Microfluidic Co-Culture Models for Dissecting the Immune Response in in vitro Tumor Microenvironments
Summary
In the age of immunotherapy and single-cell genomic profiling, cancer biology requires novel in vitro and computational tools for investigating the tumor-immune interface in a proper spatiotemporal context. We describe protocols to exploit tumor-immune microfluidic co-cultures in 2D and 3D settings, compatible with dynamic, multiparametric monitoring of cellular functions.
Abstract
Complex disease models demand cutting-edge tools able to deliver physiologically and pathologically relevant, actionable insights, and unveil otherwise invisible processes. Advanced cell assays closely mimicking in vivo scenery are establishing themselves as novel ways to visualize and measure the bidirectional tumor-host interplay influencing the progression of cancer. Here we describe two versatile protocols to recreate highly controllable 2D and 3D co-cultures in microdevices, mimicking the complexity of the tumor microenvironment (TME), under natural and therapy-induced immunosurveillance. In section 1, an experimental setting is provided to monitor crosstalk between adherent tumor cells and floating immune populations, by bright field time-lapse microscopy. As an applicative scenario, we analyze the effects of anti-cancer treatments, such as the so-called immunogenic cancer cell death inducers on the recruitment and activation of immune cells. In section 2, 3D tumor-immune microenvironments are assembled in a competitive layout. Differential immune infiltration is monitored by fluorescence snapshots up to 72 h, to evaluate combination therapeutic strategies. In both settings, image processing steps are illustrated to extract a plethora of immune cell parameters (e.g., immune cell migration and interaction, response to therapeutic agents). These simple and powerful methods can be further tailored to simulate the complexity of the TME encompassing the heterogeneity and plasticity of cancer, stromal and immune cells subtypes, as well as their reciprocal interactions as drivers of cancer evolution. The compliance of these rapidly evolving technologies with live-cell high-content imaging can lead to the generation of large informative datasets, bringing forth new challenges. Indeed, the triangle ''co-cultures/microscopy/advanced data analysis" sets the path towards a precise problem parametrization that may assist tailor-made therapeutic protocols. We expect that future integration of cancer-immune on-a-chip with artificial intelligence for high-throughput processing will synergize a large step forward in leveraging the capabilities as predictive and preclinical tools for precision and personalized oncology.
Introduction
The evolution of different branches of medicine as experimental disciplines has depended on the ability to manipulate cell population and organ functions under controlled conditions1. Such ability has its roots in the availability of measurable models able to recapitulate processes happening in our body.
In the age of immunotherapy and single-cell genomic profiling2, cancer biology needs to take advantage of emerging in vitro and computational models for investigating the tumor-immune interface in a proper spatiotemporal context2,3.
The tumor microenvironment4 (TME) is a complex tissue where cancer cells continuously interact and dynamically co-evolve with the other cellular (immune, stromal, and endothelial cells) and non-cellular (the extracellular matrix, ECM) components. The dynamic nature of this complex landscape dictates whether immune cells play as friends or foes of malignant cells, thus strongly affecting both disease progression and response to therapy. Nowadays, great efforts from onco-immunologists, bioinformaticians, and systems biology experts are converging to address the clinical significance of cancer heterogeneity5,6, either in the space (i.e., in distinct tumoral regions) and time (i.e., at distinct tumor progression stages)5,6, and to characterize cancer and immune cell phenotype and function at a single-cell level. As an example of this synergy, advanced computer-vision techniques are now routinely used for spatial mapping of immune infiltrate in histological samples7,8.
On the front of experimental models, bridging animal studies and traditional in vitro methods, advances in microfluidics and co-culturing techniques give access to different classes of micro-engineered cellular models such as organoids, micro-physiological systems9,10,11 (MPS), and organs-on-chip12,13,14 (OOC). They share the common trait to zoom in the 'big picture' view of the cellular ecosystems and expanding the in vitro potential to control microenvironmental factors while exploiting high-content microscopy15 and image processing approaches.
Nowadays, state-of-the-art- MPS and OOC systems have begun to include immunological aspects , incorporating different subtypes of immune cells in existing tissues- and co-cultures, so to explore and measure a variety of processes like inflammatory diseases, wound healing, mucosal immunity, and response to toxins or daily food products16. TME-on-a-chip models10,11,12,13,14,15,16,17, also integrated with perfusable microvessels18,19,20,21, have been developed to investigate cell-type-dependent interactions, physical and chemical perturbations, and the cytotoxic activity of infiltrating lymphocytes22, as well as clinically relevant immunomodulatory agents23.
Here, we provide versatile protocols, spanning from loading cells in chips to image processing tools, to exploit advanced tumor-immune microfluidic co-cultures in 2D (section 1) and 3D (section 2) settings16, compatible with dynamic, multiparametric24 monitoring and visualization of cellular functions. This is achieved maintaining easiness of use and flexibility both in sample management and data analysis, taking advantage of Fiji freeware software and its toolboxes25,26.
The microfluidic device, described in section 1, is designed to perform 2D co-cultures of adherent cancer and floating immune cells. This platform was validated for the in vitro measurement of immune cell behavior in the presence of genetic mutations27 and/or immunodeficiencies28. Here, we illustrate steps for tracking immune cells in time-lapse bright-field images, by exploiting a semi-automatic method based on Trackmate (a plugin implemented in Fiji software). This procedure enables the extraction of kinematic descriptors of immune migration 29 and response (i.e., interaction times) to target cancer cells, treated or not with immunogenic cell death inducers27.
Importantly these parameters, extracted from time-series images, can be processed with advanced mathematical machinery. As an example of the potentiality of this approach, our groups recently published an analysis based on mathematical methods from stochastic processes and statistical mechanics to model cellular network properties and provide a parametrized description of immune cell behavior (i.e., biased or uncorrelated random walk, highly or not coordinated motion30,31).
The 3D setting, provided in the second section, is based on a co-culture protocol to recreate more complex immunocompetent TMEs embedded in two gel regions with different combinations of cell types and drugs in a competitive fashion. Here, image processing steps are described to measure, at different timepoints, the infiltration of stained immune cells in human A375M melanoma cells cultivated within Matrigel, to evaluate antitumor agent combinations32. A375M line, an A375P derived cell line characterized by a highly metastatic phenotype was chosen to evaluate their metastatic capability in the presence of immune cells32.
The described models can be fully compliant with different cell sources (murine and human immortalized or primary cell lines, organoids, xenografts, among the others). In recent studies of our lab, by combining high-content video microscopy with image analysis, the competitive 3D layout was applied to investigate: i) an anti-tumoral (antibody-dependent cell-mediated cytotoxicity, ADCC) immune response and dissect the role of fibroblasts in resistance to trastuzumab therapy in HER2+ breast cancer on-chip models33; ii) the action of myeloid cells (i.e., cancer-associated macrophages) in mechanisms of tumor evasion and recruitment of T cells34; iii) the efficacy of immunotherapeutic regimes, specifically based on Interferon-α-conditioned dendritic cells (IFN-DCs), cultivated with drug-treated colon cancer cells in collagen matrices, and to evaluate efficient motion and the succeeding phagocytosis events35; iv) the chemotactic migration of bone marrow-derived eosinophils towards IL-33 treated or untreated melanoma cells36.
These advanced models could serve as observation windows for understanding the role of immune contexture in cancer metastasis and resistance mechanisms, but efforts are required to translate findings into the clinics, closing the gap with basic research37.
As an emerging scenario, harnessing the power of automated high-content microscopy coupled to the use of more physiologically-relevant microsystems is opening novel potential challenges for the handling, processing, and interpretation of hundreds, and even thousands, of Gigabytes of multiparametric data, which can be generated from a single experimental campaign. This implies a direct link of OOC experiments with artificial intelligence38,39,40,41,42 (AI)-based algorithms both for advanced automated analysis, and generation of features which can feed in turn in silico models of cancer-immune interplay43, with exciting new applications at the horizon, such as the development of predictive drug screening assays44.
An ever-expanding flow of efforts is focused on the design of disease models jointly with the optimization of strategies to implement the large-scale perturbation screens with single-cell multi-omics readouts. This will undoubtedly help the development and, hopefully, the clinical implementation, accompanied by an appropriate degree of method standardization, of a systematic onco-immunology-on-a-chip approach to gain novel insights into immune disorders and cancer dissemination mechanisms.
Protocol
1. Chip design for adherent and floating cells 2D co-cultures
NOTE: The 2D co-culture layout (Figure 1A-C) is characterized by three chambers (100 µm high) interconnected by two sets of microchannel arrays (500 x 12 x 10 µm3, L×W×H). The intermediate chamber forms two closed dead-end compartments which block floating immune cells overflowing into the tumor site during the loading step 2.5. This device type is useful for real-time bidimensional measurements of single-cell (either adherent or floating) motility, and of cell-cell interactions16,27,28,30,31. A typical cell migration study (conducted from several hours to several days) combines live-cell microscopy with image-processing algorithms45, in order to translate the acquired image sequences into numerical features25. Based on the migratory patterns, several biophysical indicators can be estimated, such as the displacement and velocity of cells, as well as the duration of immune cell and target cell interactions24.
- Preparation of cancer and PBMC cells
- Cancer cell culture
NOTE: MDA-MB-231 triple-negative [estrogen receptor (ER)-, progesterone receptor (PR)-, and human epidermal growth factor receptor 2 (HER2)-] human breast adenocarcinoma cells are routinely grown in a Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM L-glutamine, 100 IU mL−1 penicillin G sodium salt and 100 µg mL−1 streptomycin sulfate (growth medium), under standard culture conditions (37 °C and 5% CO2).- To optimize cell culture growth, plate MDA-MB-231 cells in 75 cm2 flasks at a density of 1 × 106 cells mL-1 in 12-to-15 mL of growth medium.
- When cells reach the 75-80% of confluence, discard the growth medium, wash cells with pre-warmed phosphate-buffered saline (PBS) to completely remove FBS, and then detach them with pre-warmed trypsin (1-to-2 min at 37 °C).
- Add growth medium to inactivate trypsin enzymatic activity and collect detached cells. Wash cells twice for 5 min at 1,100 x g at room temperature (RT).
- Count cells in a cell counting slide by means of Trypan Blue dye exclusion test and then reseed them either for maintenance culture (for no more than 6 passages from thawing) or for experimental procedures.
- For microfluidic experiments, seed 1 × 106 cells in 6-well plates in 3 mL of growth medium and either treat with 25 µM doxorubicin (DOXO) or an equal volume of DOXO solvent (PBS) as control.
- Four-to-6 hours after, wash DOXO-treated cells twice with pre-warmed PBS for 5 min at 1,100 x g at RT.
- Count DOXO-treated and PBS-treated control cells as above (see step 1.1.1.5) and set the co-culture with peripheral blood mononuclear cells (PBMCs) in microfluidic devices.
- PBMC isolation
- Collect venous whole blood (10 mL approximately) from healthy volunteers in heparinized vials and gently mix by inverting the tube 2-to-4 times47.
- Dilute blood 1:1 with PBS and layer over 10 mL of density gradient medium Lymphoprep in a 50 mL tube.
