Detection of Trypanosoma brucei Variant Surface Glycoprotein Switching by Magnetic Activated Cell Sorting and Flow Cytometry
Summary
African trypanosomes grown in vitro undergo antigenic variation at a low rate, such that populations are made up of parasites expressing a dominant variant surface glycoprotein (VSG) type and a small population of “switched” variants. This protocol describes a fast method for detecting and quantifying these populations.
Abstract
Trypanosoma brucei, a protozoan parasite that causes both Human and Animal African Trypanosomiasis (known as sleeping sickness and nagana, respectively) cycles between a tsetse vector and a mammalian host. It evades the mammalian host immune system by periodically switching the dense, variant surface glycoprotein (VSG) that covers its surface. The detection of antigenic variation in Trypanosoma brucei can be both cumbersome and labor intensive. Here, we present a method for quantifying the number of parasites that have ‘switched’ to express a new VSG in a given population. The parasites are first stained with an antibody against the starting VSG, and then stained with a secondary antibody attached to a magnetic bead. Parasites expressing the starting VSG are then separated from the rest of the population by running the parasites over a column attached to a magnet. Parasites expressing the dominant, starting VSG are retained on the column, while the flow-through contains parasites that express a new VSG as well as some contaminants expressing the starting VSG. This flow-through population is stained again with a fluorescently labeled antibody against the starting VSG to label contaminants, and propidium iodide (PI), which labels dead cells. A known number of absolute counting beads that are visible by flow cytometry are added to the flow-through population. The ratio of beads to number of cells collected can then be used to extrapolate the number of cells in the entire sample. Flow cytometry is used to quantify the population of switchers by counting the number of PI negative cells that do not stain positively for the starting, dominant VSG. The proportion of switchers in the population can then be calculated using the flow cytometry data.
Introduction
The protozoan parasite, Trypanosoma brucei, is a causative agent of diseases that affect humans (via Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense) and animals (via Trypanosoma brucei brucei) throughout Sub-Saharan Africa. It is transmitted to the mammalian host through the saliva of the tsetse fly vector. Both Human and Animal African Trypanosomiasis cause a severe economic burden in endemic regions, and few drugs are available or in development to treat either disease. Understanding mechanisms of immune evasion is critical for the development of drugs for trypanosomiasis. Antigenic variation of the dense, Variant Surface Glycoprotein (VSG) coat that covers the parasite is one of the primary means by which T. brucei evades the mammalian antibody response. Approximately 2,000 variants of the VSG gene exist in the trypanosome genome, but only one is transcribed at any given time from one of ~15 expression sites. 'Switching' of the expressed VSG can occur either by copying a VSG gene into the active expression site, or by transcriptional activation of a previously silent expression site (reviewed in1).
Though much work has been devoted to understanding antigenic variation in T. brucei, the factors that influence switching frequency are still poorly understood. Studies to date have been hampered by the fact that while switching occurs stochastically in vitro, switching frequencies are very low, on the order of 1 in around 106 cells2. This makes it difficult to measure whether a given factor increases or decreases switching frequency, since switching is hard to detect in the first place. Prior to 2009, methods for isolating switchers in a given population were lengthy and labor intensive. These included passaging trypanosomes through mice immunized against the dominant VSG and then harvesting cells a day later3, or counting hundreds of cells by immunofluorescence4,5. Another strategy relies on selection for drug resistance to isolate switchers6. Because African trypanosomes grown in vitro are typically made up of a large population expressing one major variant, and a much smaller population of switchers expressing alternate variants, we refer to the major variant throughout this paper as the dominant, starting VSG. In doing so, we in no way wish to imply that this major variant has greater fitness than other variants in the population.
Here we describe a method that can reliably measure the number of trypanosomes expressing a non-dominant VSG in a given population in 3 – 4 hr. This method is particularly useful for when one wants to ascertain whether a given genetic manipulation or drug treatment increases the number of switched cells in a population. Instead of ridding the population of cells expressing the dominant, starting VSG through drugs or by immunological means, these cells are eliminated by first coating them with magnetic beads coupled to antibody against the dominant VSG and then isolating them on a magnetic column. The switched population is then collected in the flow-through and stained again with a fluorophore labeled anti-VSG antibody to identify contaminants. Quantification is achieved by adding a defined number of absolute counting beads to each sample so that the ratio of beads to cells can be determined and used to quantify the number of switchers in the population7.
