.
4. Sanger sequencing
- Perform Sanger sequencing to confirm potential causative variants and determine variant segregation in the family using an ABI 3500 sequencer23.
- Design primer sequences for the variant in the MYH7 gene (NM_000257) as follows: 5'-TTCAAACACAGAGACCTGCAGG-3' and 5'- CGGACTTCTCTAGCGCCTCTT -3'.
- Confirm a variant, screen all the available family members to determine variant segregation within the family23.
5. Generation of C57BL/6N-MYH7em1(G823E) knockin mice
- Use the CRISPR/Cas9 system to generate C57BL/6N-Myh7em1(G823E) knockin mice. The mouse Myh7 gene (GenBank accession number: NM_001361607.1; Ensembl: ENSMUSG00000053093) is located on mouse chromosome 14 and has 41 exons. The ATG start codon in exon 4 and TAG stop codon in exon 41, and the G823E is located on exon 23. Select exon 23 as the target site.
- Design the sequence of gRNA targeting vector and donor oligo (with targeting sequence, flanked by 134 bp homologous sequences combined on both sides).
- Log in to the NCBI website and click on the BLAST online primer design function on the right.
- Click on the Primer-BLAST function at the bottom of the page to design primers.
- Paste the MYH7 NCBI reference sequence number NM_000257.4 into the PCR template box.
- Amplify the target fragment (MYH7 cDNA exon 23 and its adjacent exons) so design the upstream primer on exon 21 (2392-2528) and the downstream primer on exon 25 (3205-3350). Enter the exon number of the upstream and downstream primers into the right range box.
- Click on the Get Primers button at the bottom to automatically generate multiple pairs of primers.
- Input the MYH7 gene into ZFIN database, introduce the p.g823e (GGG to GAG) mutation site in the donor oligonucleotide and the silent mutation p.r819 (AGG to CGA) into exon 23. The silent mutation prevents the binding and re-cutting of the sequence by gRNA after homology-directed repair.
- Design gRNA according to the general sequence, the sequences at both ends are TAATACGACTCACTATA- and -GTTTTAGAGCTA. The middle of the sequence is the target site mentioned above.
- Co-inject Cas9 mRNA, gRNA generated by in vitro transcription and donor oligo into fertilized eggs for KI mouse production.
- Use the Cas9/gRNA target efficiency detection kit to transcribe the designed gRNA target into gRNA in vitro and detect the activity of the transcript (see the VK007 kit instructions for specific detection methods) according to manufacturer's protocol.
- Thaw the T7 ARCA mRNA kit components, mix, and pulse-spin in a microfuge to collect solutions to the bottoms of the tubes. Assemble the reaction at room temperature in the following order: 2x ARCA/NTP mix to 10 µL, 1 µg of template DNA, 2 µL of T7 RNA polymerase mix, and nuclease-free water to 20 µL.
- Mix thoroughly and pulse-spin in a microfuge. Incubate at 37 °C for 30 min.
- Remove the template DNA by adding 2 µL of DNase I, mix well, and incubate at 37 °C for 15 min to obtain mRNA.
- Perform intraperitoneal injection of serum gonadotropin and human chorionic gonadotropin into pre-prepared C57BL/6 female mice at about 4 weeks old with a 0.5 mL syringe at a dose of approximately 5 U per mouse with an interval between the two injections of 48 h.
- Eighteen hours after dosing, sacrifice C57BL/6 female mice and one 8-12-week-old C57BL/6 male mouse by cervical dislocation after hormone injection. Collect the eggs and sperm separately.
- The sperm is capacitated in the capacitation fluid. Take and drop the sperm at the edge of the fluid into the short-lived egg cells for in vitro fertilization for 3-4 h.
- Dilute the successfully transcribed gRNA and Cas9 mRNA in vitro with RNase-free water to 25 ng/µL and 50 ng/µL. Introduce into the cytoplasm of mouse fertilized eggs by microinjection. Transplant fertilized eggs in good condition into the enlarged oviduct of the female mice. Co-cage with ligated male mice.
