Sedimentation Equilibrium of a Small Oligomer-forming Membrane Protein: Effect of Histidine Protonation on Pentameric Stability
Summary
Sedimentation equilibrium (SE) can be used to study protein-protein interactions in a physiological environment. This manuscript describes the use of this technique to determine the effect of pH on the stability of a homo-pentamer formed by the small hydrophobic (SH) protein encoded by the human syncytial respiratory virus (hRSV).
Abstract
Analytical ultracentrifugation (AUC) can be used to study reversible interactions between macromolecules over a wide range of interaction strengths and under physiological conditions. This makes AUC a method of choice to quantitatively assess stoichiometry and thermodynamics of homo- and hetero-association that are transient and reversible in biochemical processes. In the modality of sedimentation equilibrium (SE), a balance between diffusion and sedimentation provides a profile as a function of radial distance that depends on a specific association model. Herein, a detailed SE protocol is described to determine the size and monomer-monomer association energy of a small membrane protein oligomer using an analytical ultracentrifuge. AUC-ES is label-free, only based on physical principles, and can be used on both water soluble and membrane proteins. An example is shown of the latter, the small hydrophobic (SH) protein in the human respiratory syncytial virus (hRSV), a 65-amino acid polypeptide with a single α-helical transmembrane (TM) domain that forms pentameric ion channels. NMR-based structural data shows that SH protein has two protonatable His residues in its transmembrane domain that are oriented facing the lumen of the channel. SE experiments have been designed to determine how pH affects association constant and the oligomeric size of SH protein. While the pentameric form was preserved in all cases, its association constant was reduced at low pH. These data are in agreement with a similar pH dependency observed for SH channel activity, consistent with a lumenal orientation of the two His residues in SH protein. The latter may experience electrostatic repulsion and reduced oligomer stability at low pH. In summary, this method is applicable whenever quantitative information on subtle protein-protein association changes in physiological conditions have to be measured.
Introduction
Analytical ultracentrifugation1-5 is one of the most important methods to study interactions of macromolecules under physiological conditions, being accessible to both weak and strong interactions. The method is label-free and uses light absorption or interference, and even fluorescence optical systems can be used to access concentration ranges over several orders of magnitude6.
This method is especially useful since most biochemical processes depend on reversible interactions. The stoichiometry and strength of these interactions have to be quantitatively characterized to understand biological processes, and a number of methods exist for this purpose 7,8. However, transient interactions are difficult to study 9.
The choice of a method to characterize macromolecular interactions depends on its static or dynamic nature. In the first case, sedimentation velocity (SV) is used, where the rate of radial transport is measured and complexes are fractionated on the basis of differences in buoyant mass and shape.
In contrast, dynamic associations that are reversible on the time scale of the experiment cannot be physically separated. In this case, self- or hetero-interactions leading to non-covalent interactions are in an equilibrium that depends on the total protein concentration. These dynamic interactions can be studied by both sedimentation equilibrium (SE) and sedimentation velocity (SV) 10. However, the first method is simpler to perform and is described here. In SE, centrifugation is performed at a sufficiently low speed so that an equilibrium is reached between diffusion and sedimentation. At this point, the equilibrium profile of an optical signal (UV-VIS) as a function of radial distance, can be analyzed using pre-set thermodynamic models for associations11.
In the present paper, a sedimentation equilibrium study is presented of the self-association of a viral membrane protein that forms ion channels. Because of its hydrophobicity, the experiment is run in presence of detergent, and in this case the density of solvent has to be matched to that of the detergent. However, the protocol described would identical in the case of a water soluble protein, except that no solvent density matching would be required.
The protein used is encoded in the human respiratory syncytial virus (hRSV), an enveloped pneumovirus in the paramyxoviridae family that causes lower respiratory tract disease in infants, elderly and immunocompromised populations worldwide 12. Up to 64 million reported cases of hRSV infection and 160,000 deaths occur each year.
The hRSV genome transcribes 11 proteins, including the three membrane proteins F, G, and small hydrophobic (SH). SH protein is involved in the pathogenesis of RSV infection. RSV lacking the SH gene (RSVΔSH) was viable, caused formation of syncytia and grew as well as the wild-type (WT) virus 13-16. However, RSVΔSH virus replicated 10-fold less efficiently than the WT in the upper respiratory tract 15,16. Also, RSVΔSH virus was attenuated in in vivo mouse and chimpanzee models 13,17.
The SH protein is a 64 (RSV subgroup A) or 65 (RSV subgroup B) amino acids long type II integral membrane protein that accumulates mostly at the membranes of the Golgi compartment 18. SH protein has a single predicted a-helical transmembrane (TM) domain 19 which is highly conserved 20,21. The C- and N-terminal extramembrane domains are oriented lumenally/extracellularly and cytoplasmically, respectively.