NOTE: Ensure to do the layering very gently and slowly to let blood and Lymphoprep form two distinct layers. - Centrifuge tubes for 30 minutes at 400 x g at 4 °C in a swing-out bucket without brakes. Four distinct layers will form: (i) plasma at the top, (ii) a white and cloudy layer containing PBMCs, (iii) Lymphoprep, and (iv) a pellet of erythrocytes and granulocytes.
- Aspirate gently PBMCs with a 2 mL pipette and immediately resuspend in warm growth medium (same used in step 1.1.1) and wash twice for 5 min at 1,100 x g at RT.
- Count pelleted PBMCs as above (see step 1.1.1.5) and either use for experimental procedures or freeze for long-term storage.
- Cancer cell culture
- Plating the cells in 2D chips
NOTE: PBMCs are not stained in this protocol. To characterize specific phenotypes on-chip, immune cell sub-populations can be isolated by immunomagnetic bead selection, stained with fluorescent cell trackers, re-mixed with the unlabeled remaining fraction, and thus confronted with target cancer cells, as reported in the on-chip experiments, described in Vacchelli et al.27, and in Racioppi et al.34.- Before starting co-culture experiments and to facilitate the addition of reagents, activate stored chips by an oxygen plasma treatment for a few seconds. Immediately fill reservoirs with deionized water or PBS to keep PDMS (polydimethylsiloxane) surfaces hydrophilic until plating steps.
NOTE: PDMS is intrinsically hydrophobic, which may result in difficulties in operating and in the entrapment of air bubbles in microchannels. See step 7 in the supplementary file providing details about oxygen plasma activation. - Sterilize under a UV cabinet for 20 min, wash 2-3 times with fresh PBS, and then incubate with culture media for 1 h. Keep chips in incubator until performing plating steps.
- Withdraw excess media from all six reservoirs. Take care to avoid sucking up media from the main culture chambers.
- Slowly apply 1 × 105 cancer cells resuspended in 10-20 µL of growth medium in the upper left-hand reservoir, and then in the lower well (Figure 1A, reservoirs 1 and 2). Wait 5 min to let cells adhere into the tumor chamber. Some cells will settle and attach in the reservoirs.
NOTE: Insert cellular suspension next to the channel openings. This procedure is applied to MDA-MB-231 cancer cells, other lines will require cell density optimization. To improve cancer cell attachment, a coating functionalization of surfaces (e.g., poly-L-lysine, fibronectin) can be performed. Please, refer to previously published protocols for coating steps16, 48,49,50. - On the right side gently pipet 1 × 106 PBMCs resuspended in 50 µL of growth medium into wells 3 and 4 (see Figure 1A, reservoirs 3 and 4).
NOTE: After flowing, PBMC will distribute into the intermediate chamber creating a "front", which represents the starting point of the experiment. - Fill all the six reservoirs with up to 100-150 µL of growth medium. Under a microscope check that cells have distributed correctly in the culture compartments as depicted in Figure 1D-E. Final volumes can vary with the size of the reservoirs. Adjust volumes to be equal in all wells.
- Place the chips back in the incubator for approximately 1 h to stabilize the system prior to time-lapse recording. Add fresh medium every 3 days, as it may be subjected to evaporation losses.
NOTE: The system is compatible with both live/dead-cell analysis and dynamic multiplex cytokine secretion profiling from conditioned media. For chemokine analyses, up to 200-250 µL aliquots of supernatants may be accessible by collecting media from the two reservoirs of each compartment. Classical ELISA and Luminex cytokine profiling assays require about 50 µL of supernatants. Please see 51, 52 examples of studies of other labs performing cytokine profiling on OOC models.
- Before starting co-culture experiments and to facilitate the addition of reagents, activate stored chips by an oxygen plasma treatment for a few seconds. Immediately fill reservoirs with deionized water or PBS to keep PDMS (polydimethylsiloxane) surfaces hydrophilic until plating steps.
- Time-lapse acquisition of unlabeled cancer and immune cells
NOTE: Typically, 3 chips are arranged on a single microscope slide (see Figure 1A for 2D chip and Figure 4B for 3D chip). Using stage holders allocating 4 slides, co-cultures can be suitable to be monitored by automated high-content microscopy to analyze large batches of experimental conditions. Chips can be easily mounted on slides with thickness equal to 1 mm or 170 microns (plastic or glass coverslips, 6-well optical bottom multi-wells) for high-resolution confocal imaging.- Record bright-field image series of unlabelled cells by means of a video microscopy setup equipped with an incubation system.
NOTE: Here time-series datasets (time window: 48 h, frame rate: 2 min) were acquired with a fluorescence microscope, equipped with a 4x objective and CMOS 1.3M pixels, optimized to fit into a standard cell culture incubator. - Warm up the microscope for at least 2 h to equilibrate to 37 °C and 5% CO2 before starting acquisition.
- Select the window of observation by centering the microchannel array between the tumor and the central compartment. This allows to visualize the dynamics of immune infiltration and the interactions within the region in which cancer cells are seeded.
- Adjust the illumination intensity and focus of cancer and immune cells.
- For launching time-lapse acquisition, optimize frame rate and time duration according to experiment and cell type under study.
NOTE: Imaging conditions must be optimized to avoid excessive photo-exposure while maintaining a good signal-to-noise ratio (SNR). As immune cells are very motile, the acquisition frame rate needs to be sufficiently high to follow the dynamic process of interest and enable easy tracking53. A compromise should be reached between the tracking algorithm, the compatibility with the size of the resulting dataset, and the viability, density, and motility of observed cells. - At the end of the time-lapse, use the function Import Image Sequence and Save as of the ImageJ software to convert the frame dataset in a 25 fps uncompressed video file.
NOTE: The generated video file is now ready for cell tracking analysis. Here, RGB (1280×1024 pixels) images were collected with a spatial resolution of 1.33 µm/pixel. A 24 h duration movie (3.5 GB stack) of a single field of view (FOV) consists of 720 frames for each condition.
- Record bright-field image series of unlabelled cells by means of a video microscopy setup equipped with an incubation system.
- Data analysis: Semi-automatic extraction of unlabeled immune tracks by Trackmate
NOTE: Here, immune motility analysis in 2D unlabeled time-lapse images is carried on using TrackMate54, an open-source toolbox available in the Fiji/ImageJ software bundle (https://imagej.nih.gov/ij/). Several algorithms are provided to perform automated single-particle tracking55 (SPT) of spot-like structures. They have been applied efficiently to fluorescent images, where objects are bright over a dark background with high SNR (i.e., sub-resolution fluorescent spots, labeled traffic vesicles, nuclei)1,25,56,57,58. SPT is mainly based on two sequential steps. First, objects are localized with identified positions in multiple frames (segmentation), as schematized in Figure 2. In the second stage (particle linking), detected spots are linked over consecutive frames to estimate motion and reconstruct their trajectories, in the shape of a track (Figure 3). Numerical features can be computed from each extracted X, Y, Z coordinates array over time. Extended documentation is reported in 54 as well as online (http://imagej.net/TrackMate), following the Getting started with TrackMate tutorial. The accuracy of the process can be inspected immediately, handling an intuitive graphical user interface (wizard-like GUI) that enables users, at every step, to readjust settings. The following part briefly depicts how to use Trackmate for image processing and quantification steps, applied to visible light images:- Drag and drop the full-time video/image stack on the Fiji toolbar.
- Calibration stack setup (Figure 2A).
- Check the dimensionality and assign the image properties by selecting Image> Properties. Fill Unit of length, Pixel dimensions and Frame interval boxes.
NOTE: To perform the calibration, use the known length of the microchannels (500 µm, Figure 1C) and divide by the corresponding measured length in pixels. For 2D time-series, make sure to swap Z/T field entering 1 as z-slice and the correct number of movie frames. If not accomplished, Trackmate quantitative outputs and parameters will be reported in pixel units and timeframes.
- Check the dimensionality and assign the image properties by selecting Image> Properties. Fill Unit of length, Pixel dimensions and Frame interval boxes.
- Pre-processing of images.
- To enhance the correct discrimination of immune cells from a noisy background, pre-process bright-field images to compensate artifacts. Ensure that datasets consist of 8-bit TIFF images (brightness range: 0-255).
NOTE: Uneven illumination, low SNR, and contamination by small debris particle in visible light images could compromise the success of a cell tracking process. Here, time-series datasets are pre-processed through background subtraction, brightness/contrast adjust function, and by local image subtraction of a Gaussian blur from original images. There are other different analysis toolkits available in ImageJ for processing and segmentation of phase-contrast or bright-field images, including Empirical Gradient Threshold (EGT)59.
- To enhance the correct discrimination of immune cells from a noisy background, pre-process bright-field images to compensate artifacts. Ensure that datasets consist of 8-bit TIFF images (brightness range: 0-255).
- First calibration panel (Figure 2A)
- With image stack selected, start Trackmate (Plugins>Tracking). Revise/confirm the dimensionality and temporal window of data (i.e., pixel-width and frame interval).
NOTE: TrackMate automatically reads in the image properties box to give the final tracking results in calibrated physical units (i.e., µm and minutes). - Define a region of interest to compute the extraction of immune tracks, by manually inserting values or by drawing a closed area over the active image, and then pressing the Refresh source button. To extract global immune migration paths, select rectangular regions respectively on the right side of microchannels (central chamber, Figure 1E) and on the left (tumor chamber, Figure 1D). To analyze interactions between cancer and immune hotspots, draw circular sub-regions by using ROI tools (go to Edit → Selection → Specify).
NOTE: When running for the first time this toolbox on a new biological application, spend the necessary time to optimize settings for reconstructing the tracks.
NOTE: Perform manual tracking of the cell trajectories (about 50-100 cells) to find empirically the right configuration and next to validate as a benchmark the reliability of automatic extraction of movements. Additionally, work initially on a smaller area to easily check the accuracy of the chosen parameters.
- With image stack selected, start Trackmate (Plugins>Tracking). Revise/confirm the dimensionality and temporal window of data (i.e., pixel-width and frame interval).
- Immune spots detection step (Figure 2B)
- Select the default Laplacian of Gaussian (LoG) detector. The LoG detector works to find bright, blob-like, roundish objects and applying a Laplacian of Gaussian filter on the image tuned for intermediate spot sizes (5-to-20 pixels in diameter).
- In Estimated Blob Diameter (here, 10-13 µm) enter a value slightly bigger than the expected spot size. Increase Threshold (here 1-3 µm) value until extra spurious background spots are reduced possibly without removing the object features. Detections below Threshold value (based on a quality metrics) will be discarded from subsequent analysis. Check the box for the median filter and sub-pixel localization to improve the quality of spot detection.
- Use the Vorschau button to view and quickly inspect identified immune cells overlaid on the images by magenta-colored circles.
NOTE: Mistakes during the detection will have a considerable impact on the linking process. Other unwanted detections can be corrected in the subsequent menus by user-defined filters (i.e., by spot intensity, size, or position).
- Once satisfied with selections, hit Next.
NOTE: These settings may vary depending on experimental setup and acquisition imaging modalities (e.g., FOV, objective magnification, bright-field or fluorescent images), cell type (adherent or floating cells), from slow or fast motility, kind of cellular behavior (interacting or not) and low/medium/high density in the observation area. - Proceed and skip Initial Thresholding menu. Select the Hyperstack Displayer window.