- Genotype the pups by PCR. Use the following PCR conditions: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 60 °C for 35 s, and 72 °C for 35 s; 72 °C for 5 min. Follow with sequence analysis.
- Cut the tails of 4-month-old mice with scissors and place them into 1.5 mL EP tube. Add 180 µL of Buffer GL, 20 µL of Proteinase K, and 10 µL of RNase A per tail piece (2-5 mm) in a microcentrifuge tube. Be careful not to cut too much tail.
- Incubate the tube at 56 °C overnight.
- Spin in a microcentrifuge tube at 1,000 x g for 2 min to remove impurities.
- Add 200 µL of Buffer GB and 200 µL of absolute ethyl alcohol with sufficient mixing.
- Place the spin column in a collection tube. Apply the sample to the spin and centrifuge at 1,000 x g for 2 min. Discard the flow-through.
- Add 500 µL of Buffer WA to the spin column and centrifuge at 1,000 x g for 1 min. Discard the flow-through.
- Add 700 µL of Buffer WB to the spin column and centrifuge at 1,000 x g for 1 min. Discard the flow-through.
NOTE: Make sure the Buffer WB has been premixed with 100% ethanol. When adding Buffer WB, add to the tube wall to wash off the residual salt. - Repeat step 5.5.7.
- Place the spin column in a collection tube and centrifuge at 1,000 x g for 2 min.
- Place the spin column in a new 1.5 mL tube. Add 50-200 µL of sterilized water or elution buffer to the center of the column membrane and let the column stand for 5 min.
NOTE: Heating sterilized water or elution buffer up to 65 °C can increase the yield of elution. - To elute DNA, centrifuge the column at 1,000 x g for 2 min. To increase the yield of DNA, add the flow-through and/or 50-200 µL of sterilized water or elution buffer to the center of the spin column membrane and let the column stand for 5 min. Centrifuge at 1,000 x g for 2 min.
- Quantify the eluted genomic DNA by electrophoresis.
6. Evaluation of the cardiac morphology and function
NOTE: Apply M-mode echocardiography to assess heart morphology and function of C57BL/6N-Myh7em1(G823E) knockin mice.
- Put the knockin mice into a closed acrylic box connected to the anesthesia machine and adjust the three-way interface to allow isoflurane (concentration: 3%) to flow into the acrylic box. After the knockin mice cease to move autonomously, remove the knockin mice.
- Place the knockin mice flat on the life monitoring platform of the small animal anesthesia machine, and continuous inhalation anesthesia mixed with oxygen (flow: 0.8 L/min) and isoflurane (concentration: 3%). Apply eye lubricant to the anesthetized mice to prevent drying of the cornea.
- Fix the knockin mice horizontally on the platform. Tilt the platform 30° caudal to the knockin mouse. Remove hair on the anterior chest wall of the knocking mouse using a depilatory cream.
- Position the probe vertically with the bump facing the animal's head. Then, rotate the probe counterclockwise, approximately 45°.
- In the parasternal long-axis view, rotate the probe 90° clockwise to observe the parasternal short-axis view. After rotating the probe 90°, adjust the y-axis displacement to get the correct slice.
- Observe the long-axis image of the heart (Figure 1C), and then select M-mode measurement data.
- Acquire ultrasound data from parasternal long-axis views of mice (Figure 1). Heart rate (HR), left ventricular ejection fraction (LVEF), cardiac output (CO), Left ventricular end-diastolic dimension (LVDd), LVDs left ventricular end-systolic dimension (LVSd), interventricular septum (IVS), and left ventricular posterior wall (LVPW) are measured.
- After the measurement, provide the mice with oxygen, and place them back into their respective cages when they regain autonomous activity.
Representative Results
Clinical profile of the families
The family pedigrees of HCM were obtained and are shown in Figure 2. All the documented family members were diagnosed with HCM at enrollment.