Both synthetic TM domain (residues 18-43) and full length SH protein have been shown to form homopentamers in a variety of detergents. The homopentameric form is responsible for channel activity in planar lipid bilayers 22,23. The correct orientation of the TM monomers in the lipid bilayer was first determined using site specific infrared dichroism 23, which showed His-22 to be in a lumenal, close to inter-helical, orientation. The same TM domain orientation was confirmed by NMR studies that reconstructed the pentameric a-helical bundle of the full-length protein in dodecylphosphocholine (DPC) micelles 22. In this ‘micelle’ model, a single a- helical TM domain was flanked N-terminally by an a-helix, and C-terminally by an extended b-hairpin. The two protonatable residues of SH protein, His-22 and His-51, are located in the TM domain (lumenally oriented), and at the tip of the extramembrane C-terminal β hairpin (far from the channel pore), respectively. In a bicellar environment, however, the TM α-helix extends up to His-51, and both His residues are accessible to the lumen of the channel24. The channel structure adopts a funnel-like architecture 22, where the narrower region (Ser-29 to Cys-45) 22 is lined with hydrophobic side chains (Ile-32, Ile-36, Ile-40 and Leu-44), and Ile-36 defines the narrowest point in the channel lumen. His-22 is located at the largest opening of this funnel, whereas His-51 is at the tip of the smallest opening.
In the present paper, analytical centrifugation in a sedimentation equilibrium mode has been used to determine if His protonation affects the stability of the SH protein pentamer. In this case, SH protein was solubilized in C14-betaine detergent, which has been used previously to show that SH protein forms pentameric oligomers22.
Protocol
This protocol is based on the following resources, which are to be referred for more details and special considerations3,25-28.
1. Density matching of detergent micelles with 2H2O
Note: The density of the buffer solution needs to be matched to the density of the detergent micelles. Common density-adjusting agents include 2H2O, H218O, 2H218O, glycerol and sucrose29. H218O has the same density as 2H2O and may be a better choice if deuteration of exchangeable protons in the protein is not desired. In this procedure, the density of 3-(N,N-dimethylmyristylammonio)-propanesulfonate (C14SB) detergent in 50 mM Tris pH 7.3, 100 mM NaCl will be matched with 2H2O. As an initial guess the following concentrations of 2H2O will be used: 10, 30, and 50% v/v.
1.1. Sample preparation
- Prepare the following stock solutions and filter sterilize through a 0.2 μm syringe filter: 50 mL 500 mM Tris pH 7.3 and 1 M NaCl (10X buffer solution); 1 mL 250 mM C14SB (50X detergent solution).
- Prepare 200 μl of sample solution by mixing 20 μl 10X buffer solution, 4 μl 50X detergent solution, 20 μl 2H2O (99.9%), and 156 μl deionized H2O. Prepare also 200 μl of reference solution by mixing 20 μl 10X buffer solution, 20 μl 2H2O (99.9%), and 160 μl deionized H2O.
- Repeat step 1.1.2 for the other 2H2O concentrations, i.e. 30% and 50%, adjusting the 2H2O and H2O amounts appropriately.
1.2. Assembly of 6-channel AUC cells and sample loading into the cells.
Note: There are two types of AUC cell depending on the sample loading method. Cells without external fill has to be loaded prior to sealing the cell, whereas external-fill cells can be loaded after the cells are sealed. Assembly of an external-fill AUC cell has been described previously3. In this protocol, the assembly of a 6-channel AUC cell without external fill is described. The main difference is that it has screw rings on both sides which need to be tightened separately, and it doesn’t need housing plugs (Fig. 1). The difference in assembly steps are highlighted below.
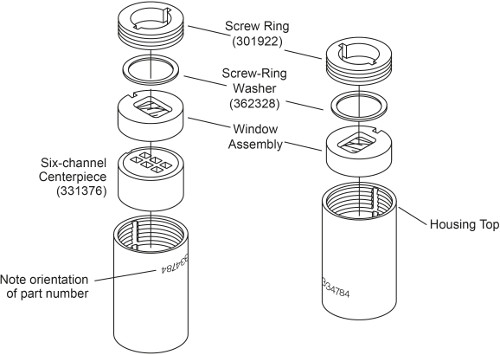
Figure 1. Exploded view of a 6-channel AUC cell without external fill. This figure has been modified from Beckman Coulter An-50 Ti and An-60 Ti Analytical Rotor, Cells, and Counterbalance user manual.
- Prepare two window assemblies for each AUC cell with sapphire window instead of quartz window (Fig. 2). Place the window gasket into the window holder. Slightly bend the window liner and place it into the window holder such that the gap is formed opposite to the window holder keyway. Place the sapphire window inside the window liner, aligning the mark with the window holder keyway.
Note: The quartz window is compressible and thus will produce more light refraction at high speed 28, 30. Therefore for interference measurements above 30000 rpm, such as in this density matching experiment, sapphire windows are used. A sapphire window is heavier than quartz window and has an “X” etched onto its side.