Protocol
NOTE: Throughout the procedure, it is necessary to keep cells on ice. Cold media should also be used throughout.
1. Sample Harvesting
- Grow Lister 427 strain bloodstream trypanosomes to a density of 0.5 – 1 million/ml. It is best to start cultures with a small number of parasites. Spin down 50 x 106 cells/sample for 10 min at 1,500 x g. Be sure to leave 1 x 106 cells in culture for later use as positive and negative antibody controls.
NOTE: This protocol can also be used on trypanosomes isolated from animal blood (see discussion for details). - Pipette or pour off most of the supernatant (leave about 750 µl). Transfer the cells to a 1.5 ml microcentrifuge tube.
- Spin cells at 4 °C for 4 min at 5,200 x g in a microcentrifuge and remove supernatant.
2. Magnetic Labeling
- Resuspend cells in 150 µl culture medium (HMI-9 with serum for example) + primary anti-VSG antibody at the appropriate dilution.
NOTE: The anti-VSG antibody is made in house and used at a dilution of 1:50. It is important to use a primary anti-VSG antibody that is not labeled with a fluorophore. - Vortex cells using a vortex adapter-60 at speed 6 – 8 in a cold room at 4 °C for 10 min. Wash with 800 – 1,000 µl cold HMI-9 with serum.
- Spin at 4 °C for 4 min at 5,200 x g and aspirate the supernatant. Wash with 1 ml HMI-9 with serum and spin as above. Aspirate the supernatant. Resuspend pellet in 100 µl of HMI-9 with serum.
- Add 110 µl of magnetic-activated cell sorting (MSC) microbeads. Use anti-mouse, anti-rabbit, or anti-biotin beads as appropriate for the primary anti-VSG antibody. Mix well. Vortex cells in a cold room at 4 °C for 10 min.
- While cells are vortexing, set up one magnetic-activated separation column/sample on a separation magnet. Be sure to have a receptacle (we use a 15 ml centrifuge tube) under the column to collect the flow-through. Set up the apparatus in a cold room.
- Add 2 ml of HMI-9 with serum to prime column. After priming place a 15 ml centrifuge tube under the column to collect flow-through from each sample.
- After the 10 min incubation in step 2.4, add 800-1,000 µl cold HMI-9 with serum. Spin at 4 °C for 4 min at 5,200 x g and remove supernatant to remove unbound antibody. Wash with 1 ml of HMI-9 with serum and repeat leaving 100 µl of the supernatant.
- Flick the centrifuge tube vigorously and visually inspect the tube to make sure that the pellet is completely resuspended. Add 1 ml of HMI-9 with serum and pipette up and down to make sure that cells are well resuspended.
3. Magnetic Separation
- Ensure that the columns are primed as described above. Apply cells to column. Collect flow-through with cells expressing the non-dominant VSG. 1 ml of medium + trypanosomes typically takes 6 – 7 min to flow through the column.
- Wash 2x with 1 ml of HMI-9 with serum and collect flow-through in the same tube as in step 3.1.
- Divide flow-through (3 ml) into two 1.5 ml microcentrifuge tubes.
NOTE: These can be combined later or half can be used to isolate RNA, etc. - Remove column from magnet. Add 3 ml of HMI-9 with serum to column and plunge column into a 15 ml centrifuge tube to obtain cells that express the starting, dominant VSG. Remove 300 µl of eluted material for later analysis. Keep these cells on ice.
- For use as a positive and a negative control, take approximately 1 million cells from the starting culture of control cells growing at 37 °C (those not expected to switch frequently) and pipette them into a 1.5 ml microcentrifuge tube.
NOTE: Positive control cells will be stained with fluorophore-labeled antibody in step 4.1. Negative control cells remain unstained throughout the rest of the protocol. - Spin flow-through samples and positive antibody control sample at 4 °C for 4 min at 5,200 x g and remove supernatant.
NOTE: Another 300 µl aliquot of eluted cells can be used as a positive control for anti-VSG staining.
4. Staining to Identify Contaminants and Switchers
- Resuspend flow-through samples and positive control cells in 100 µl of HMI-9 with serum + primary, fluorophore-labeled, anti-VSG antibody at the appropriate dilution.