In the family (Figure 2A), the proband was patient III-7, who was diagnosed with HCM and left ventricular outflow tract obstruction (LVOTO) at 46 years old and underwent cardiac surgery. Patient III-3 had minor HCM that did not require surgical treatment. Patient IV-3 also had minor HCM, which was similar to his father, Patient III-3. Patient II-5 had HCM and underwent surgery to repair the defect at the age of 51 due to LVOTO. Patient I-1 and patient II-2 died due to cardiac accidents at 57 and 46 years old, respectively. Patient I-1's medical history was unavailable. Patient II-7 and patient III-9 presented with a shortness of breath and were diagnosed with HCM.
Exome sequence analysis and segregation of variants
In this family, exome sequencing of the five individuals generated a mean of a total of 19,978,731 pairs of sequenced reads with an average read length of 125 bp. In total, 98.72% of sequenced reads passed the quality assessment and were mapped to 98.66% of the human reference genome. Even after filtering, more than 42 variants (including single nucleotide substitutions and indels) were shared by these four patients. Of these, 27 were missense SNVs, 15 were predicted to alter splicing. Finally, according to ACMG rating guidelines, a heterozygous c.G2468A; p.G823E variant of MYH7 (NM_0002571) was observed in proband III-7 as well as the other three patients.
Identification of a pathogenic mutation
Sanger sequencing confirmed the same MYH7 p.G823E variant in all patients but not in the healthy individuals in the families and 174 controls (Figure 2B). The heterozygous MYH7 p.G823E completely co-segregated in this family. In this family, patients IV-3 who harbored this variant inherited it from patient III-3. Patients III-7 and III-8 carried the same variant, which was inherited from their father. The information for patient III-9 was unavailable.
This variant, previously described in HCM by a sporadic patient24, has not been reported in the 1000 G, ESP6500, ExAC, HGMD, ClinVar databases or control subjects. The glycine residue at codon 823 in the neck domain region of MYH7 is highly conserved in all the available vertebrate myosin sequences (Figure 2C). MYH7 is a known HCM-causative gene and plays an important role in cardiac development or structure/function. Based on ACMG standards and guidelines, the MYH7 p.G823E variant was predicted to be a pathogenic variant (PVS1 + PS3 + PS4 + PM2). Taken together, these results support that this variant is detrimental and contributes to the pathogenesis of HCM in these families.
C57BL/6N-Myh7em1(G823E) knockin mice developed severe cardiac hypertrophy
To further verify the pathogenesis of MYH7 p.G823E, we generate C57BL/6N-Myh7em1(G823E) knockin mice. C57BL/6N-MYH7em1(G823E) knockin mice developed an age-dependent cardiac hypertrophy after birth (Figure 2A,B). Echocardiography revealed that IVS and LVPW developed with increased HR in C57BL/6N-Myh7em1(G823E) knockin mice (Table 1). These results were likewise validated by histological analysis (Figure 3). There were no obvious differences in LVDd, LVDs, EF, or CO between wild-type and heterozygous mice (Table 1).
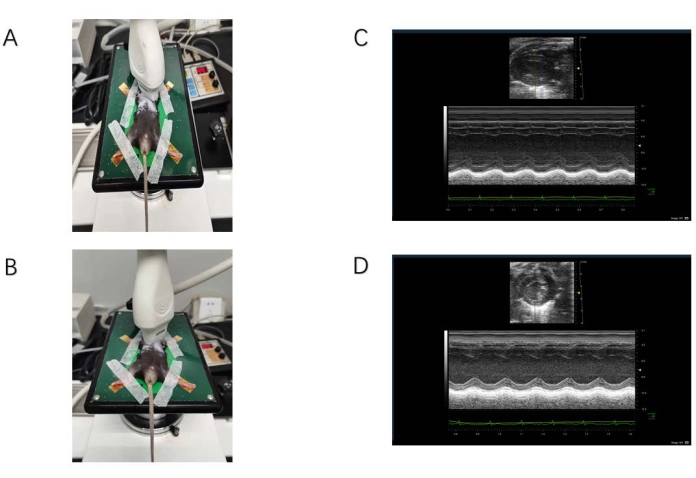
Figure 1: Ultrasound data acquisition. (A) The probe is located on the long axis of the sternum. (B) The probe is located on the short axis of the sternum. (C) M-mode ultrasound image of the long-axis view of the sternum. The yellow arrow indicates the location of the interventricular septum. The red arrow indicates the floating valve. (D) M-mode ultrasound image of the short-axis view of the sternum. The yellow arrow indicates the location of the anterior papillary muscle, and the bulge below is the posterior papillary muscle. Please click here to view a larger version of this figure.