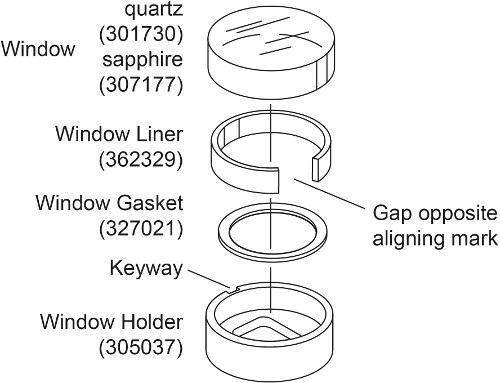
Figure 2. Exploded view of the window assembly. This figure has been modified from Beckman Coulter An-50 Ti and An-60 Ti Analytical Rotor, Cells, and Counterbalance user manual
- Place the cell housing with the part number upside-down. With the keyways aligned with the housing key, slide into the cell housing firstly a 6-sector centerpiece with beveled side down, followed by one window assembly with the window facing down (Fig. 1, left).
- Lightly coat the screw ring threads and screw ring washer with spinkote. Place a screw ring washer on top of the window assembly. Install the screw ring into the window housing with the word “OUT” facing outside. Hand-tighten the screw ring by using the cell aligning tool.
- Using the torque wrench, tighten the screw ring to only 60 inch-pounds.
- Place the cell with the part number upright and positioned at 12-noon. Load 120 μl reference into the left rows and 110 μl sample into the right rows. Ensure that each sample and reference is correctly paired.
Note: Exact sample volume is not critical, but the reference needs to have slightly more volume than the sample (5-10 μl) so that the sample meniscus will be distinct. - Carefully slide into the cell housing one window assembly with the window facing down (Fig. 1, right). Take care not to disturb the cell excessively and spill the contents.
- Repeat step 1.2.3 and 1.2.4, tightening the second screw ring to 120 inch-pounds. Invert the cell and re-tighten the first screw ring to 120 inch-pounds.
- Load the cells into the rotor, install the rotor into the centrifuge and install the monochromator according to the manufacturer’s instructions 28.
Note: Details on this step can also be found in this reference 3.
1.3. Setting up interference measurement
- Start up the user interface software for the AUC instrument and perform laser setup and radial calibration for each cell at 3000 rpm, according to the manufacturer’s instructions, which will be briefly summarized in the following step.
- 1.3.1.1 After sufficient vacuum has been reached (< 100 microns), run the centrifuge at 3000 rpm. Preview the interference pattern in the user interface software and adjust the laser parameters to obtain the highest contrast.
- Create a new set up file (File | New File) specifying “Equilibrium” and “Interference” measurement. Set up a sedimentation equilibrium method (“Method” button) to run at 45000 rpm or the highest speed anticipated for protein samples, whichever is higher, with run temperature at 20°C and collect 1 scan every 15 minutes. Monitor equilibrium progress by opening the data files in HeteroAnalysis and selecting “Match” function after at least 12 hours (approximately overnight, Fig. 3).
Note: a similar function is also available in SEDFIT (Options | Loading Options | Test Approach to Equilibrium).
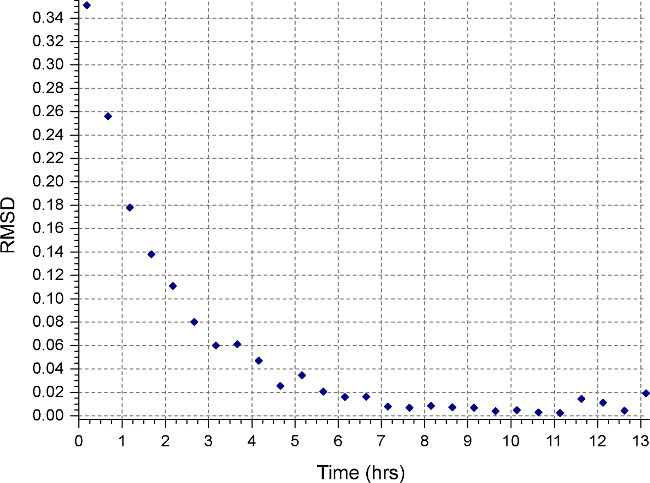
Figure 3. Result from HeteroAnalysis Match function. The Match function can be used to monitor equilibrium progress by comparing RMSD between successive scans and the last scan. This example shows attainment of equilibrium after 8 hr as indicated by RMSD values asymptotic to X-axis.
1.4. Data analysis
- For each set of samples, plot the slope of the radial distribution profile against the 2H2O concentration.
Note: The distribution will be a very shallow exponential that approaches linearity. X-axis intercept corresponds to the matching 2H2O concentration. - For more accurate results, perform the experiment in several replicates. Alternatively, repeat the experiment with a narrower range of 2H2O concentration.
2. Sedimentation equilibrium of SH in C14SB micelles
2.1. Run parameters
- Calculate buffer density and viscosity, protein partial specific volume and centrifugation speed by using SEDNTERP. To calculate buffer density and viscosity, select Compute in “Buffer Data Select” section and enter the buffer components accordingly, including the D2O concentration.