NOTE: The labeled anti-VSG antibody is made in house and used at a dilution of 1:1,000. The fluorophore-labeled, anti-VSG can be from the same source as the unlabeled antibody in the MACS separation step.There should be 3 ml of flow-through sample in two microcentrifuge tubes following step 3.5. - To analyze all of the flow-through by flow cytometry, combine the pellets in the two microcentrifuge tubes while re-suspending the samples in 100 µl of HMI-9 with serum + anti-VSG antibody. Mix well.
- Vortex cells in a cold room at 4 °C for 10 min. Add 800 – 1,000 µl cold HMI-9 with serum. Spin samples at 4 °C for 4 min at 5,200 x g and remove supernatant to remove unbound antibody.
- Wash cells with 1 ml of HMI-9 with serum. Spin samples at 4 °C for 4 min at 5,200 x g and remove supernatant. During this last spin, spin down the eluted-samples from step 3.4 and the negative control sample.
- Resuspend all flow-through and eluted samples in: 148.5 µl HMI-9 with serum, 25 µl absolute counting beads and 1.5 µl Propidium Iodide staining solution (175 µl total). Resuspend positive and negative antibody control samples in 175 µl HMI-9 with serum. Be sure to record the number of absolute counting beads/25 µl for the particular lot used.
- Run all samples through a flow cytometer and collect data. Dead cells stain as PI+. Contaminants stain as PI–VSG+. Switchers stain as PI–VSG– 8.
- Calculate switching frequencies using flow cytometry data.
NOTE: For instructions on how to calculate switching frequencies, please refer to the supplemental file Flow Cytometry Calculations.docx and Table 1.
Representative Results
The method described here was used to demonstrate that double-strand breaks within the VSG expression site increased the number of switchers in a population8. Here, we show representative results from a population of trypanosomes that have been similarly induced to generate a double-strand break at the expression site. We compare these trypanosomes to those that have not been induced to generate a double-strand break. Figure 1 shows representative flow cytometry plots from un-induced and induced cells collected in the flow-through from the magnetic activated cell sorting column. For this particular experiment the dominant starting VSG was VSG2. The left-hand panels of Figure 1A show forward and side scatter, which allows gates to be drawn around the absolute counting beads and cells that show a forward and side scatter signature that is characteristic of live cells. Note that gates can be drawn to include more cells because dead cells can be eliminated with PI staining; however, the gate must be kept the same for all the samples. The right-hand panels of Figure 1A show only those cells that fall within the 'Live' gate on the left-hand panels. Cells that stain positively for PI (Q1 and Q2) are dead cells. Cells that stain positively for the dominant VSG and negatively for PI in Q3 are contaminants. Cells that stain negatively for both PI and the dominant VSG in Q4 are live cells that have switched. One can observe that the percent of contaminants in Q3 is lower for those cells where a double-strand break has been induced. However, the percentages in Q3 cannot be used to infer whether there are more switchers in a given population. It is only by comparing the ratio of the number of cells in Q4 to the number of beads collected that one can calculate switching frequencies. Table 1 shows the numbers obtained from the flow cytometry plots and the method to calculate the percent switchers in these populations. These calculations were performed on 3 biological samples to obtain the observed switching frequencies displayed in Figure 1B. The level of stochastic switching in vitro is quite low, as calculated here at an average of 9.53 x 10-5, while inducing a break results in 1 – 6 x 10-3 cells that have switched to the non-dominant VSG.