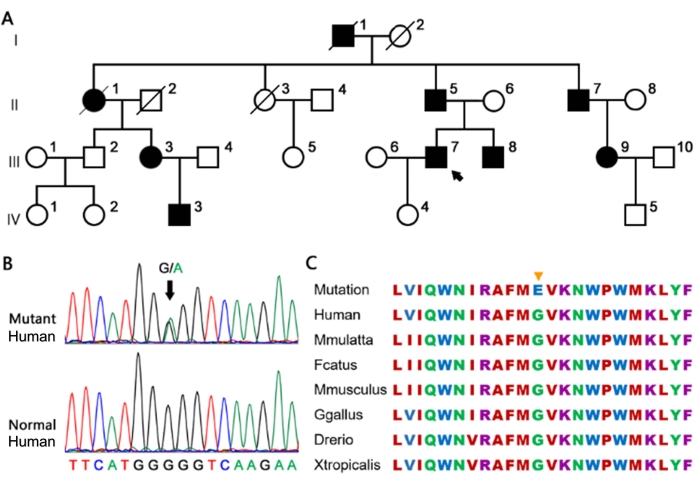
Figure 2: The large family carrying the heterozygous MYH7 G823E variant. (A) The proband is marked by an arrow. Full and open circles and squares indicate affected and normal individuals, respectively. (B) The G823E variant in MYH7 confirmed by Sanger sequencing. (C) Conservation of the MYH7 G823E site in different species. The yellow arrow represents the site of G823E in the amino acid sequence of different species, which is highly conserved. Please click here to view a larger version of this figure.
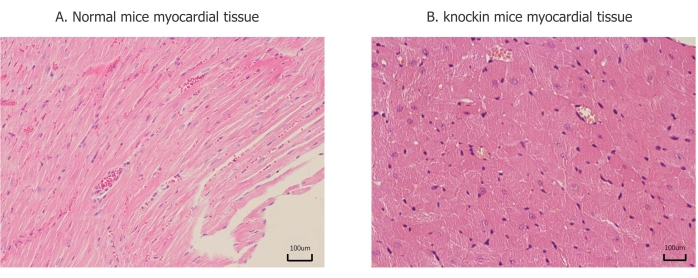
Figure 3: Pathological changes of myocardial tissue. (A) Myocardial fibers are arranged in an orderly manner, and there is no significant difference between each part. (B) Ventricular muscle fiber hypertrophy, disordered arrangement, and lesions are mainly concentrated in the posterior wall of the left ventricle and interventricular septum. Please click here to view a larger version of this figure.
| C57BL/6N-Myh7em1(G823E) knockin mice group | Control group | ||
| (n=4) | (n=6) | ||
| HR (/min) | 451.25 ± 25.786 | 413.83 ± 12.77 | P = 0.015 |
| LVDd (mm) | 4.12 ± 0.33 | 3.95 ± 0.20 | P = 0.330 |
| LVDs (mm) | 2.95 ± 0.44 | 2.85 ± 0.20 | P = 0.626 |
| EF(%) | 55.02 ± 9.52 | 54.31 ± 5.11 | P = 0.881 |
| CO (mL) | 18.46 ± 3.05 | 15.30 ± 2.39 | P = 0.102 |
| IVS (mm) | 1.13 ± 0.20 | 0.67 ± 0.07 | P = 0.001 |
| LVPW (mm) | 1.40 ± 0.60 | 0.70 ± 0.06 | P = 0.000 |
Table 1: Morphology and function analysis for p.G823E and wild type. Data were analyzed using SPSS. Continuous variables are expressed as mean ± standard deviation (SD). The Student's t or Wilcoxon rank sum tests for continuous variables. P-values below 0.05 were considered statistically significant.