- 2.1.1.1 To calculate protein partial specific volume, select Compute in “V-bar” section and enter the protein amino acid sequence. Specify the highest expected oligomeric size in “Make an oligomer from this monomer: N=” field, in this case N = 5. Calculate the speed by entering values in the RPM field on the main window until σ ≈ 1; this is a rule of thumb to ensure a good exponential shape of the radial distribution profile25.
Note: The values calculated for this experiment was as follows: ρ = 1.03839 g/ml, η = 1.0267 cP = 0.7569 ml/g, ω1 = 16000 rpm. - Calculate subsequent speeds to follow to ensure sufficient difference between the distribution profile at one speed and the next25.
Note: this can also be done from the function "Estimate equilibrium rotor speeds function" in SEDFIT, which takes into account the solution column (filling volume).
2.2. Sample preparations
- Prepare 1 ml reference solution with 5 mM C14SB and 32.3% 2H2O as determined from density matching experiment (section 1), by mixing 100 μl 10X buffer solution (step 1.1.1), 20 μl 50X detergent solution (step 1.1.1), 323 μl 2H2O (99.9%) and 527 μl deionized H2O.
- Dissolve lyophilized, HPLC-purified SH peptides (expression and purification described previously31) in appropriate solvent such as methanol or 50% v/v aqueous acetonitrile. Measure A280 of the dissolved peptides in a microlitre-scale UV/Vis spectrophotometer and aliquot for three samples to give A280, 12mm = 0.3, 0.5, and 0.8 (A280, 10mm = 0.25, 0.417, and 0.67) each when diluted to 130 μl. Lyophilize the samples overnight and resuspend in 130 μl reference solution (step 2.2.1) to give the sample solutions.
Note: SH protein can be detected from UV/Vis absorbance at 280 nm because it contains Trp and Tyr residues. Proteins without aromatic residues can be detected by tagging them with a suitable chromophore, using a Trp-containing mutant, or by using interference measurements instead of absorbance. - Follow steps in section 1.2 to assemble a 6-channel AUC cell with quartz windows. Load the highest concentration sample (A280, 12mm = 0.8) in the channel nearest to the rotor center and lowest concentration sample (A280, 12mm = 0.3) furthest from the rotor center.
2.3. Setting up absorbance measurements
- Create a new set up file (File | New File) specifying “Equilibrium” and “Absorbance” measurement. Specify 280 nm as the detector wavelength.
- Perform radial calibration at 3000 rpm by checking “Radial calibration before first scan” in Scan Options, specifying data collection at low resolution, e.g. with Radial step size = 0.01 cm, Replicates = 3 (low resolution, fast), and executing a Single Scan. After the scan is completed, uncheck the option.
- Set up a sedimentation equilibrium method (“Method” button) to run at the first speed calculated in step 2.1.3, at 20°C, and collect 1 scan every 30 minutes. In each cells’ “Detail”, specify data collection at low resolution as in step 2.3.2. Monitor equilibrium progress by opening the data files in HeteroAnalysis and selecting “Match” function after at least 18 hours (approximately overnight, Fig. 4).
Note: attainment of equilibrium may take substantially longer time for the first speed, whereas subsequent speeds will take less time.
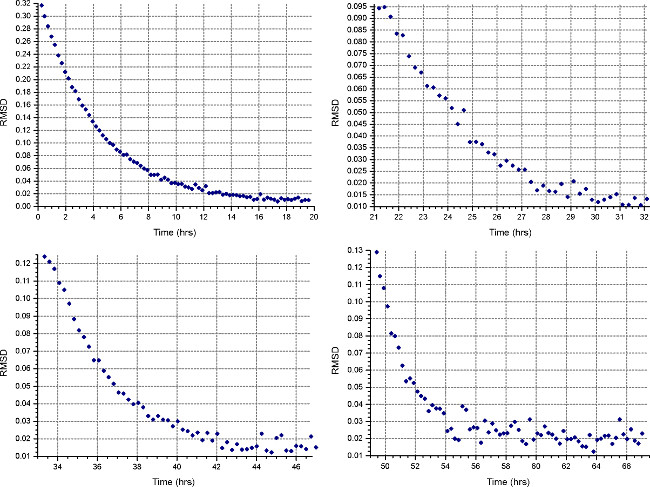
Figure 4. Results from HeteroAnalysis Match function. The first and second speeds (top left and right) appear to have reached equilibrium, but it is better to wait a few more hours to be sure. In comparison, the third and fourth speeds (bottom left and right) have clearly reached equilibrium in a shorter time. Please click here to view a larger version of this figure.
- Once equilibrium has been reached, collect a single scan at high resolution, e.g. with Radial step size = 0.001 cm, Replicates = 10 (high resolution, slow).
- After the scan is completed, repeat step 2.3.3 and 2.3.4 for the next speed.
- Optionally, when the time required to reach equilibrium for each speed is known (calculated or from experience), set up the sedimentation equilibrium method scan to include all the speeds calculated in step 2.1.3 and to collect 1 scan after the equilibration time for each speed. In this case, specify high resolution data collection in each cells’ “Detail”.