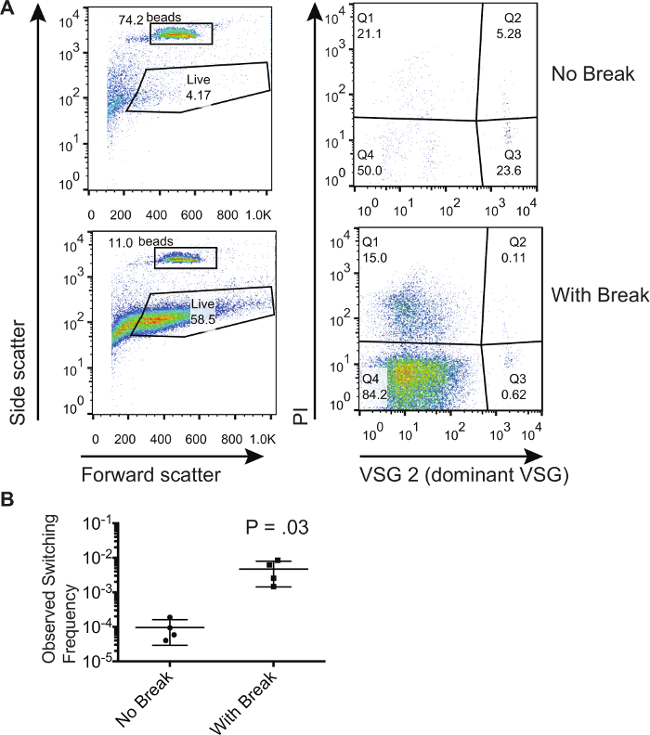
Figure 1. Isolation and Quantification of Switched Trypanosome Populations. (A) Flow cytometry showing fractions of trypanosomes in the flow-through fraction following selection of non-switchers on a magnetic activated cell sorting column. A population of cells with an induced I-SCEI double-strand break is compared to a population of cells without an induced break. (B) Calculated switching frequencies for cells that have or have not been induced with an I-SCEI double-stranded break. The number on the plot represents the results of a two-tailed, unpaired T test. Error bars represent SD. Please click here to view a larger version of this figure.
Supplemental File 1. Flow Cytometry Calculations. Please click here to download this file.

Table 1. Calculation of Switching Frequencies. Please click here to view a larger version of this table.
Discussion
With respect to experimental technique, the most critical component of the protocol is keeping all the samples cold. Trypanosomes very rapidly internalize antibody bound to their surface9, but this process is motility dependent, and does not influence the assay as long as the cells are kept at 4 °C. All samples should always be kept on ice, and pipetting should be done quickly to minimize exposure to the 25 °C lab environment. Cold HMI-9 with serum should be available at the start of the experiment and should be kept on ice throughout. Isolation of the trypanosomes by running them over the column must be done in a cold room, as it takes 5 – 7 min to run a 1 ml sample through the column. If there are more samples than magnetic separation units available, magnetic isolation can be done in sequence, as long as samples not being actively separated are kept on ice.
It is also critical that samples be very well mixed after the last wash following incubation with the microbeads and prior to isolation on the magnet. This can be accomplished by aggressive flicking of the centrifuge tube or light vortexing. No pellet or clumps of cells should be visible prior to addition of the sample to the separation column, which can be checked by looking at the light microscopy level. If clumps are present, the level of contamination in the flow-through sample of cells expressing the dominant VSG is usually much higher than desirable. When performing the calculation for the number of switchers in the sample at the end of the protocol, it is important to take into account whether all the flow-through was used for flow cytometric analysis, or whether half was used. If only half was used, the total number of switchers calculated should be multiplied by 2.
When removing the supernatant during the wash steps, it is not necessary to pipette off every last drop, and typically 15 – 25 µl are left surrounding the pellet of cells. This is especially critical during the final stages of staining and washing, because at this point pellets are rarely seen in the flow-through samples because there are so few switched cells.
While the final anti-VSG stain is typically performed with purified, fluorophore-labeled anti-VSG antibody, the first anti-VSG stain prior to magnetic isolation can be done with purified antibody or serum. We have also used bioreactor supernatant derived from hybridomas expressing anti-VSG antibodies. Regardless, the material used for the initial stain must be titrated to ensure good separation of cells that do or do not express the dominant, starting VSG.
It is typical for the population of cells in the flow-through to be contaminated with cells expressing the dominant VSG. While this contamination should be minimal, in our experience it is rare for the population to be completely devoid of cells expressing the dominant VSG. It is also important to note that cells that have gone through the isolation procedure do not stain as brightly with anti-VSG antibody as those that have not been through the procedure. We hypothesize that two things could account for this difference. The first is that spinning trypanosomes leads to shedding of VSG, and there are many spins involved in the isolation procedure. This would be expected to decrease the intensity of the signal for the final anti-VSG stain. The second is that the first step of the protocol involves staining with a primary anti-VSG antibody. If the epitopes recognized by the antibody used in this first step are the same as or occlude the epitopes recognized by the antibody in the final stain, one would expect the intensity of the signal to be lower than if only one antibody were used. For this reason it is important to titrate the antibody that is used in the final step, such that the signal from a positively stained cell is roughly two orders of magnitude higher than for a negatively stained cell. This way, even if the intensity of signal from a positively staining cell is decreased in cells that have undergone the isolation procedure, these positive cells will still be well separated from the negative cells on the flow cytometry plot. If auto-compensation is being used, single stain control samples should be prepared. Antibodies that we have generated are available for purchase from the Memorial Sloan Cancer Center monocolonal antibody facility. We have had the best success with using unlabeled monoclonal IgG antibody. We have carried out the protocol with polyclonal VSG antibodies, but polyclonal antibodies can occasionally recognize other VSGs so monoclonal antibodies are preferred. VSG identity can be determined by sequencing VSG cDNA generated from amplification of conserved sequences in the VSG 3'UTR and the spliced leader. A particular antibody can be tested for specificity on previously isolated switched cells who's VSG identity has been determined as just described.
