Multi-step Preparation Technique to Recover Multiple Metabolite Compound Classes for In-depth and Informative Metabolomic Analysis
Summary
The reliability of results in metabolomics experiments depends on the effectiveness and reproducibility of the sample preparation. Described is a rigorous and in-depth method that enables extraction of metabolites from biological fluids with the option of subsequently analyzing up to thousands of compounds, or just the compound classes of interest.
Abstract
Metabolomics is an emerging field which enables profiling of samples from living organisms in order to obtain insight into biological processes. A vital aspect of metabolomics is sample preparation whereby inconsistent techniques generate unreliable results. This technique encompasses protein precipitation, liquid-liquid extraction, and solid-phase extraction as a means of fractionating metabolites into four distinct classes. Improved enrichment of low abundance molecules with a resulting increase in sensitivity is obtained, and ultimately results in more confident identification of molecules. This technique has been applied to plasma, bronchoalveolar lavage fluid, and cerebrospinal fluid samples with volumes as low as 50 µl. Samples can be used for multiple downstream applications; for example, the pellet resulting from protein precipitation can be stored for later analysis. The supernatant from that step undergoes liquid-liquid extraction using water and strong organic solvent to separate the hydrophilic and hydrophobic compounds. Once fractionated, the hydrophilic layer can be processed for later analysis or discarded if not needed. The hydrophobic fraction is further treated with a series of solvents during three solid-phase extraction steps to separate it into fatty acids, neutral lipids, and phospholipids. This allows the technician the flexibility to choose which class of compounds is preferred for analysis. It also aids in more reliable metabolite identification since some knowledge of chemical class exists.
Introduction
Biological reactions generate metabolites as end products of cellular processes. Metabolomics is a collection of all the compounds present in an organism as a result of these processes. It provides a picture of the physiology of cells and reflects an organism’s response to external or internal stimuli1,2. Such stimuli could be environmental, toxicological, pharmacological, dietary, hormonal, or related to disease. Many metabolomic applications have and are currently being studied by researchers and include biomarker discovery3, nutrition studies4, food science5, and drug testing6. Regardless of the application, variations in data, contamination, and presence of false positives need to be reduced or preferably removed. In biomarker discovery or in the case of determining differences between a control and a disease group, or investigating effects of drugs on subjects, a biological fluid is chosen based on the questions being asked and the types of metabolites being investigated7. For example, if studying the immediate effects of an inhaled drug on the lungs of asthmatics, then exploring metabolites in bronchoalveolar lavage fluid (BALF) samples before and following administration would be preferential. To ensure that observed differences are due to actual biological variation rather than improper sample preparation technique, standardized and consistent laboratory protocol is essential8. Sample information must be carefully documented to ensure that variables such as biological fluid, animal strain, sampling time, subject age, gender, to name a few, are all considered and factored into the study9. In addition, to reduce the possibility of contamination or false positives, it is recommended that solvent blanks and instrument blanks be analyzed10.
For this protocol, the term “metabolites” will be used to refer to the actual compounds identified. Using vendor software, an initial peak finding algorithm is used to detect mass spectral peaks. These peaks are aligned based on mass-to-charge (m/z) ratio and retention time. A second algorithm is then used to combine multiple features into a single compound. This includes such features as sodium, potassium, or ammonium adducts in the positive ionization mode, and chloride in the negative ion mode. Additional options in the software include features such as dimers and other adducts. Using glucose as an example, with peaks at 181.0707 m/z (M+H), 198.0972 m/z (M+NH4), and 203.05261 m/z (M+Na), there would be three peaks corresponding to the same compound using the first algorithm. However when the second algorithm, which is based on molecular formula, is applied these three adducts become grouped together resulting in one compound.