2.4. Data analysis in SEDFIT and SEDPHAT
Note: For further details and considerations in data analysis reader is referred to the following website: www.analyticalultracentrifugation.com.
- Open high resolution scans in SEDFIT (Data | load sedimentation equilibrium data) and split the data into 3 channels (these correspond to different concentrations for each sample; Options | Loading Options |Save 6-channel Raw Data in 3 Subsets).
- Re-open data files that belong to the same sample and same concentration but different speeds in SEDFIT. Adjust the meniscus (vertical red line), cell bottom (vertical blue line) and fitting limits (vertical green lines), and export the data for use in SEDPHAT (Data | Export Data to SEDPHAT). Input the parameters calculated in step 2.1.1, as well as rotor type and centerpiece type as requested. Repeat this step for every sample and concentration.
- Open all data from the same sample (all concentrations and speeds) in SEDPHAT and fill in Experiment Parameters; an example is shown in Fig. 5.
Note: when D2O is added in the buffer, deuteration of exchangeable protons could significantly alter the molecular weight of the protein, especially for water-soluble proteins. Membrane proteins, especially small ones like SH protein, are less affected because membrane-embedded regions are protected from exchange. To correct this, input the “buffer D mol fraction”.
Note: At this step it is recommended to save the edited dataset separately by selecting menu Data | Copy All Data And Save As New Config. - Select a Model and fill in Global Parameters for that model.
Note: As an example, the “Monomer-n-Mer Self-Association” model and its parameters are shown in Fig. 6.
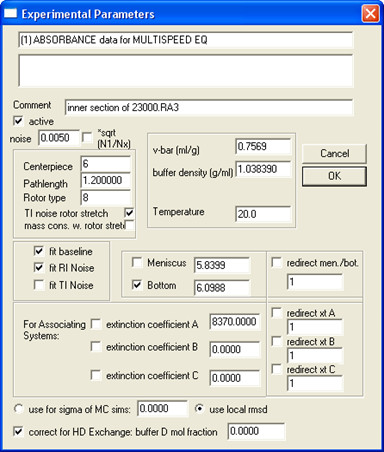
Figure 5. An example on how to fill in Experimental Parameters.
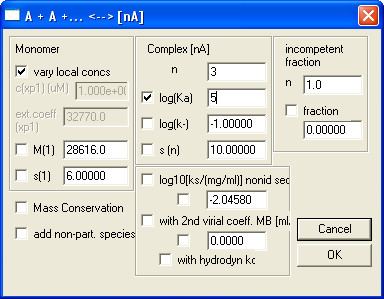
Figure 6. An example on how to fill in Global Parameters for Monomer-n-Mer Self-Association model.
- Run a Global Fit by selecting menu Fit | Global Fit and wait until the fit converges. Note down (or take a screenshot of) the fitting results, especially the global reduced chi-square and log Ka values. Extract other data such as fit data and fitting residuals from menu Copy and Display | Display Thermodynamic Information.
- Return to Global Parameters and check M(1) to fit the monomer molecular weight and repeat step 2.4.5. Note down the fitted molecular weight and global reduced chi-square.
- Repeat step 2.4.3 to 2.4.6 for each model to be tested and compare the fit quality of each model by comparing the reduced global chi-square value as well as fitting residuals.
Note: Small and random fitting residuals generally indicates a good fit, and the model that fits best would have the smallest global reduced chi-square. The fitted monomer molecular weight and its chi-square value should not differ substantially from that of the fixed (theoretical) molecular weight. - Calculate the confidence interval for the obtained log Ka by value by first selecting Statistics | Critical chi-square for error surface projections and inputting the desired confidence interval. Next, go to Statistics | Generate 1-dimensional error surface projection and unselect log Ka in Global Parameters dialog to obtain the chi-square values for log Ka.
Note: readers are advised to consult the following sources (http://www.analyticalultracentrifugation.com/ sedphat/statistics.htm) for more details on the method 32 as well as illustration of this method 33.
Representative Results
The radial distribution profile of C14SB detergent micelles in 50 mM Tris, 100 mM NaCl pH 7.3 forms a very shallow exponential that could be fitted to a linear model (Figure 7A). The slope of this distribution is inversely correlated to D2O concentration (Figure 7B). The point where the slope is zero, i.e., the matching D2O concentration, was found to be 32.3%.
See Figure 7 Below.
The same experiment was repeated for different buffers: 50 mM sodium phosphate, 100 mM NaCl pH 5.5 and 50 mM phosphate-citrate, 100 mM NaCl pH 3 to obtain matching D2O concentrations of 30.3% and 41.0%, respectively.