The protocol presented here has a number of advantages. It can be used both to isolate switchers for later analysis and to quantify the number of switchers in a given population. It also can be used to isolate a particular variant of interest, as long as an antibody is available for that variant. It is also possible to carry out this protocol using trypanosomes isolated from animals, provided that red blood cells are eliminated using anti-Ter119 coated-magnetic beads according to the manufacturer's protocol. We have not tested the lower limit for the number of trypanosomes required, but we have successfully carried out the protocol using 2.5 – 10 million trypanosomes from 250 µl of blood. Finally, the protocol can be used in conjunction with fluctuation analysis to obtain the frequency of switching in a given population.
The isolation procedure is quite fast, taking 3 – 4 hr from start to finish depending on the number of samples, and thus is more efficient than previous methods that required prior immunization of mice or specialized strains containing drug resistance markers. The method is limited by the fact that antibodies against the starting VSG are required, however. Without such reagents, an alternate method such as VSG-seq might be a more appropriate choice to gauge which VSGs are being expressed in a given population10.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge George Cross for general advice on trypanosome biology. This work was also supported by a Bill and Melinda Gates Foundation GCE grant to DS, a NSF Graduate Research Fellowship (DGE-1325261) to MRM and an NIH/NIAID (grant #AI085973) to FNP. We thank Galadriel Hovel-Miner for use of the strain containing the I-SCEI gene and recognition site.
Materials
| unlabeled anti-VSG antibody | n/a | n/a | VSG antibody is made in house |
| fluorophore labeled anti-VSG antibody | n/a | n/a | VSG antibody is made in house |
| HMI-9 media | n/a | n/a | HMI-9 is made in house |
| Propidium Iodide | BD Pharmingen | 556463 | |
| CountBright absolute counting beads | Thermofisher Scientific | C36950 | |
| LD columns | Miltenyi Biotech | 130-042-901 | |
| MACS magnet | Miltenyi Biotech | 130-042-303 | |
| MACS magnetic separator | Miltenyi Biotech | 130-042-302 | |
| vortex adapter-60 | ThermoFisher scientific | AM10014 | |
| flow cytometer | Coulter | n/a | |
| flow cytometer analysis software | FloJo | n/a |
Referenzen
- Horn, D. Antigenic variation in African trypanosomes. Molecular & Biochemical Parasitology. 195 (2), 123-129 (2014).
- Lamont, G. S., Tucker, R. S., Cross, G. A. Analysis of antigen switching rates in Trypanosoma brucei. Parasitology. 92 (Pt 2), 355-367 (1986).
- Rudenko, G., McCulloch, R., Dirks-Mulder, A., Borst, P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 80 (1), 65-75 (1996).
- Turner, C. M., Barry, J. D. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 99 (Pt 1), 67-75 (1989).
- Miller, E. N., Turner, M. J. Analysis of antigenic types appearing in first relapse populations of clones of Trypanosoma brucei. Parasitology. 82 (1), 63-80 (1981).
- Horn, D., Cross, G. A. Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol Biochem Parasitol. 84 (2), 189-201 (1997).
- Figueiredo, L. M., Janzen, C. J., Cross, G. A. M. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 6 (7), e161 (2008).
- Boothroyd, C. E., Dreesen, O., et al. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 459 (7244), 278-281 (2009).
- Engstler, M., Pfohl, T., et al. Hydrodynamic Flow-Mediated Protein Sorting on the Cell Surface of Trypanosomes. Cell. 131 (3), 505-515 (2007).
- Mugnier, M. R., Cross, G. A. M., Papavasiliou, F. N. The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science. 347 (6229), 1470-1473 (2015).