- Set filters on spots panel (Figure 2C).
- Select: Uniform Color. Filters, as shown in Figure 2C, can be added to retain labeled spots with feature values, displayed in a histogram, above or below a reversible threshold.
- Tracker selection stage (Figure 3A). Choose the Simple LAP tracker, as particle linking algorithm asking for three fields to fill (in this case, "Linking max distance": 30-50 µm, "Gap-closing max distance": 25-50 µm, "Gap-closing max frame gap": 4-6). This detector manages gap-closing events, with cost linking calculation solely based on their respective distance.
NOTE: The maximal allowed linking distance limits the spatial search range for candidate matching spots, corresponding to the maximally allowed displacement traveled by between two subsequent frames (Figure 3D).- Provide larger values of maximal displacement when the fragmentation of tracks of highly motile particles is noticed.
NOTE: Two links will not be connected if the frame-to-frame movement is larger than the given maximal distance value. If segments bridge badly two different cells, decrease the value of maximal displacement. - Try to reconnect missing spots, varying the values of "the max distance for gap closing" and "the maximal frame gap".
NOTE: These parameters deal with gap-closing events in non-adjacent frames. Spot disappearance may occur for some frames (i.e., out of focus particles, cells leaving out and in the FOV, segmentation failures in a noisy image).
NOTE: To handle splitting or merging events, opt for LAP linker as detector which introduces linking cost matrix penalties.
- Provide larger values of maximal displacement when the fragmentation of tracks of highly motile particles is noticed.
- Click Next to run the tracking computation. Press Next.
- Filtering tracks panel (Figure 3B). Change the color of immune paths selecting, from the drop-down menu, "Track ID" or other track features. At this point, choose optionally to set interactive filters functional, to improve the quality of the outcome and revisit the procedure.
NOTE: Spurious spots arise from noise in the image and loss of feature quality. This will generate short segments while cells of interest can be tracked over many frames.- To remove short paths, try to filter out, based on the number of spots they contain. Additionally, sort tracks using a combination of options such as Track displacement, Track duration or Minimal/Mean/MaximalVelocity to exclude false or unwanted tracks (with fewer frames respect to overall duration of time-lapse or involving dirty or not moving particles) from further post-processing.
NOTE: The choice of filters can vary depending on the specific application and biological system.
- To remove short paths, try to filter out, based on the number of spots they contain. Additionally, sort tracks using a combination of options such as Track displacement, Track duration or Minimal/Mean/MaximalVelocity to exclude false or unwanted tracks (with fewer frames respect to overall duration of time-lapse or involving dirty or not moving particles) from further post-processing.
- Examine all tracks in the Display Options interface, scroll through time, and verify how accurate tracks match cell migration paths. The drop-down menu provides color codes for spots and paths for easy visualization and filtering by several modalities (e.g., kinetic parameters, intensity, temporal or spatial position).
NOTE: For tracking high-density cultures, or high-motile cells, increase the acquisition frame rate minimizing cell displacement traveled in consecutive time intervals. - Manual correction of segmentation and linking mistakes (Figure 3E).
- To enhance further the quality of results, edit manually spots (debris particles, stationary cells) and remove erroneous tracks deriving from detected tumor boundaries when analyzing tumor-immune interaction ROIs.
- First, select the TrackMate tool in the ImageJ toolbar. For eliminating an existing spot throughout the whole stack, press shift and create by mouse cursor a ROI mask over the target spot (edited in a green circle), and then hit the DEL key.
- For adding a new spot (in case of missing tracks due to spots disappearance) press the A key, laying the mouse at the pointed location. Repeat the tracks-linking computation process after this step.
- When satisfied, select Analysis in the Display Options panel to generate three text files (Figure 3C and 3F). The table in "Spots in tracks statistics" provides the spatiotemporal coordinates of immune spots (X-Y-Z positions of the cells labeled with the associated frame and track number). "Links in tracks statistics" and "Track statistics" contain information relative to the tracks: track durations, number of detected gaps or spots, track initial and stop-frame, etc. Save and export for each dataset.
NOTE: When clicking on a row within the result windows, the respective spot, link, or track is activated within the time-lapse video for visual inspection. Repeat the filtering steps to select/remove tracks. All future exported data will be updated. TIP: Track initial and stop-frame and track duration values can be exploited to calculate times of contact between cancer and immune cells when processing ROIs of interaction. - Press the Save button to generate a resulting XML file containing all the parameter values, the path to images, and spot positions in time. The ''Load TrackMate file'' command (Plugins> Tracking) restores the whole process session for each movie file individually.
- Move to the last panel of the GUI called Select an action. In the list, use Captureoverlay > Execute function to produce a video with tracks overlaid. TIP: "Plot N-spots vs time" option may be used to compute the spatial density of immune cells in a ROI (Figure 6B, right panel).
- Post-processing analysis and migration statistics
- Analyze the raw positional data directly in Trackmate or export data to calculate comprehensive kinetic parameters29 (i.e., total trajectory length, Euclidean distance, confinement ratio, mean-squared displacement56, average or instantaneous track velocity, arrest coefficient, distribution of angles of migration, Forward migration index, mean straight-line speed) to classify immune cell migration behavior (e.g., directed or diffusive motion30,31) and response to target cancer cells (e.g., treated vs control).
NOTE: Additional useful plugins such as The Chemotaxis and Migration Tool (http://ibidi.com/software/chemotaxis_and_migration_tool/) provides various graphs (e.g., Rose or sector plots, such as depicted in Figure 6) and statistical tests for advanced analysis and visualization of experimental migration and chemotaxis data. Combining cell tracking and cell segmentation algorithms24,25,45 may enable measurements of morphological metrics at the single-cell level (i.e., cell surface area, the major and minor axis length, and the cell aspect ratio).
- Analyze the raw positional data directly in Trackmate or export data to calculate comprehensive kinetic parameters29 (i.e., total trajectory length, Euclidean distance, confinement ratio, mean-squared displacement56, average or instantaneous track velocity, arrest coefficient, distribution of angles of migration, Forward migration index, mean straight-line speed) to classify immune cell migration behavior (e.g., directed or diffusive motion30,31) and response to target cancer cells (e.g., treated vs control).
2. 3D immuno-competent cancer on-chip model in a competitive assay
NOTE: The 3D chip design, depicted in Figure 4, consists of 5 major compartments: a central one for the floating immune cells intake, two side regions for embedding tumor cells in hydrogel matrices (150-250 µm high), and media perfusion chambers. Immune and tumor chambers are connected by two sets of narrow arrays of microchannels (200×12×10 µm3, L×W×H, Figure 4E). Regularly 100 µm-spaced trapezoidal isosceles micropillars (about 25-30 interfaces for each side gel region, Figure 4C) work as barriers to confine gel solution during injection exploiting the balance between surface tension and capillary forces60,61 and connect tumor regions to the two lateral additional media chambers in order to set a gel-liquid interface (Figure 5). The detailed features of the 3D competitive assay are shown in Figure 4. Preferential migration of immune cells towards the two hydrogel compartments hosting tumor cells that have undergone different treatments can be monitored and quantified. The particular competitive layout can be applied to investigate a plethora of different cancer biology phenotypes (e.g., drug-resistant vs aggressive, primary or metastatic, responders vs non-responders). Additionally, the gel embedded regions can be easily integrated with different cell populations to recreate more heterogenous TMEs, including stromal components (fibroblasts, endothelial cells)23 or to simulate specific immunosuppressive milieu34 (e.g., macrophages) for dissecting mechanisms of drug resistance and tumor evasion.
NOTE: Nuclear and active caspase staining, by using commercial kits for Live/dead assays (e.g., Thermo Fisher Scientific, Incucyte reagents), can be implemented to assess mitotic or apoptotic death events, as reported in Nguyen et al.33.
- Preparation of matrix solution with cells and Loading in the device
NOTE: In the following experimental setting, the two gel regions contain mixtures of human A375M melanoma cell lines, grown in matrix solution (e.g., Matrigel), exposed to therapeutic agents used as monotherapy or in combination. This setting allowed us to evaluate the efficacy of a combination of two drugs with respect to single ones in a competitive fashion and to quantify their ability to attract PBMCs.- Defrost a stock of matrix solution (e.g., Matrigel) by placing on ice into a 4 °C refrigerator one day before the experiment.
NOTE: Do not expose the product to multiple freeze-thaw cycles as it becomes "clumpy". Other synthetic or natural hydrogels protocols can be suitable to be used in this setting. Please refer to 33,34,35 for the preparation of cancer cells in collagen matrices. - Resuspend A375 human melanoma cells, stained with live-compatible PKH67 Green Fluorescent Cell Linker in matrix solution (2 mg mL-1). Where indicated, add 5-aza-2'-deoxycytidine (DAC; 2.5 µM), referred as DAC, and/or IFN-α2b ,referred as IFN, at the proper doses32.
NOTE: Match the Lot # on the matrix bottle spec sheet. Based on the concentration calculate the volume of medium needed to make up to 2 mg mL-1 Please adjust the optimal protein concentration and cancer cell suspension concentration accordingly to your application of interest. - Pipette up and down carefully to avoid the generation of bubbles. Keep the microcentrifuge tube on ice while mixing to prevent any unwanted polymerization.
- After sterilization, place the devices on ice (using an ice bucket and lid) to avoid matrix solution solidification during the whole procedure of cell loading.
- Slowly inject the two IFN and DAC/IFN Matrigel/tumor cell mixtures (2-4 µL) into the left and right gel port, respectively with 10-µL micropipette using cold tips (Figure 5A). Apply gentle pressure to push matrix solution from one side until reaches the opposite one.
NOTE: The volume of matrix solution was chosen to avoid overflowing into adjacent channels. Do not exert excessive pipetting pressure to prevent the solution from leaking into the media and central channels. If during loading gel path is blocked along the channel, try to insert the solution from the other inlet until the gel fronts meet. When removing the micropipette from the inlets, hold the plunger, otherwise the negative pressure will aspirate matrix solution. - Place the device in an incubator in upright position at 37 °C and 5% CO2 for 30 minutes to allow gelation of the matrix solution to take place (Figure 5B). Handle with care chips with embedded unpolymerized gel to prevent leaking out of the gel channel.
- In the meantime, resuspend PKH67-labelled PBMCs (1×106 cells) in 10 µL of complete DMEM (Dulbecco's modified Eagle's medium).
- After matrix gelation, fill media channels with (50-100 µL) the same aliquot of culture medium in all six reservoirs to prevent gel drying in the chips. Keep in incubator until immune cell suspension seeding.
NOTE: Check under a microscope the correct and homogeneous distribution of tumor cells in the gel and the integrity of the polymerized gel barriers. Partially or not uniform gelled regions or bubbles in the mixture lead to the premature flowing of PBMCs in gel media channels at the starting point of the experiment due to pressure initial fluctuations. - Aspirate media from the six wells and position the tip near the inlet of a media channel to inject gently with moderate pressure PBMC cell suspension. The loading temporal sequence is depicted in Figure 5C:
- PBMCs in 10 µL medium into the upper central well.