Samples of SH wild type (wt) in detergent were exposed to pH 3, 5.5 and 7.3 (total 6 samples), followed by centrifugation at 15,000, 19,000, 23,000, 28,000, 34,000 and 42,000 rpm. Data obtained at lower speed could not be reliably fitted, possibly because equilibrium had not been attained, therefore data obtained from the four higher speeds were used. At pH 7.3, SH WT was found to form pentamers (Figure 8 and Table 1) with apparent log Ka = 21.35 (Table 2). The association constant did not change at pH 5.5, but it was significantly reduced at pH 3. This is consistent with previous reports24, which reported a decrease of conductance at lower pH, with a pK of 4.5.
See Figure 8 Below.
See Table 1 Below.
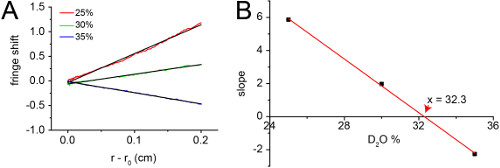
Figure 7. Density matching of C14SB with D2O. (A) Radial distribution profile formed by C14SB detergent micelles in 50 mM Tris, 100 mM NaCl pH 7.3 with 25, 30, and 35% D2O. The data was separately fitted to a linear model (black). (B) The slopes were plotted against D2O concentration (black squares) and fitted to a linear model (red line). The matching D2O concentration is indicated (red arrow).
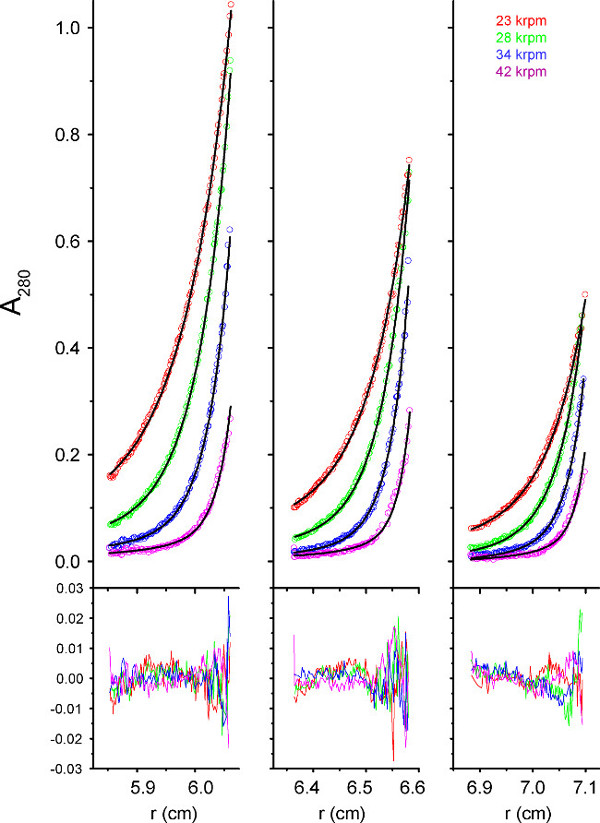
Figure 8. Fitting of SH in C14SB to monomer-pentamer model. Radial distribution profile of SH in C14SB at pH 7 (open circles) was found to fit best to the monomer-pentamer self-association model (black solid line). Fitting residual is shown below. Please click here to view a larger version of this figure.
| Model (n-mer) | Chi-square (fixed MW) | Chi-square (fitted MW) | Fitted MW |
| 3 | 15.444 | 1.0492 | 13477 Da |
| 4 | 3.8094 | 1.0469 | 9889 Da |
| 5 | 1.0499 | 1.0497 | 7822 Da |
| 6 | 2.5994 | 1.547 | 6504 Da |
| 7 | 6.1667 | 1.2112 | 5743 Da |
Table 1. Comparison of global reduced chi-square values of different monomer-n-mer models.
| pH | Lower limit (1σ) | Log Ka | Upper limit (1σ) |
| 3 | 17.432 | 17.576 | 17.737 |
| 5.5 | 20.064 | 20.419 | 20.839 |
| 7.3 | 20.687 | 21.052 | 21.492 |
Table 2. Comparison of apparent log Ka values at different pH.
Discussion
This paper provides an experimental protocol for sample preparation and analysis of oligomerization of a small membrane protein in detergent using equilibrium sedimentation. The protocol described is equally valid –and simpler- for soluble proteins, as the density matching step is not required. Indeed, the system is constituted by a mixture of detergent and protein. To conduct sedimentation studies, the detergent must be invisible to the gravitational field so that it does not contribute to the particle flotation. Thus, the density of the detergent has to be carefully matched by addition of D2O to the buffer, with the limitation that if the detergent used is too dense, e.g., SDS, not even 100 % D2O can match it. The density matching step is not required when working with a water soluble protein, since the sample does not have detergent.
In our application, a small viral protein that forms ion channels has been used. Like all viroporins, SH protein has an α-helical transmembrane domain, and therefore has to be studied in a detergent that will not disrupt its native oligomeric size.
Prior to these studies, therefore, suitable detergents have to be screened. For example, in previous SE studies SH protein formed pentameric oligomers in DPC, C8E5 and C14-betaine, and pentamers were also observed during electrophoresis in the mild detergent PFO22. All these detergents are suitable for SE, since their density can be matched by addition of D2O. Samples should also be tested by sedimentation velocity (SV), which provides the minimum number of species present34.