Metabolites can cause interferences within samples due to the complexity of compounds present. The presence of thousands of metabolites in one sample causes signal suppression particularly of the lower abundance metabolites. Sample cleanup to remove interfering proteins, and subsequent separation into multiple fractions reduces the complexity of the sample thereby improving peak separation, increasing resolution, and reducing metabolite coelution. Therefore, sample cleanup and improved separation of compounds is required. It has been shown that protein precipitation alone, even with the use of various polarity solvents, cannot resolve this issue11,12. However, by combining a strong organic solvent such as MTBE with a subsequent fractionation step, the metabolite coverage is increased. Yang et al.12 reported an increase in metabolites from 1,851 or 2,073 with methanol or methanol-ethanol precipitation alone respectively, to 3,806 metabolites using combined MTBE solvent extraction followed by solid-phase extraction (SPE) steps. Reduced metabolite overlap, improved peak separation and increased metabolite abundance was observed with this method.
Contamination from non-metabolites, such as polymers, can result from sample collection, solvents, or instrument noise, and can result in signal suppression of potentially significant metabolites. It is recommended that the technician(s) and those who collect the samples prior to sample preparation consistently use the same brand, type and size of sample collection vials, pipette tips and any other tubes used during the collection and preparation of the samples. This allows the data analyst to have full confidence that the observed changes are real and not due to background differences from other sources. Treatment effectiveness, variations between disease and control groups, or any other metabolic analyses can then be investigated with increased confidence.
The method discussed here focuses on combined sample preparation methods13-15 which can be applied to plasma, BALF, or cerebrospinal fluid (CSF) samples for non-targeted metabolomic profiling for liquid chromatography-mass spectrometry (LCMS) based analysis. Both liquid chromatography (LC) and ultra-performance liquid chromatography (UPLC) separation techniques can be coupled to MS subsequent to this procedure. Many researchers performing metabolomic studies use either a protein precipitation technique and/or a liquid-liquid extraction technique16,17. In our studies, this resulted in fewer metabolites being detected. The method described here12 enables the detection and identification of a greater number of metabolites, covering a wider range of the metabolome. This increase is due to the higher purity of the samples and reduced matrix effects caused by prior separation of the metabolite classes.
An initial protein precipitation step is performed using cold methanol (MeOH) to remove protein from the sample. Liquid-liquid extraction (LLE) using methyl tert-butyl ether (MTBE) and water is used to separate the hydrophilic and hydrophobic compounds. Then solid-phase extraction (SPE) is performed on the hydrophobic layer to separate the hydrophobic compounds into three classes – fatty acids, neutral lipids, and phospholipids. The hydrophobic fractions are reconstituted in 100% methanol, while the hydrophilic fraction is reconstituted in 5% acetonitrile in water. The solid-phase extraction (SPE) step provides an added level of confidence in the results by reducing the number of coeluting compounds which would otherwise be present had a separation step not been performed.
Protocol
1. Initial Considerations, Preparation of Instruments and Standards
- Always use glass for storing (glass culture tubes, glass autosampler vials) or transferring (glass pipettes) lipids and organic solvents.
- Minimize exposure of all lipid samples and standards to air. Seal polytetrafluoroethylene (PFTE) caps tightly to avoid air exposure and evaporation. Immediately resuspend dried lipids in the next solvent or keep them in a steady stream of nitrogen.
- Use 100 µl of sample. In instances where more (or less) sample is available, adjust volumes accordingly. However during the lipid fractionation on the NH2 SPE column, stated volumes must remain as indicated.
- Turn on centrifuge and set to 0 °C prior to start of sample preparation.
- This technique uses many volatile solvents so keep all solvents capped during sample preparation.
- Prepare two types of standards per recommendation.
- Prepare a negative control comprised of spiked-in standards all at constant concentration. This is spiked into all samples as well as a pooled sample which is used as a batch QC (spiked pooled sample used to monitor sample preparation reproducibility on separate days) and instrument QC (spiked pooled samples previously prepared, sub-aliquoted, and used to monitor instrument conditions/fluctuations during analysis and on separate days).