Supplemental File 1. Please click here to download this File.
Discussion
In this study, we describe one Chinese Han families with HCM. Genetics analysis revealed that a heterozygous MYH6 mutation p.G823E co-segregates with the disease in family members with autosomal dominant inheritance. To validate the pathogenicity of G823E mutation and discuss the underlying mechanisms, we created a C57BL/6N mouse model with G823E at mouse Myh7 locus by CRISPR/Cas9-mediated genome engineering.
Phenotypic characteristics of C57BL/6N-Myh7em1(G823E) knockin mice were evaluated by echocardiography. Multiple trabeculation and a higher ratio of non-compacted to compacted myocardial layer were found in the C57BL/6N-Myh7em1(G823E) knockin mice compared to the controls. The transgenic mice also showed LVPW and IVS hypertrophy and an increase in HR. However, the function of the heart showed no significant change between the two groups. These results suggested that an increased HR may exhibit a compensatory effect on maintenance ventricle function during cardiac remodeling. Further studies are needed to observe and analyze the morphology and function of the heart.
Myosin-7 (MYH7) belongs to the family of myocardial sarcomeric structural proteins. MYH7 encoded by the MYH7 gene (14q11.2; OMIM: 160760), which is found in the first pathogenic gene associated with HCM in 1990. To date, more than 300 variants in the MYH7 gene have been associated with HCM25,26. However, the pathogenicity of great majority of variation remains elusive. Using this protocol, we created the model mice successfully. When planning and designing the model mouse, there are three main issues under consideration. First, the amino acid sequence of the mouse MYH7 protein is highly homologous with human. Second, MYH7 is highly expressed in ventricles, as well as MYH7 in humans. The glycine residue at codon 823 in the MYH7 is highly conserved in all available myosin sequences.
MYH7 contains a conserved head motor domain at the N-terminus, which binds actin and has actin-activated Mg·ATPase activity and a neck region, as well as a myosin tail, which contains a coiled-coil (CC) region at the C-terminus. The glycine residue at codon 823 in the neck domain region of MYH7 suggests that this variation may impact on lever arm rotation in contraction27,28,29,30.
WES mainly focuses on the exons of genes, which only account for 1.5%8 of the total genome DNA. However, most of the human genetic diseases that have been identified are related to exons31. This makes WES widely recognized and applied in clinical practice. There are certain ethical issues with the widespread application of WES. The study of this kind of human genetic disease often involves cross-border discussions. Although researchers try to remove personally identifiable information as much as possible, the huge information contained in DNA can still re-identify the research individuals, and even their families. Only standardized information exchange measures can ensure the rational application of WES. In the future, WES is of great significance for medical strategies for specific gene mutations and individualized diagnosis and treatment measures for multiple diseases.
In summary, we identified a MYH7 heterozygous variant, p.G823E, in a large Chinese Han family with HCM using WES. The C57BL/6N-Myh7em1(G823E) knockin mice were found to have thicker IVS and LVPW, and faster HR than wild-type mice. The p.G823E variant may impair lever arm rotation, suggesting that this mutation plays an important role in familial HCM. Our work broadens the information on the mutation spectrum of the MYH7 gene associated with HCM and provides better clarity into the appropriate diagnosis and genetic counseling of affected families.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Medical Research Fund project of Guangdong Province (A2022363) and the major project of the Guangdong Committee of Science and Technology, China (grant no.2022).
We would like to thank Qingjian Chen of the University of Maryland, College Park for the help during the preparation of this manuscript.