Also, unsuitable detergents will produce multiple species in SV, and multiple bands in PFO electrophoresis, indicative of multiple non-specific association. Useful information from SE is obtained when the predominant number of species present in the system is not higher than two; in the case of SH protein, the data was fitted to an equilibrium between monomers and pentamers.
It is important to note that the model chosen to fit the SE data, should also be as simple as possible, i.e., a single species of unknown molecular weight should be tried first, followed by reversible equilibria between monomer and oligomers of increasing size. More complex models have associated a higher ambiguity. Also, small populations of other smaller or larger oligomers cannot be detected, and the model will just indicate what are the predominant species present. In the case of viroporins, this is especially evident because oligomeric size changes depending on subtle experimental conditions, e.g., detergent used, pH, protein concentration, centrifugation speed or mutations, e.g, in Hepatitis C Virus p7 35, influenza A M2 36 or SH protein 24.
The results obtained for WT SH protein clearly show that the association constant of the pentamer is reduced at pH 3. These results match channel activity measurements obtained with SH protein in synthetic lipid bilayers, where conductance was markedly reduced at pH 3 (pK ~4.5), whereas it remained constant between pH 7 and 524. Based on these results, it may be possible that SH channel activity is somewhat regulated by lower pH when present in native biological membranes. Although in the Golgi lumen the pH is only one unit below that of the cytoplasm, intravesicular pH drops along the endocytic pathway from pH 6.0–6.5 in early endosomes to pH 4.5–5.5 in late endosomes and lysosomes 37. In the infected cell, the pK for conductance changes or pentameric stability may be higher than those obtained in vitro herein and previously24, therefore pH could indeed play a role in modulating channel activity during the life cycle of the virus. Mutation of protonatable His residues in the context of the infected cell is an interesting avenue for future experiments38.
Finally, the study of SH protein is facilitated by the presence of Trp in its sequence, facilitating UV absorption measurements. However, knowledge of the structure of the oligomer can enable introduction of Trp at insensitive locations, e.g., lipid –or solvent- facing parts of the protein in membrane – or soluble- proteins. Alternatively the protein may be tagged with a Visible-absorbing or fluorescent label.
In summary, this protocol describes the application of SE to determine oligomeric size and association constants of a defined viral channel, when experimental parameters are changed. In this case, pH has been varied to study the effect on stability of His protonation, but many other hypothesis can be tested, such as effects of mutations on the structural integrity of these oligomers.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work has been funded by the National Research Foundation grant NRF-CRP4-2008-02 (J.T.) and Tier 1 grant RG 51/13.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| 3-(N,N-dimethylmyristylammonio)propanesulfonate | Sigma | T0807 | |
| Deuterium oxide 99.8% | Cambridge Isotope | DLM-4-99.8 | |
| An-50 Ti Rotor, Analytical, 8-Place | Beckman Coulter | 363782 | |
| An-60 Ti Rotor, Analytical, 4-Place | Beckman Coulter | 361964 | |
| Cell housing | Beckman Coulter | 334784 | |
| 12 mm six-channel centerpiece, epon charcoal-filled | Beckman Coulter | 331376 | |
| Window holder | Beckman Coulter | 305037 | |
| Window gasket | Beckman Coulter | 327021 | |
| Window liner | Beckman Coulter | 362329 | |
| Sapphire window | Beckman Coulter | 307177 | |
| Quartz window | Beckman Coulter | 301730 | |
| Screw-ring washer | Beckman Coulter | 362328 | |
| Screw ring | Beckman Coulter | 301922 | |
| Spinkote | Beckman Coulter | 306812 | |
| Torque stand assembly | Beckman Coulter | 361318 | |
| Counterbalance | Beckman Coulter | 360219 | |
| Cell alignment tool | Beckman Coulter | 362340 | |
| SEDNTERP | http://bitcwiki.sr.unh.edu/index.php/Main_Page | ||
| HeteroAnalysis | http://www.biotech.uconn.edu/auf/?i=aufftp | ||
| SEDFIT | http://www.analyticalultracentrifugation .com/sedfit.htm |
||
| SEDPHAT | http://www.analyticalultracentrifugation .com/sedphat/default.htm |
Referenzen
- Laue, T. M., Stafford, W. F. Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 28, 75-100 (1999).
- Lebowitz, J., Lewis, M. S., Schuck, P. Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 11, 2067-2079 (2002).
- Balbo, A., Zhao, H., Brown, P. H., Schuck, P. . Assembly, Loading, and Alignment of an Analytical Ultracentrifuge Sample Cell. , e1530 (2009).
- Rivas, G., Stafford, W., Minton, A. P. Characterization of heterologous protein-protein interactions using analytical ultracentrifugation. Methods-a Companion to Methods in Enzymology. 19, 194-212 (1999).