- 50-100 µL medium into each of four wells of lateral channels.
- 40-90 µL medium into the upper central well.
- 50-100 µL medium into the lower central well.
- Ensure under a microscope that the PBMCs distribution remains confined in the central chamber after the loading step. (Figure 7A).
NOTE: If it is not optimal, adjust the concentration if needed and repeat the seeding steps, using pristine chips. When calculating volumes for the planned experimental conditions, refer to an excess number of chips (15-20%) to take in account of potential errors and adjustments. Volumes and concentrations should be optimized according to the specific application. - Place assembled devices on a level surface in the incubator at 37 °C and 5% CO2 for subsequent fluorescence imaging acquisition. Handle with care chips after loading immune cells which are floating.
NOTE: Compensate evaporative losses of volumes in reservoirs by replacing media every 2-3 days. For chemokine profiling, up to 100 µL from each of two wells of culture compartments can be aspirated, please refer to step 2.7, in step 1.
- Defrost a stock of matrix solution (e.g., Matrigel) by placing on ice into a 4 °C refrigerator one day before the experiment.
- Automated Counting of recruited PBMCs in single-channel fluorescent images in ImageJ
NOTE: Classical methods of immunofluorescence for confocal high-resolution imaging can be applied to on-chip operations as endpoint measurements. The basic staining procedure involves cell on-chip fixation, permeabilization, blocking, antibody binding, staining of nuclei with washing steps in between. Unlabeled immune cells infiltrated in 3D gels regions with embedded cancer microenvironments can be fixed at desired time-points and stained for expression markers of activation/exhaustion /maturation (e.g., for CD8 cells, monitoring of CD69, CD95, PD1, TIM3 markers). In Parlato et al.35, phagocytosis of SW620 apoptotic cells was evaluated by confocal microscopy, using devices mounted on 170 µm-thick coverslips. IFN-DCs were stained adding on-chip anti-human HLA-DR-FITC Ab aliquots.
To calculate the extent of infiltrated fluorescently stained live immune cells challenged with competitive signals, a common image analysis workflow is set as follows (Figure 7D-G):- Acquire, at specific time endpoints, phase contrast, and red/green channels fluorescence microphotographs of the left and right gel regions containing tumor cells respectively exposed to single or combinations of pharmacological regimes.
NOTE: Here images were captured by an EVOS-FL fluorescence microscope after cell loading (0 h), after 48 h and after 72 h of incubation (Figure 7A-B). A 4x-10x magnification was used to acquire the central chamber, the microchannel arrays, and the two juxtaposed side channels containing A375 plus IFN and A375 plus DAC/IFN.- When performing acquisition operations, consider the parameters to measure and avoid saturation of features to be counted. Optimal segmentation results depend on the nature of the acquired images, due to variability in the biological samples themselves, quality of staining, and the microscopy techniques utilized for user-oriented applications.
- Load fluorescence single-channel data (in this case red = PBMCs), in Fiji by dragging it into the main window (Figure 7D). Duplicate the image to avoid overwriting the raw data during the selection of pre-processing filters until final segmentation.
- If the image is a color image (RGB), hit Image > Type 8 or 16-bit to convert to greyscale. Check that Edit > Options > Conversions is set to scale when converting.
- Preprocessing raw data via cleaning-up of noise and artifacts.
- Go to Process > Subtract Background menu by applying the rolling ball algorithm to correct uneven noise background with large spatial variations of intensities. Set the radius to at least the size of the largest foreground particle. Pick the Vorschau box for trial and error procedure to yield optimal results. Too small values may incorrectly remove structures of interest.
- In Brightness&Contrast command drag Minimum/Maximum sliders to change the range of intensities in the histogram. Shift the Maximum slider to the left to increase the brightness, without wash-out features. Move the Minimum slide to the right to increase the contrast of the image avoiding disappearance of less visible features in the background. Click Apply to fix changes.
- Image Enhancement.
- Go to Process > Filters and experiment with the Median, Gaussian filters on images (Figure 7E).
NOTE: Pre-filtering radius should be adapted to the image noise pixels. The non-linear Median filter replaces pixel value with the median value of neighbours, to reduce salt-and-pepper noise. "Gaussian Blur" is used to smooth a digital picture, by replacing pixels with a weighted average of surrounding pixels. The weights come from the Gaussian probability distribution, so the nearest pixels are more influential. - Go optionally to Process > Math > Gamma with ticked Vorschau box to increase the contrast.
NOTE: Intensities set in the B&C panel are scaled between the two min and max limits. Values < 1.0 accentuate differences between low intensities while values > 1.0 accentuate differences between high intensities. Gamma correction is functional to find a display range, showing the dimmest objects without saturating the brightest.
- Go to Process > Filters and experiment with the Median, Gaussian filters on images (Figure 7E).
- Creation of a binary image mask.
- Go to Image > Adjust > Threshold. The most simply employed method to determine thresholds relies on histogram analysis of intensity levels, as shown in Figure 7F. In the drop-menu, play with different global thresholding methods (in our case, Otsu is applied).
- Manually scroll or type a known range of pixel intensities in the histogram panels, observe the change of the red pattern overlaying the image which mostly resembles the actual cell area. The Reset button removes the overlay. Once satisfied, click Apply to generate a binary version of the image. Check Process > Binary > Options to control how thresholded images are displayed and how objects are identified by the Particle Analyzer.
- Use the menu command Process/Binary/Watershed Particles to divide partially overlapping or merged during the threshold. Watershed can often accurately cut them apart by adding a 1-pixel thick line. Perform morphological operations such as Dilate or Erode operations to either grow or remove pixels from under or over- saturated pixels.
NOTE: For more information see the Menu Commands section or refer to MorphoLibJ, an integrated library based on mathematical morphology to process binary data. - Quantitative Image Feature Description.
- Once obtained satisfactory object recognition, open the Particle Analyzer from Analyze > Analyze Particles. Particles can be excluded by their size and circularity, expressed in pixels or in a calibrated unit of measurement (check the correct scale relative to microscope settings under Image > Properties). To include everything, keep the default of 0-Infinity and circularity default range at 0.00 – 1.00 (0 = straight line, 1 = perfect circle).
- To filter small "noise" pixels or features of not interest, set the minimum and maximum range. Tick Include Holes, Show, Outline and Display Results options in the window field. Exclude on Edges will discard particles detected on the borders of the image. Add to Manager adds the obtained selection to the ROI manager for further analysis keeping the position information of the particle.
NOTE: In the "ROI Manager", it is possible to correct automatic segmentation recorded output (merge split cells, split merged cells). Select, by using selection tools in the Fiji Toolbar, ROIs inside both gel regions where cancer cells are embedded to estimate immune infiltration.
- Export the obtained values to a spreadsheet to perform statistical analysis as shown in Figure 7G. "Results" lists a data table relative to numbered outlined particle properties identified. "Summarize" opens a window with the name of the image, total counts, and other information for the whole image.
- Go to Analyze > Set Measurements to include a wide range of parameters.
- Record optionally a macro (by selecting Plugins > Macros > Record) to automate the processing workflow and save time analysis on large datasets.
- Use the same processing routine to analyze green fluorescence channel in the same regions for analyzing morphological changes of tumor cells in 3D regions16
- Acquire, at specific time endpoints, phase contrast, and red/green channels fluorescence microphotographs of the left and right gel regions containing tumor cells respectively exposed to single or combinations of pharmacological regimes.
Representative Results
Tumor immune infiltration is a parameter of the host anti-tumor response. Tumors are heterogeneous in the composition, density, location, and functional state of infiltrating leukocytes which interactions with cancer cells can underlie clinically relevant information to predict disease course and response to therapy. In this sense, microfluidic technologies could be used as complementary and privileged in vitro tools to explore the immune contexture of tumors, as well as to monitor the response to anticancer therapies. The coupling of the microfluidic assay, live-cell imaging, and tracking software may establish reliable quantification methods to quantify how immune cells adjust their migration pattern in different contexts. In this chapter, we have reported steps for setting up versatile 2D or 3D co-cultures of immune and target cancer cells in ad hoc microfluidic devices realized by standard soft-lithographic procedures. In Section 1 microfluidic devices were employed to allow chemical and physical contacts between adherent (MDA-MB-231 cancer cells) and non-adherent (PBMCs) populations. Some chemotherapeutic agents (e.g., anthracyclines among the others) can induce "immunogenic" apoptosis of malignant cells, thus enhance their visibility in immunocompetent hosts. Cancer immunogenic cell death (ICD) is characterized by the release of membrane-bound and soluble signals delivered by dying cells functioning as alarmins for immune cells. To provide a quantitative validation of ICD response, we employ data collected in the microfluidic platform where leukocytes can move through suitably built microchannel bridges toward their target cells. Time-lapse recordings were performed after co-loading PBMCs, from healthy donors (WT, wild type), with human MDA-MB-231 breast cancer cells pre-treated or not with the anthracycline DOXO. Microphotographs were generated every 2 min, for two successive time intervals of 24 and 48 h. (Exemplary Movie S1 of 0-24 h interval). Tracking analysis of individual PBMCs challenged with dying (DOXO-treated) or live (PBS-treated) cancer cells was done using Trackmate plug-in, as seen in Figure 6A (left panel). Relevant chemotaxis values and migration plots were automatically generated by using the Chemotaxis and Migration Tool, as detailed in62. The cell trajectories were all extrapolated to (x, y) = 0 at time 24 h. The results indicated a different migratory profile of the immune cells when co-loaded with breast cancer cells exposed to DOXO or PBS. When PBMCs were confronted with apoptotic cancer cells, they crossed microchannels towards dying/dead cells (but not to live untreated cells). Migration X/Y spider and rose plots, presented in left panel Figure 6B-C, were mapped and compared to highlight the impact of immunogenic inducers agents on differences in immune dynamics. Rose plots demonstrate that PBMCs migrated mainly in nearly all directions in the control experiment, only a negligible fraction is guided up the gradient generated by proliferating cancer cells. Conversely, the paths of individual cells highlight a strong bias movement along the direction of apoptotic breast cancer cells (negative x-direction). To evaluate directed immune cell migration towards DOXO-treated or PBS-treated cancer cells, several chemotaxis parameters are computed, including: a) the center of mass (spatial averaged point of all endpoints); b) the directionality; c) the forward migration index (i.e., the average cell displacement in a direction of interest which means towards target tumor site, in our case). The latter values represent a measurement of the efficiency of a cell to migrate in the direction given chemotactic stimuli. After reaching the left chamber, a fraction of leukocytes with an increasing density over 24-48 h exhibited long-term (>60 min) contacts with DOXO-treated MDA-MB-231. PBMCs fail to massively migrate and engage in such long-term and persistent interactions with live cancer cells (as shown by representative microphotographs of approaching PBMCs in Figure 6A, right). For quantifying differences in tumor-immune interplay, the tumor region was delineated with a fixed circle of diameter ranging 20-80 microns, named "hotspot". The quantification and analysis from representative FOVs are shown in Figure 6 (Right panel, B-C).