Materials
| 0.5×TBE | Shanghai Sangon | ||
| 2× Taq Master Mix (Dye Plus) | Nanjing Novizan Biotechnology Co., Ltd. | ||
| Agarose | Regu | ||
| Anesthesia machine for small animals | Reward Life Technology Co., Ltd. | R500 | |
| BEDTools | 2.16.1 | ||
| Cas9 in vitro digestion method to detect gRNA target efficiency kit | Viewsolid Biotechnology Co., Ltd. | VK007 | |
| DNA Marker | Thermo Fisher Scientific | ||
| DNA stabilizer | Shanghai Seebio Biotechnology Co., Ltd. | DNAstable LD | prevent DNA degradation |
| Electric paraffin microtome | Shenyang Hengsong Technology Co., Ltd. | HS-S7220-B | |
| GATK | v3.5 | ||
| Gentra Puregene blood kit | Santa Clara | ||
| Glass slide, coverslip | Jiangsu Invotech Biotechnology Co., Ltd. | ||
| Hematoxylin staining solution, Eosin staining solution | Shanghai Biyuntian Biotechnology Co., Ltd. | C0107-500ml, C0109 | |
| HiSeq X-ten platform | Illumina | perform sequencing on the captured libraries | |
| Injection of chorionic gonadotropin | Livzon Pharmaceutical Group Inc. | ||
| Injection of pregnant mare serum gonadotropin | Livzon Pharmaceutical Group Inc. | ||
| Isoflurane | Local suppliers | inhalation anesthesia | |
| Microinjection microscope | Nikon | ECLIPSE Ts2 | |
| NanoDrop | Thermo Fisher Scientific | 2000 | |
| Paraffin Embedding Machine | Shenyang Hengsong Technology Co., Ltd. | HS-B7126-B | |
| Picard | (2.2.4) 20 | ||
| Proteinase K | Merck KGaA | ||
| samtools | 1.3 | ||
| Sequencer | Applied Biosystems | ABI 3500 | |
| Stereomicroscope | Nikon | SMZ745T | |
| SureSelect Human All Exon V6 | Agilent Technology Co., Ltd. | exome probe | |
| T7 ARCA mRNA Kit | New England BioLabs, Inc. | NEB-E2065S | |
| Temperature box | BINDER GmbH | KBF-S Solid.Line | |
| Trizma Hydrochloride Solution | Sigma, Merck KGaA | No. T2663 | |
| Veterinary ultrasound system | Royal Philips | CX50 |
Referenzen
- Toepfer, C. N., et al. Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation. 141 (10), 828-842 (2020).
- Writing Committee Members et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: A report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. The Journal of Thoracic and Cardiovascular Surgery. 162 (1), 23-106 (2021).
- Elliott, P., McKenna, W. J. Hypertrophic cardiomyopathy. Lancet. 363 (9424), 1881-1891 (2004).
- Maron, B. J., Maron, M. S. Hypertrophic cardiomyopathy. Lancet. 381 (9862), 242-255 (2013).
- Inoue, T., Orgel, L. E. A nonenzymatic RNA polymerase model. Science. 219 (4586), 859-862 (1983).
- Sanger, F., Nicklen, S., Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 74 (12), 5463-5467 (1977).
- Sachidanandam, R., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 409 (6822), 928-933 (2001).
- Ng, S. B., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 461 (7261), 272-276 (2009).
- Wong, K. M., Hudson, T. J., McPherson, J. D. Unraveling the genetics of cancer: genome sequencing and beyond. Annual Review of Genomics and Human Genetics. 12, 407-430 (2011).
- Mattivi, C. L., et al. Clinical utility of a phenotype-enhanced MYH7-specific variant classification framework in hypertrophic cardiomyopathy genetic testing. Circulation. Genomic and Precision Medicine. 13 (5), 453-459 (2020).