- Howlett, G. J., Minton, A. P., Rivas, G. Analytical ultracentrifugation for association and assembly the study of protein. Curr. Opin. Chem. Biol. 10, 430-436 (2006).
- MacGregor, I. K., Anderson, A. L., Laue, T. M. Fluorescence detection for the XLI analytical ultracentrifuge. Biophys. Chem. 108, 165-185 (2004).
- Phizicky, E. M., Fields, S. Protein-Protein Interactions – Methods for Detection and Analysis. Microbiol. Rev. 59, 94-123 (1995).
- Alexandrov, A. A facile method for high-throughput co-expression of protein pairs. Mol. Cell. Proteomics. 3, 934-938 (2004).
- Nooren, I. M. A., Thornton, J. M. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 325, 991-1018 (2003).
- Ebel, C. Sedimentation velocity to characterize surfactants and solubilized membrane proteins. Methods. 54, 56-66 (2011).
- Minton, A. P. Quantitative characterization of reversible macromolecular associations via sedimentation equilibrium: an introduction. Exp. Mol. Med. 32, 1-5 (2000).
- Dowell, S. F. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174, 456-462 (1996).
- Bukreyev, A., Whitehead, S. S., Murphy, B. R., Collins, P. L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71, 8973-8982 (1997).
- Fuentes, S., Tran, K. C., Luthra, P., Teng, M. N., He, B. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81, 8361-8366 (2007).
- Jin, H. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 273, 210-218 (2000).
- Karron, R. A. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B. Proc. Nat. Acad. Sci. USA. 94, 13961-13966 (1997).
- Whitehead, S. S. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73, 3438-3442 (1999).
- Rixon, H. W. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85, 1153-1165 (2004).
- Collins, P. L., Mottet, G. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J. Gen. Virol. 74, 1445-1450 (1993).
- Collins, P. L., Olmsted, R. A., Johnson, P. R. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J. Gen. Virol. 71, 1571-1576 (1990).
- Chen, M. D., Vazquez, M., Buonocore, L., Kahn, J. S. Conservation of the respiratory syncytial virus SH gene. J. Infect. Dis. 182, 1228-1233 (2000).
- Gan, S. W. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J. Biol. Chem. 287, 24671-24689 (2012).
- Gan, S. W., Ng, L., Xin, L., Gong, X., Torres, J. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein Sci. 17, 813-820 (2008).
- Li, Y. Inhibition of the Human Respiratory Syncytial Virus Small Hydrophobic Protein and Structural variations in a bicelle environment. J. Virol. 88 (22), 11899-914 (2014).
- Burgess, N. K., Stanley, A. M., Fleming, K. G. Determination of membrane protein molecular weights and association equilibrium constants using sedimentation equilibrium and sedimentation velocity. Meth. Cell. Biol. 84, 181-211 (2008).
- Cole, J. L., Lary, J. W., Moody, T. P., Laue, T. M. Analytical Ultracentrifugation: Sedimentation Velocity and Sedimentation Equilibrium. Meth. Cell. Biol. 84, 143-179 (2008).
- Fleming, K. G. Determination of membrane protein molecular weight using sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Prot. Sci. 53, 17.12.11-17.12.13 (2008).
- . . An-50 Ti and An-60 Ti Analytical Rotor, Cells, and Counterbalance. , (2005).
- Mayer, G. Studying membrane proteins in detergent solution by analytical ultracentrifugation: Different methods for density matching. Prog. Colloid Polym. Sci. 113, 176-181 (1999).
- Laue, T. Ch. 20.3. Current Protocols in Protein Science. 20, 20.23.21-20.23.13 (2001).
- Gan, S. W. The Small Hydrophobic Protein Of The Human Respiratory Syncytial Virus Forms Pentameric Ion Channels. J. Biol. Chem. 287, 24671-24689 (2012).
- Bevington, P. R., Robinson, D. K. . Data reduction and error analysis for the physical sciences. 336, (1969).
- Schuck, P., Radu, C. G., Ward, E. S. Sedimentation equilibrium analysis of recombinant mouse FcRn with murine IgG1. Molecular Immunology. 36, 1117-1125 (1999).
- Gan, S. W., Vararattanavech, A., Nordin, N., Eshaghi, S., Torres, J. A cost-effective method for simultaneous homo-oligomeric size determination and monodispersity conditions for membrane proteins. Anal. Biochem. 416, 100-106 (2011).
- Montserret, R. NMR structure and ion channel activity of the p7 protein from hepatitis C virus). J. Biol. Chem. 285, 31446-31461 (2010).
- Stouffer, A. L., DeGrado, W. F., Lear, J. D. Analytical Ultracentrifugation Studies of the Influenza M2 Homotetramerization Equilibrium in Detergent Solutions. Progr Colloid Polym Sci. 131, 108-115 (2006).
- Sorkin, A., von Zastrow, M. Signal transduction and endocytosis: Close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3, 600-614 (2002).
- Gan, S. W., Ng, L., Lin, X., Gong, X., Torres, J. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein science : a publication of the Protein Society. 17, 813-820 (2008).