In Section 2 a novel 3D immuno-competent tumor model was described to quantify the recruitment of immune cells in response to anti-cancer combinations of epigenetic drugs32 (DAC/IFN vs IFN alone), allowing the comparison of two different treatment conditions of A375 melanoma cells simultaneously. Thus, A375M melanoma cells, labeled with PKH67 green fluorescent dye were embedded in Matrigel matrices in the presence of DAC and/or IFN into each gel chamber, whereas PKH26 red-labeled PBMCs were distributed homogenously into the central fluidic chamber at the starting point (Figure 7A). We compared simultaneously the two tumor masses in 3D matrices for their capacity to attract PBMCs. At 48 and 72 h, PBMCs are guided massively into the right-side microchannels, as observed in Figure 7B.
A preferential homing of PBMCs toward the gel matrix containing DAC/IFN-treated, rather than IFN-treated, melanoma site was clearly observed, whereas poor migration rate was visible toward A375 cells exposed to single treatment (left chip side). The competitive setting is applicable to investigate a plethora of different cancer biology phenotypes (e.g., drug-resistant vs aggressive, primary or metastatic (e.g., A375P vs A375M melanoma cells), and responders vs non-responders). Gel chambers can be comprised of complex co-cultures of malignant cells and multiple non-cancerous tumor-associated cells, such as endothelial cells, immune cells, and fibroblasts33 to characterize TME-level drug responses. Events of mitosis and apoptotic death of cancer cells can be monitored by staining with live dyes33. Immunostaining procedures on 3D microfluidic devices could be adapted to evaluate states of activation of infiltrated immune cells in tumor sites by confocal microscopy35. In this sense, these 3D microfluidic systems may mimic complex tumor structures and multicellular interactions and therefore are valuable platforms for more reliable preclinical drug testing.
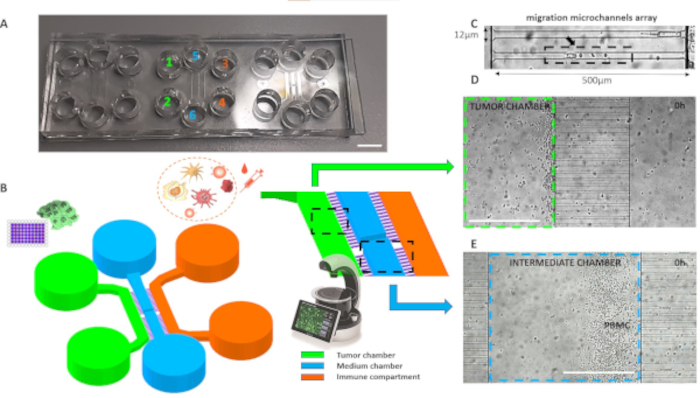
Figure 1. Planimetry of the microfluidic device for assembling 2D tumor-immune co-cultures. (A) Real microphotograph of 3 chips assembled into a single microscope slide. The reservoirs are numbered depending on the loading sequence and are color-coded as the corresponding culture chambers listed in the legend. Scalebar, 6 mm. (B) 3D CAD of chip consisting of three main cell culture areas connected by microgrooves. C) Details of the narrow microgrooves bridge, showing the dimensions tailored to cancer cells and PBMCs size. D-E) Visible light microphotograph was acquired before the beginning of the time-lapse, showing the distribution of cancer cells and PBMCs respectively in the left and the intermediate chamber. Scalebar, 500 µm. Please click here to view a larger version of this figure.
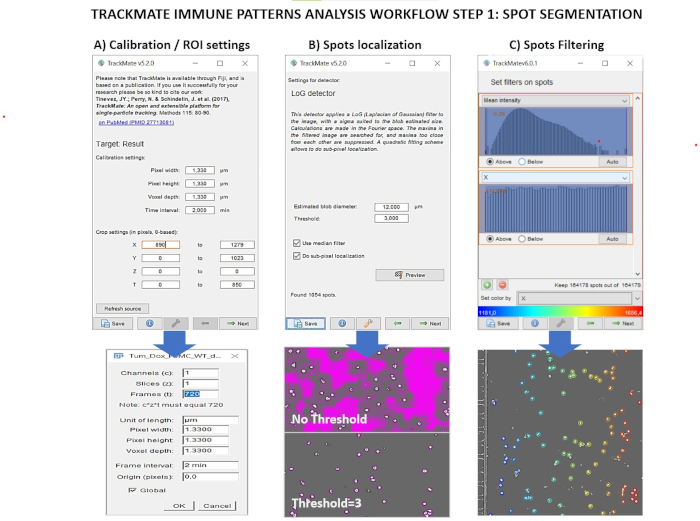
Figure 2. Trackmate analysis pipeline for localization of immune spots in time-series images. A) Upper panel: Screenshot of Trackmate image calibration menu. Lower panel: Fiji "Image properties menu" to set time and spatial units. B) Upper panel: Screenshot of Trackmate "Spots detection" menu. Lower panel: Output image with applied different values of "Threshold". C) Upper panel: Screenshot of Trackmate "Spot filtering" menu illustrating some filters. Lower panel: Exemplary time-lapse image, depicting immune spots filtered by position along X in the intermediate chamber. Please click here to view a larger version of this figure.
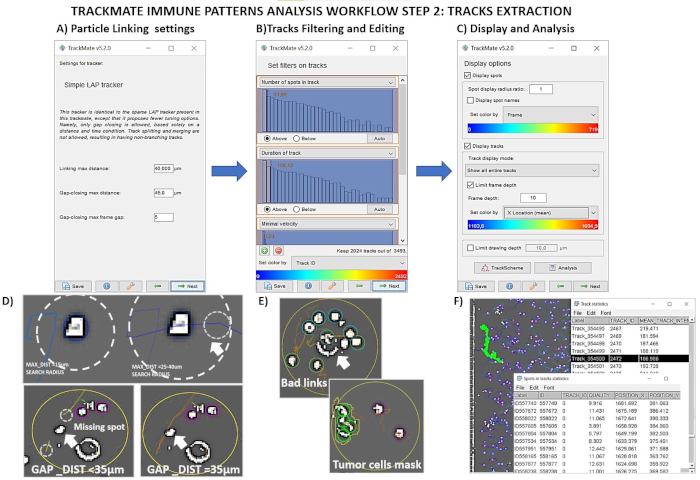
Figure 3. Trackmate Analysis pipeline for reconstructing immune tracks in time-series image sequences. A-C) Screenshots of Trackmate menus for tracks building, tracks filtering, and for exporting final data. D) Examples of generated tracks in pre-processed time-lapse images varying the linking detector settings. E) Example of false tracks and bad links derived from the detection of tumor cells borders. F) Tracks are highlighted in green by selecting rows in the file .txt of results. Please click here to view a larger version of this figure.
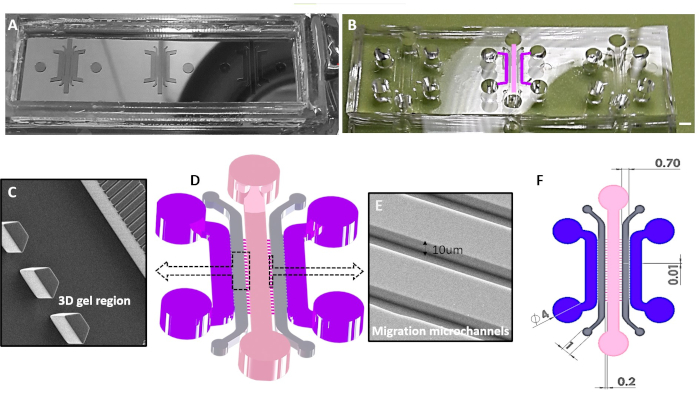
Figure 4. Schematic overview for 3D immunocompetent tumor-on chips. A) Example of a micro-structured silicon master. Stamps for PDMS were patterned in SU-8 negative resist in a cleanroom facility equipped with e-beam and optical lithography. B) PDMS replicas were fabricated by standard soft-lithography methods. The central unit has the chambers colored as the ones drawn in 3D rendering of the chip, depicted in the panel D. C) Scanning electron microscopy (SEM) enlarged view of the array of micropillars suited for hydrogel solution entrapment. D)3D CAD showing the chambers, connected by microgrooves and the loading wells. Dashed boxes are referred to SEM photographs of details (panel C, and E). E) SEM photograph of 10 µm high connecting microgrooves. F) 2D CAD layout depicting the dimensions of microstructures. Please click here to view a larger version of this figure.
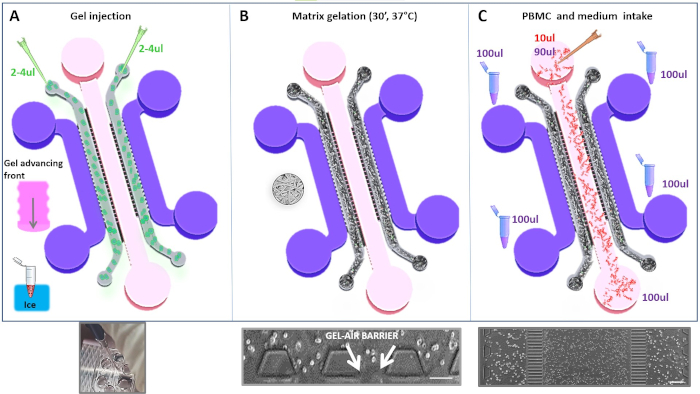
Figure 5. Schematic workflow of main loading protocol steps for assembling 3D co-cultures. A) Upper panel: Schematics of injection step of Matrigel solution in each gel side region. Lower panel: Real photograph of the chip showing gel front advancement along the channel during loading step. B) Schematics of Matrigel polymerization step. Lower panel: Phase-contrast microscopic view of the gel-air interface formed after gelation step. Scalebar, 100 µm. C)Upper panel: Drawing, depicting the loading of cells and media with exemplary volumes used in the protocol. Lower panel: A 4X Phase-contrast image of the assembled co-culture at 0h is shown. Scalebar, 200 µm. Please click here to view a larger version of this figure.