- Jiang, J., Wakimoto, H., Seidman, J. G., Seidman, C. E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 342 (6154), 111-114 (2013).
- Hayashi, T., et al. Genetic background of Japanese patients with pediatric hypertrophic and restrictive cardiomyopathy. Journal of Human Genetics. 63 (9), 989-996 (2018).
- Gil, W. S., Ávila Vidal, L. A., Vásquez Salguero, M. A., Cajiao, M. B., Peña, C. V. Genetic variant affecting the myosin light chain 2 related to familial hypertrophic cardiomyopathy. Intractable & Rare Diseases Research. 9 (4), 229-232 (2020).
- Berge, K. E., Leren, T. P. Genetics of hypertrophic cardiomyopathy in Norway. Clinical Genetics. 86 (4), 355-360 (2014).
- McNamara, J. W., Schuckman, M., Becker, R. C., Sadayappan, S. A novel homozygous intronic variant in TNNT2 associates with feline cardiomyopathy. Frontiers in Physiology. 11, 608473 (2020).
- Wang, W., et al. Comparative transcriptome analysis of atrial septal defect identifies dysregulated genes during heart septum morphogenesis. Gene. 575, 303-312 (2016).
- Andersen, P. S., et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Human Mutations. 30 (3), 363-370 (2009).
- Nakashima, Y., et al. Lifelong clinical impact of the presence of sarcomere gene mutation in Japanese patients with hypertrophic cardiomyopathy. Circulation Journal. 84 (10), 1846-1853 (2020).
- Hu, L. R., Kontrogianni-Konstantopoulos, A. Proteomic analysis of myocardia containing the Obscurin R4344Q mutation linked to hypertrophic cardiomyopathy. Frontiers in Physiology. 11, 478 (2020).
- Girolami, F., et al. Novel alpha-actinin 2 variant associated with familial hypertrophic cardiomyopathy and juvenile atrial arrhythmias: a massively parallel sequencing study. Circulation. Cardiovascular Genetics. 7 (6), 741-750 (2014).
- Salazar-Mendiguchia, J., et al. The p.(Cys150Tyr) variant in CSRP3 is associated with late-onset hypertrophic cardiomyopathy in heterozygous individuals. European Journal of Medical Genetics. 63 (12), 104079 (2020).
- Teekakirikul, P., Zhu, W., Huang, H. C., Fung, E. Hypertrophic cardiomyopathy: An overview of genetics and management. Biomolecules. 9 (12), 878 (2019).
- Crossley, B. M., et al. Guidelines for Sanger sequencing and molecular assay monitoring. Journal of Veterinary Diagnostic Investigation. 32 (6), 767-775 (2020).
- Song, L., et al. Mutations profile in Chinese patients with hypertrophic cardiomyopathy. Clinica Chimica Acta. 351 (1-2), 209-216 (2005).
- Marian, A. J., Braunwald, E. Hypertrophic cardiomyopathy: Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circulation Research. 121 (7), 749-770 (2017).
- Cann, F., et al. Phenotype-driven molecular autopsy for sudden cardiac death. Clinical Genetics. 91 (1), 22-29 (2017).
- Lafreniere-Roula, M., et al. Family screening for hypertrophic cardiomyopathy: Is it time to change practice guidelines. European Heart Journal. 40 (45), 3672-3681 (2019).
- Winkelmann, D. A., Forgacs, E., Miller, M. T., Stock, A. M. Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity. Nature Communications. 6, 7974 (2015).
- García-Giustiniani, D., et al. Phenotype and prognostic correlations of the converter region mutations affecting the β myosin heavy chain. Heart (British Cardiac Society). 101 (13), 1047-1053 (2015).
- Moore, J. R., Leinwand, L., Warshaw, D. M. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circulation Research. 111 (3), 375-385 (2012).
- Majewski, J., Schwartzentruber, J., Lalonde, E., Montpetit, A., Jabado, N. What can exome sequencing do for you. Journal of Medical Genetics. 48 (9), 580-589 (2011).