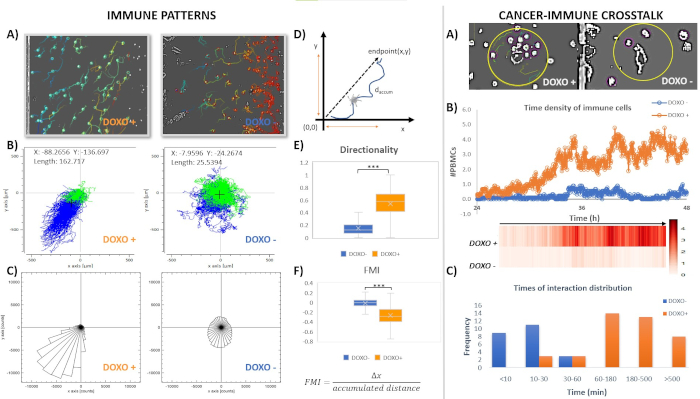
Figure 6. Analysis of migratory profiles and interaction behavior of PBMC towards dying/live tumor chamber in the time window of 24-48 h. Left panel. A) Screenshots of time-lapse pre-processed images overlaid with immune colored tracks, extracted by Trackmate. The immune paths are obtained by a single FOV in the indicated experimental conditions in the interval 24-to-48 h in the intermediate chamber. B-C) Representative migration tracks and rose plots of PBMCs cultivated in the two different conditions (n = 1550 PBMCs versus DOXO-treated cancer cells, DOXO+ or n =1434 PBMCs versus control cancer cells, DOXO-). Each line inside x-y plots depicts a single PBMC trajectory and each circle represents the final position of a single cell with respect to the initial position. Starting points are set to (0,0) using a coordinate transformation. The coordinates and displacement of the center of mass of cell migration are displayed in the indicated experimental conditions. The center of mass coordinates and displacement (computed as average Euclidean distance for the X and Y components) provide indication of the average direction in which the group of cells primarily travelled and magnitude of the overall cell movement in condition groups. Blue and green lines respectively mark individual tracks by Euclidean distance value greater/smaller than a threshold value (100 µm). D) Scheme of numerical data extracted by a cell track. E-F) Box & Whiskers plot depicting respectively directionality (p<0,0001 Unpaired t test with Welch's correction) and FMI (p<0,0001 Unpaired t test with Welch's correction). The horizontal line in boxes represents median values. The FMI values are the average for all cell-tracks in each condition group.Right panel. A) Screenshots of Trackmate extracted ROIs showing differential interactions between PBMCs and DOXO or PBS-treated cancer cells. B) Time density of PBMCs around DOXO-treated or control MDA-MB-231 cancer cells. Number of PBMCs present into selected ROI around cancer cells for each condition. Values reported are the average over 9 selected ROI from a single time-lapse FOV. Cancer hotspots are defined in a ROI (80 µm in diameter). Corresponding density heatmap is displayed. Dots represent mean number of cells calculated over 9 cancer hotspots at indicated time points. C) Distribution of times (min) of contacts for each experimental group. Please click here to view a larger version of this figure.
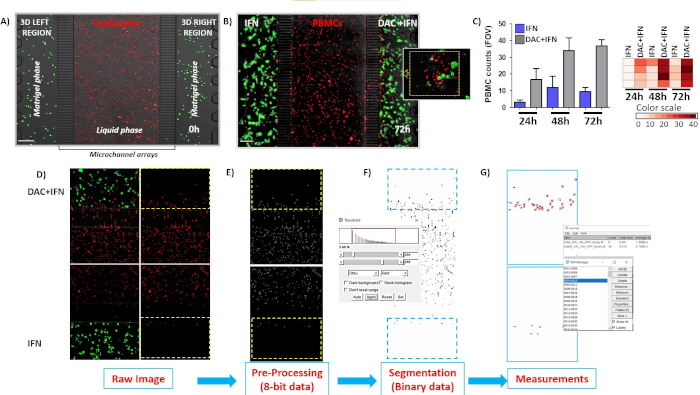
Figure 7. Preferential recruitment of PBMCs in response to DAC plus IFN drug combinations in a competitive 3D immuno-competent melanoma on chip model. A) Distribution of initially loaded PBMCs in the central chamber of microfluidic devices. Microphotographs are acquired by EVOS-FL fluorescence microscope during 0-72 h interval. Red fluorescence (PKH67-labeled cells) represents PBMCs from healthy donors. PKH67-labeled (green) human melanoma cells embedded in Matrigel containing single or double combinations treatments were plated in lateral chambers. B) A375 cells plus IFN on left versus A375 plus DAC/IFN on the right. Fluorescence images were shown at 72 h of co-culture. Discontinued yellow box depicts visibly massive recruitment in A375 plus DAC/IFN side. C) PBMC counts in four different ROIs from IFN chambers vs DAC+IFN chambers. Histograms represent cell counts +/- S.D.; heatmap enumerated values from each ROI. Scalebars, 200 µm. D-G) Schematics of segmentation and quantification steps of infiltrated PBMCs in gel matrices applied to single channel fluorescence images. Please click here to view a larger version of this figure.
Supplementary File. Microfabrication protocol Please click here to download this file.
Supplementary Movie 1. Time-lapses sequences in a 2D tumor-immune on-chip co-culture. Microphotographs were acquired every 2 min, in a time interval of 0-24 h by means of a compact microscope placed into a standard cell culture incubator. Left panel. Movie of PBMCs from healthy donors (WT, wild type), plated with MDA-MB-231 control breast cancer cells. Right panel. Movie of a massive migration of PBMCs WT towards the chip chamber where MDA-MB-231 DOXO-treated cancer cells, are loaded. 2D chip layout is described in detail in section 1of Protocol and shown in Figure 1. Please click here to download this movie.
Discussion
The described methods try to design a general approach to recapitulate, with modulable degree of complexity, two significant aspects in the field of onco-immunology, which can benefit from the adoption of more relevant in vitro models. The first one involves the tumor cell population side, where tackling single cell characteristics may lead to a better description of heterogeneity and correlated biological and clinical significance including resistance to therapy, propension to metastasis, stem cell and differentiation grade. The other side of the story is represented by the TME, including noncancerous components (immune and stromal cells, blood vessels) and chemical/physical landscape (ECM constituents, chemokines and other soluble factors released) that can profoundly shape both the characteristics of the disease and individual's response to therapy63, particularly to immunotherapy. It is worth to note that exporting the described approach to other research fields requires a deep understanding of limitations and challenges brought by the development and adoption of new models.
Quoting a maxim applied in engineering modelling ("model the problem not the system"), it must be optimized which is the minimum number of (cellular, physical and chemical) components and conditions needed to make own on-chip model biologically relevant and stable along the whole experiment. Every combination of cell types/microenvironmental/experimental setting must, thus, be accurately chosen, evaluated, and continuously monitored along single experiments and from session to session. These checks include, as mentioned in the protocol sections, strict control of parameters like volumes in the microfluidic devices that can be modified by humidity conditions or heating from illumination sources, possible movements and non-planarity of stages causing uncontrolled liquid drifts, but also some endpoint verification of cell state, viability and phenotypic characterization. One critical step concerns the decision to use perfusion systems to create vascularized (like) structures, or for on-chip drug delivery33, since this may affect the experiments in terms of increasing complexity, cell culture duration, and modulation of the chemical factors in presence of floating cells like the immune ones.
In the following, we summarize the main critical and relevant issues found in the experimental settings and in the most recent literature.
ECM and chemical landscape definition
TME is deeply affected also by the mechanical64,65 properties of the surroundings63, 66. For this reason, the choice of the culturing matrix is fundamental, especially when dealing with immune cells that, in certain conditions, could respond to signals released by the matrix itself. Relevant advances are expected in the next future also from design and development of new materials and hydrogels (featured by different stiffness, porosity, presence of soluble factors, among the other parameters) that are enabling an increasingly refined and controllable ECM, mimicking specificities of different tissues today, different patients tomorrow. On the other side is also critical to monitor the changes induced by the different cell populations in the ECM and define strategies to include this information in dynamic and phenotypic analyses.
Data management
The path toward deployment of tumor on-chip techniques in a clinical workflow is going to benefit from a huge experimental work that is taking place along several directions. Organ-on-chip models, as many in vitro techniques, are functional, at least potentially, to perform high-throughput/high-content measurements. Parallelization of experimental conditions, including positive and negative controls, and technical/biological replicates is made possible by integrating several chips on the same plate. The increase in the quantitative throughput plays a crucial role in translating and validating these systems in fast drug or gene screening pipeline for immunotherapies. Indeed, several companies have already developed platforms in a multi-well format, and different imaging approaches are being tested to allow for monitoring a high number of devices simultaneously14. In the above described protocols, we used microscope slides allocating up to 3 chips, with the possibility to visualize up to 12 experimental conditions in parallel, using a standard microscope multi-slide tray. This setup is compatible with hand pipetting and suitable for custom oriented adjustments performed in our microfabrication facility. On the contrary, when a strong parallelization is necessary (multiple screening tests and controls), setup optimizations to set a higher level of automation (i.e., use of pipetting robots, plastic multi-wells) are required.
When designing experiments, a trade-off between spatial and temporal resolution must be taken in account.
The huge amount of data, produced by high content microscopy, constitutes a limiting factor in terms of storage, transfer and analysis. These issues are being addressed with computational approaches implementing machine learning tools and hardware/software resources, which may foster the future possibility to link on-chip/in-silico experiments67,68 in choosing therapeutic strategies, and that represents an ambitious yet non-deferrable opportunity.
Beyond Fluorescence Imaging
Fluorescence labelling, by dyes or reporter genes, due to its high specificity, undoubtedly represents the gold standard method to identify different cell populations in co-culture conditions and to resolve molecular properties with high SNR. In OOC models this approach is commonly used as a reference and particularly in heterogeneous tumor-on-chip microenvironments, it can be valuable to characterize the phenotype of immune cells infiltrated and interacting.
Nevertheless, there is increasing evidence that effects induced by fluorophores, laborious staining procedures and illumination routines must be monitored and minimized69 because can deeply affect cell behavior and state70,71. This risk seems particularly true for fragile systems, such as immune cells72,73. Phototoxic reactions may introduce limitations in defining the temporal window acquisition of live cells when monitored at high spatial and temporal resolution. Moreover, the richness in the variety of immune sub-populations make unfeasible to discriminate among them only by fluorescent staining, considering the limited number of filters commonly available on microscopes.
To address this issue, from an instrumental point of view, novel label free74 microscopy techniques such as holographic56 or hyperspectral microscopy57 are now appearing on the stage. They promise advanced cellular process classification strategies beyond fluorescence and can be particularly valuable for studying sensitive samples. From a data analysis point of view, advanced computational approaches, based on deep learning algorithms75 (trained by fluorescence images datasets), are door-openers to perform the so called "in silico labeling"76,77. They have been successfully applied to predict fluorescent markers from bright-field images78, generating labeled images, without staining cells, thus increasing bright field microscopy informative power. This strategy can also be useful to save time and fluorescence channels for other markers. We believe that OOC community will benefit from these new techniques allowing a less invasive study of cell populations interaction.
Data analysis
Mechanisms of interactions between immune and target cancer cells can be investigated through tracking of cell movements28,79. Time-lapse tracking experiments in complex and heterogeneous co-cultures are extremely valuable to extract cell migration patterns, morphological and state changes, and comprehensive lineage information. Performing manual analysis is practically only feasible for short sequences with few cells but not for high-throughput, systematic experiments. Consequently, the development of computational tools for cell tracking, either fully or partly automated, is a vital field of research in image analysis24. Typically, conventional cell tracking requires relatively high frequency of sampling and spatial resolution to correctly perform segmentation tasks, which can be challenging in many experimental conditions. To localize cells, there are available open-access segmentation and tracking tools (e.g., ImageJ software22) as presented in this protocol or dedicated proprietary software. In large-scale studies, we applied a proprietary software called Cell Hunter16,31, developed by University of Tor Vergata in Rome, to distinguish in a fully automatic way cancer and immune cells in multi-population context. Tailor-made solutions based on machine learning and Neural network approaches80,81,82 are beginning to be today implemented in microscopy software packages83, from improving SNR to managing critical acquisition parameters or segmentation steps. Machine learning can be exploited to recognize common cellular patterns (e.g., motion styles) in order to characterize the biological response with respect to microenvironmental factors.
In Comes et al.41, a pre-trained Deep Learning Convolutional Neural Network architecture was applied to classify if cancer cells are or not exposed to a drug treatment by using as a "marker" the motility of the immune cells, tracked from time-lapse data of co-cultures of breast cancer cells and PBMCs in collagen matrices in microdevices, as described in33.
Single-cell omics methods and ontologies development
We point out the need for a strategic alliance between single-cell omics technologies84 (e.g., proteomics, metabolomics, genomics) and on-chip methods: the molecular detailed characterization, conjugated to functional dynamic information could enhance comprehension of basic mechanisms and clinical description. In this case new instruments for linking the two worlds are at their beginning85. The first challenge is to implement single-cell omics approaches directly on the onco-immunology chips22. Moreover, organ-on-chips could be exploited as platforms to test potential targets identified by genomics and proteomics analyses86,87. A second linking tool, which is still largely missing, is a structured, standardized way of annotating and storing measured system results88,89. But this is exactly what we need in the future to build systematic databases of cellular quantitative results and measured characteristics, to mine, infer and correlate them with the inherent biological information. In a word, the need for standardization and systematic analysis of heterogeneous experimental datasets calls for an ontology framework.
Personalizing models
Organs-on-chip technology is suitable for personalization12, since cells and tissues from single patients (or classes of patients) can be used in the devices under controlled conditions, leading to clinically relevant readouts, useful to inform therapeutic or prevention strategies. Some examples are starting to appear in literature90. Of course, for tumor-on-chip models, this challenge poses several technical and scientific questions to be solved36 (like TME characteristics control such as oxygen concentration, cytokine gradients, etc). Importantly, the prospect of making oncology and onco-immunology therapies more effective while reducing harmful effects for each patient is attractive, from a quality of life perspective as for the optimization of healthcare resources.
In conclusion, it is apparent that this field is an authentically interdisciplinary one and as such, requires a great effort in establishing a common language and shared goals between researcher, clinicians, industry, but also from different disciplines (engineering, biology, data science, medicine, chemistry) finding good balance and new solutions91.
Offenlegungen
The authors have nothing to disclose.
Materials
| Cell culture materials | |||
| 50 mL tubes | Corning-Sigma Aldrich, St. Louis, MO | CLS430828 | centrifuge tubes |
| 5-aza-2'-deoxycytidine DAC | Millipore-Sigma; St. Louis, MO | A3656 | DNA-hypomethylating agent |
| 6-well plates | Corning-Sigma Aldrich, St. Louis, MO | CLS3506 | culture dishes |
| 75 cm2 cell culture treated flask | Corning, New York, NY | 430641U | culture flasks |
| A365M | American Type Culture Collection (ATCC), Manassas, VA | CVCL_B222 |
human melanoma cell line |
| Doxorubicin hydrochloride | Millipore-Sigma; St. Louis, MO | D1515 | anthracycline antibiotic |
| Dulbecco's Modified Eagle Medium DMEM | EuroClone Spa, Milan, Italy | ECM0728L | Culture medium for SK-MEL-28 cells |
| Dulbecco's Phosphate Buffer Saline w/o Calcium w/o Magnesium | EuroClone Spa, Milan, Italy | ECB4004L | saline buffer solution |
| Fetal Bovine Serum | EuroClone Spa, Milan, Italy | ECS0180L | ancillary for cell culture |
| Ficoll | GE-Heathcare | 17-1440-02 | separation of mononuclear cells from human blood. |
| hemocytometer | Neubauer | Cell counter | |
| Heparinized vials | Thermo Fisher Scientific Inc., Waltham, MA | Vials for venous blood collection | |
| interferon alpha-2b | Millipore-Sigma; St. Louis, MO | SRP4595 | recombinant human cytokine |
| L-Glutamine 100X | EuroClone Spa, Milan, Italy | ECB3000D | ancillary for cell culture |
| Liquid nitrogen | |||
| Lympholyte cell separation media | Cedarlane Labs, Burlington, Canada | Separation of lymphocytes by density gradient centrifugation | |
| Lymphoprep | Axis-Shield PoC AS, Oslo, Norway | ||
| Matrigel | Corning, New York, NY | 354230 | growth factor reduced basement membrane matrix |
| MDA-MB-231 | American Type Culture Collection (ATCC), Manassas, VA | HTB-26 | human breast cancer cell line |
| Penicillin/ Streptomycin 100X | EuroClone Spa, Milan, Italy | ECB3001D | ancillary for cell culture |
| Pipet aid | Drummond Scientific Co., Broomall, PA | 4-000-201 | Liquid handling |
| PKH26 Red Fluorescent cell linker | Millipore-Sigma; St. Louis, MO | PKH26GL | red fluorescent cell dye |
| PKH67 Green fluorescent cell linker | Millipore-Sigma; St. Louis, MO | PKH67GL | green fluorescent cell dye |
| RPMI-1640 | EuroClone Spa, Milan, Italy | ECM2001L | Culture medium for MDA-MB-231 cells |
| serological pipettes (2 mL, 5 mL, 10 mL, 25 mL, 50 mL) | Corning- Millipore-Sigma; St. Louis, MO | CLS4486; CLS4487; CLS4488; CLS4489; CLS4490 | Liquid handling |
| sterile tips (1-10 μL, 10-20 μL, 20-200 μL, 1000 μL) | EuroClone Spa, Milan, Italy | ECTD00010; ECTD00020; ECTD00200; ECTD01005 | tips for micropipette |
| Timer | |||
| Trypan Blue solution | Thermo Fisher Scientific Inc., Waltham, MA | 15250061 | cell stain to assess cell viability |
| Trypsin | EuroClone Spa, Milan, Italy | ECM0920D | dissociation reagent for adherent cells |
| Cell culture equipment | |||
| EVOS-FL fluorescence microscope | Thermo Fisher Scientific Inc., Waltham, MA | Fluorescent microscope for living cells | |
| Humified cell culture incubator | Thermo Fisher Scientific Inc., Waltham, MA | 311 Forma Direct Heat COIncubator; TC 230 | Incubation of cell cultures at 37 °C, 5% CO2 |
| Juli Microscope | Nanoentek | ||
| Laboratory refrigerator (4 °C) | FDM | ||
| Laboratory Safety Cabinet (Class II) | Steril VBH 72 MP | Laminar flow hood | |
| Optical microscope | Zeiss | ||
| Refrigerable centrifuge | Beckman Coulter | ||
| Thermostatic bath | |||
| Microfabrication materials | |||
| 3-Aminopropyl)triethoxysilane (Aptes) | Sigma Aldrich | A3648 | silanizing agent for bonding PDMS to plastic coverslip |
| Chromium quartz masks / 4"x4", HRC / No AZ | MB W&A, Germany | optical masks for photolithography | |
| Glass coverslip, D 263 M Schott glass, (170 ± 5 µm) | Ibidi, Germany | 10812 | |
| Hydrogen Peroxide solution 30% | Carlo Erba Reagents | 412081 | reagents for piranha solution |
| Methyl isobutyl ketone | Carlo Erba Reagents | 461945 | PMMA e-beam resist developer |
| Microscope Glass Slides (Pack of 50 slides) 76.2 mm x 25.4 mm | Sail Brand | 7101 | substrates for bonding chips |
| Miltex Biopsy Punch with Plunger, ID 1.0mm | Tedpella | dermal biopsy punches for chip reservoirs | |
| PMMA 950 kDa | Allresist,Germany | AR-P. 679.04 | Positive electronic resists for patterning optical masks |
| Polymer untreated coverslips | Ibidi, Germany | 10813 | substrates for bonding chips |
| Prime CZ-Si Wafer, 4”, (100), Boron Doped | Gambetti Xenologia Srl, Italy | 30255 | |
| Propan-2-ol | Carlo Erba Reagents | 415238 | |
| Propylene glycol monomethyl ether acetate (PGMEA) | Sigma Aldrich | 484431-4L | SU-8 resists developer |
| SU-8 3005 | Micro resist technology,Germany | C1.02.003-0001 | Negative Photoresists |
| SU-8 3050 | Micro resist technology,Germany | C1.02.003-0005 | Negative Photoresists |
| Suite of Biopunch, ID 4.0 mm, 6.0 mm, 8.0 mm | Tedpella | 15111-40, 15111-60, 15111-80 | dermal biopsy punches for chip reservoirs |
| Sulfuric acid 96% | Carlo Erba Reagents | 410381 | reagents for piranha solution |
| SYLGARD 184 Silicone Elastomer Kit | Dowsil, Dow Corning | 11-3184-01 | Silicone Elastomer (PDMS) |
| Trimethylchlorosilane (TMCS) | Sigma Aldrich | 92360-100ML | silanizing agent for SU-8 patterned masters |
| Microfabrication equipment | |||
| 100 kV e-beam litography | Raith-Vistec EBPG 5HR | ||
| hotplate | |||
| Optical litography system | EV-420 double-face contact mask-aligner | ||
| Reactive Ion Etching system | Oxford plasmalab 80 plus system | ||
| Vacuum dessicator |
Referenzen
- Abbas, A. K., L, A. H., P, S. . Cellular and Molecular Immunology, Ninth Edition. , (2018).
- Eisenstein, M. Cellular censuses to guide cancer care. Nature. , (2019).
- Cancer Cell. Models for Immuno-oncology Research. Cancer Cell. , (2020).
- Zhang, Z., et al. Morphology-based prediction of cancer cell migration using an artificial neural network and a random decision forest. Integrative biology quantitative biosciences from nano to macro. 10 (12), 758-767 (2018).
- Dagogo-Jack, I., Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology. 15 (2), 81-94 (2018).
- Milo, I., et al. The immune system profoundly restricts intratumor genetic heterogeneity. Science Immunology. 3 (29), (2018).
- Mlecnik, B., et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Science Translational Medicine. , (2016).
- Sbarrato, T., et al. 34th Annual Meeting & Pre-Conference Programs of the Society for Immunotherapy of Cancer (SITC 2019): part 1. Journal for ImmunoTherapy of Cancer. 7, 282 (2019).
- Miller, C. P., Shin, W., Ahn, E. H., Kim, H. J., Kim, D. -. H. Engineering Microphysiological Immune System Responses on Chips. Trends in Biotechnology. 38 (8), 857-872 (2020).
- Ma, C., Harris, J., Morales, R. -. T. T., Chen, W. Microfluidics for Immuno-oncology. Nanotechnology and Microfluidics. , 149-176 (2020).
- Mengus, C., et al. In vitro Modeling of Tumor-Immune System Interaction. ACS Biomaterials Science & Engineering. 4 (2), 314-323 (2018).
- Van Den Berg, A., Mummery, C. L., Passier, R., Van der Meer, A. D. Personalised organs-on-chips: functional testing for precision medicine. Lab on a Chip. , (2019).
- Ingber, D. E. Reverse Engineering Human Pathophysiology with Organs-on-Chips. Cell. , (2016).
- Huh, D., et al. Microfabrication of human organs-on-chips. Nature Protocols. , (2013).
- Mazzarda, F., et al. Organ-on-chip model shows that ATP release through connexin hemichannels drives spontaneous Ca2+ signaling in non-sensory cells of the greater epithelial ridge in the developing cochlea. Lab Chip. , (2020).
- Mencattini, A., et al. High-throughput analysis of cell-cell crosstalk in ad hoc designed microfluidic chips for oncoimmunology applications. Methods in Enzymology. 632, 479-502 (2020).
- Maharjan, S., Cecen, B., Zhang, Y. S. 3D Immunocompetent Organ-on-a-Chip Models. Small Methods. , 2000235 (2020).
- Phan, D. T. T., et al. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab on a Chip. , (2017).
- Jeon, J. S., Zervantonakis, I. K., Chung, S., Kamm, R. D., Charest, J. L. In vitro Model of Tumor Cell Extravasation. PLoS ONE. , (2013).
- Jeon, J. S., et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proceedings of the National Academy of Sciences of the United States of America. , (2015).
- Chen, M. B., Whisler, J. A., Fröse, J., Yu, C., Shin, Y., Kamm, R. D. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nature Protocols. , (2017).
- Sade-Feldman, M., et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. , (2018).
- Di Modugno, F., Colosi, C., Trono, P., Antonacci, G., Ruocco, G., Nisticò, P. 3D models in the new era of immune oncology: Focus on T cells, CAF and ECM. Journal of Experimental and Clinical Cancer Research. , (2019).
- Svensson, C. -. M., Medyukhina, A., Belyaev, I., Al-Zaben, N., Figge, M. T. Untangling cell tracks: Quantifying cell migration by time lapse image data analysis. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 93 (3), 357-370 (2018).
- Arena, E. T., Rueden, C. T., Hiner, M. C., Wang, S., Yuan, M., Eliceiri, K. W. Quantitating the cell: turning images into numbers with ImageJ. Wiley interdisciplinary reviews. Developmental biology. 6 (2), (2017).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. , (2012).
- Vacchelli, E., et al. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science. 350 (6263), 972-978 (2015).
- Businaro, L., et al. Cross talk between cancer and immune cells: exploring complex dynamics in a microfluidic environment. Lab on a Chip. 13 (2), 229-239 (2013).
- Beltman, J. B., Marée, A. F. M., de Boer, R. J. Analysing immune cell migration. Nature Reviews Immunology. 9 (11), 789-798 (2009).
- Agliari, E., et al. Cancer-driven dynamics of immune cells in a microfluidic environment. Scientific Reports. 4 (1), 6639 (2014).
- Biselli, E., et al. Organs on chip approach: a tool to evaluate cancer -immune cells interactions. Scientific Reports. 7 (1), 12737 (2017).
- Lucarini, V., et al. Combining Type I Interferons and 5-Aza-2′-Deoxycitidine to Improve Anti-Tumor Response against Melanoma. Journal of Investigative Dermatology. 137 (1), 159-169 (2017).
- Nguyen, M., et al. Dissecting Effects of Anti-cancer Drugs and Cancer-Associated Fibroblasts by On-Chip Reconstitution of Immunocompetent Tumor Microenvironments. Cell Reports. 25 (13), 3884-3893 (2018).
- Racioppi, L., et al. CaMKK2 in myeloid cells is a key regulator of the immune-suppressive microenvironment in breast cancer. Nature Communications. 10 (1), 2450 (2019).
- Parlato, S., et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Scientific Reports. 7 (1), 1093 (2017).
- Andreone, S., et al. IL-33 Promotes CD11b/CD18-Mediated Adhesion of Eosinophils to Cancer Cells and Synapse-Polarized Degranulation Leading to Tumor Cell Killing. Cancers. 11 (11), 1664 (2019).
- Bray, L. J., Hutmacher, D. W., Bock, N. Addressing Patient Specificity in the Engineering of Tumor Models. Frontiers in Bioengineering and Biotechnology. 7, 217 (2019).
- Fetah, K. L., et al. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small. 15 (50), 1901985 (2019).
- Jabbari, P., Rezaei, N. Artificial intelligence and immunotherapy. Expert Review of Clinical Immunology. 15 (7), 689-691 (2019).
- Mak, K. -. K., Pichika, M. R. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today. 24 (3), 773-780 (2019).
- Mencattini, A., et al. Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Scientific Reports. , (2020).
- Isozaki, A., et al. AI on a chip. Lab Chip. , (2020).
- Makaryan, S. Z., Cess, C. G., Finley, S. D. Modeling immune cell behavior across scales in cancer. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. , (2020).
- Mak, K. K., Pichika, M. R. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today. , (2019).
- Masuzzo, P., Van Troys, M., Ampe, C., Martens, L. Taking Aim at Moving Targets in Computational Cell Migration. Trends in Cell Biology. 26 (2), 88-110 (2016).
- Meijering, E., Dzyubachyk, O., Smal, I. Chapter nine – Methods for Cell and Particle Tracking. Imaging and Spectroscopic Analysis of Living Cells. 504, 183-200 (2012).
- Riedhammer, C., Halbritter, D., Weissert, R. Peripheral blood mononuclear cells: Isolation, freezing, thawing, and culture. Methods in Molecular Biology. , (2015).
- Harris, J., et al. Fabrication of a microfluidic device for the compartmentalization of neuron soma and axons. Journal of Visualized Experiments. , (2007).
- Shin, Y., et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nature Protocols. , (2012).
- Park, J. W., Vahidi, B., Taylor, A. M., Rhee, S. W., Jeon, N. L. Microfluidic culture platform for neuroscience research. Nature Protocols. , (2006).
- Gjorevski, N., et al. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab on a Chip. , (2020).
- Jenkins, R. W., et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discovery. , (2018).
- Comes, M. C., et al. The influence of spatial and temporal resolutions on the analysis of cell-cell interaction: a systematic study for time-lapse microscopy applications. Scientific Reports. 9 (1), 6789 (2019).
- Tinevez, J. -. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Ulman, V., et al. An objective comparison of cell-tracking algorithms. Nature Methods. , (2017).
- Tinevez, J. -. Y., Herbert, S. . The NEMO Dots Assembly: Single-Particle Tracking and Analysis BT – Bioimage Data Analysis Workflows. , 67-96 (2020).
- Jacquemet, G., Hamidi, H., Ivaska, J. Filopodia quantification using filoquant. Methods in Molecular Biology. , (2019).
- Caldas, P., Radler, P., Sommer, C., Loose, M. Computational analysis of filament polymerization dynamics in cytoskeletal networks. Methods in Cell Biology. , (2020).
- Chalfoun, J., Majurski, M., Peskin, A., Breen, C., Bajcsy, P., Brady, M. Empirical gradient threshold technique for automated segmentation across image modalities and cell lines. Journal of Microscopy. 260 (1), 86-99 (2015).
- Huang, C. P., et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab on a Chip. , (2009).
- Farahat, W. A., et al. Ensemble analysis of angiogenic growth in three-dimensional microfluidic cell cultures. PLoS ONE. , (2012).
- Zengel, P., Nguyen-Hoang, A., Schildhammer, C., Zantl, R., Kahl, V., Horn, E. μ-Slide Chemotaxis: A new chamber for long-term chemotaxis studies. BMC Cell Biology. , (2011).
- Henke, E., Nandigama, R., Ergün, S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Frontiers in Molecular Biosciences. , (2020).
- Wirtz, D., Konstantopoulos, K., Searson, P. C. The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nature Reviews. , (2011).
- Northcott, J. M., Dean, I. S., Mouw, J. K., Weaver, V. M. Feeling stress: The mechanics of cancer progression and aggression. Frontiers in Cell and Developmental Biology. , (2018).
- Wan, L., Neumann, C. A., LeDuc, P. R. Tumor-on-a-chip for integrating a 3D tumor microenvironment: chemical and mechanical factors. Lab Chip. 20 (5), 873-888 (2020).
- Braun, E., Bretti, G., Natalini, R. Mass-preserving approximation of a chemotaxis multi-domain transmission model for microfluidic chips. ArXiv. , (2020).
- Mahlbacher, G. E., Reihmer, K. C., Frieboes, H. B. Mathematical modeling of tumor-immune cell interactions. Journal of Theoretical Biology. , (2019).
- Magidson, V., Khodjakov, A. Circumventing photodamage in live-cell microscopy. Methods in Cell Biology. , (2013).
- Jensen, E. C. Use of Fluorescent Probes: Their Effect on Cell Biology and Limitations. Anatomical Record. , (2012).
- Skylaki, S., Hilsenbeck, O., Schroeder, T. Challenges in long-term imaging and quantification of single-cell dynamics. Nature Biotechnology. , (2016).
- Abbitt, K. B., Rainger, G. E., Nash, G. B. Effects of fluorescent dyes on selectin and integrin-mediated stages of adhesion and migration of flowing leukocytes. Journal of Immunological Methods. , (2000).
- Smith, E., et al. Phototoxicity and fluorotoxicity combine to alter the behavior of neutrophils in fluorescence microscopy based flow adhesion assays. Microscopy Research and Technique. , (2006).
- Suman, R., et al. Label-free imaging to study phenotypic behavioural traits of cells in complex co-cultures. Scientific Reports. , (2016).
- Brent, R., Boucheron, L. Deep learning to predict microscope images. Nature Methods. , (2018).
- Christiansen, E. M., et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell. , (2018).
- Waibel, D. J. E., Tiemann, U., Lupperger, V., Semb, H., Marr, C. In-silico staining from bright-field and fluorescent images using deep learning. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). , (2019).
- Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F., Johnson, G. R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nature Methods. , (2018).
- Diehl, M. I., Wolf, S. P., Bindokas, V. P., Schreiber, H. Automated cell cluster analysis provides insight into multi-cell-type interactions between immune cells and their targets. Experimental Cell Research. 393 (2), 112014 (2020).
- Chen, H., Engkvist, O., Wang, Y., Olivecrona, M., Blaschke, T. The rise of deep learning in drug discovery. Drug Discovery Today. , (2018).
- Angermueller, C., Pärnamaa, T., Parts, L., Stegle, O. Deep learning for computational biology. Molecular Systems Biology. , (2016).
- Moen, E., Bannon, D., Kudo, T., Graf, W., Covert, M., Van Valen, D. Deep learning for cellular image analysis. Nature Methods. , (2019).
- Bock, C., Farlik, M., Sheffield, N. C. Multi-Omics of Single Cells: Strategies and Applications. Trends in Biotechnology. , (2016).
- Lin, A., et al. 3D cell culture models and organ-on-a-chip: Meet separation science and mass spectrometry. Electrophoresis. , (2020).
- Ingber, D. E. Developmentally inspired human ‘organs on chips.’. Development. , (2018).
- Low, L. A., Mummery, C., Berridge, B. R., Austin, C. P., Tagle, D. A. Organs-on-chips: into the next decade. Nature Reviews Drug Discovery. , (2020).
- Mangul, S., et al. Systematic benchmarking of omics computational tools. Nature Communications. , (2019).
- Burek, P., Scherf, N., Herre, H. Ontology patterns for the representation of quality changes of cells in time. Journal of Biomedical Semantics. 10 (1), 16 (2019).
- Benam, K. H., et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nature Methods. , (2016).
- Horning, S. J. A new cancer ecosystem. Science. , (2017).