- Prepare positive controls at 1x, 2x, and 4x standard spikes. The positive controls are added to all samples as well as a pooled plasma sample and are used as quality control compounds for both the sample preparation steps and instrumental analysis steps to analyze and identify the fold change differences during data analysis to ensure that all aspects of the preparation and instrumental analysis were performed correctly.
- Store standards at -20 °C and store samples at -80 °C. Certain internal standards which are prone to degradation following long term storage should be stored at -80 °C.
- This procedure requires the use of hazardous, flammable or volatile solvents such as MTBE. Perform all steps in a fume hood.
2. Internal Standards
- Choose internal standards (ISTD) based on individual projects and their specific experimental design. For biofluids such as plasma, BALF, or urine, isotopically labeled ISTDs which are similar in nature to those found biologically in these samples would be ideal. These would include but are not limited to amino acids, hormones or lipids. For plant metabolomic samples, labeled standards such as flavonoids, or carotenoids could be used. The same applies to other metabolomic studies whereby the investigator should choose an internal standard which is representative of the sample type being analyzed.
- Ensure that the chosen ISTD covers a wide range of the chromatogram; for example if the acquisition time is 20 min, standards that elute every 5 min could be used.
- Create hydrophilic stock solutions at 2 mg/ml using a variety of isotopically labeled standards and/or other polar compounds which are exogenous to the sample being analyzed. From each stock solution, create one solution in 1:1 methanol:water with all standards to a final concentration of 25 µg/ml creatinine-D3, 100 µg/ml lysine-D4, and 200 µg/ml valine-D8.
- Create hydrophobic stock solutions using a variety of isotopically labeled standards and/or other non-polar compounds which are exogenous to the sample being analyzed; stock concentrations of 17:0 fatty acid (4 mg/ml), 19:1 fatty acid (4 mg/ml), 17:0 ceramide (2 mg/ml), 17:0 PE (1.75 mg/ml), 15:0 PC (2 mg/ml), and testosterone-D2 (1 mg/ml). From each stock, create one solution in 1:1 chloroform:methanol with all standards to a final concentration of 50 µg/ml 17:0 ceramide, 100 µg/ml 15:0 PC, 100 µg/ml testosterone-D2, 200 µg/ml 17:0 fatty acid, 200 µg/ml 19:1 fatty acid, and 200 µg/ml 17:0 PE in the standard mix.
- Test the degree of ionization of each standard ahead of time using at least five different concentrations for each standard to determine the limit of detection and linearity of the instrument for these compounds. The concentration of the stock solution for each individual standard will vary depending on experimental design, and the concentration of each standard in the combined spiked standard may range from 20 μg/ml to 2 mg/ml depending on how well they ionize.
- Create a spike mix of positive controls and add them to the samples at 1x, 2x, or 4x concentration levels to monitor quantitatively the strength of the sample preparation and the accuracy of the instrumental data. Final concentration of positive controls can be: 2 mg/ml D-glucose, 100 µg/ml alanine-D3, 200 µg/ml methylmalonic acid-D3, 20 µg/ml triglyceride-D5, and/or other hydrophobic and hydrophilic standards. Adjust the standard concentrations based on the sensitivity of the MS and HPLC instrumentation used for analysis.
3. Protein Precipitation
- Thaw samples to RT and spike 10 µl of ISTD to each sample as described below.
- Spike 10 µl of both hydrophilic and hydrophobic standard solutions (created from the stock in steps 2.3 and 2.4) to each sample. Adjust the standard concentrations as necessary based on the sensitivity of the MS and HPLC instrumentation used for analysis
- Spike 10 µl of either 1x, 2x, or 4x positive control solution (created from the stock in step 2.6) to each sample. Adjust the standard concentrations as necessary based on the sensitivity of the MS and HPLC instrumentation used for analysis
- Vortex each sample for 10 sec prior to the protein precipitation step
- Add 400 µl of ice cold methanol (stored at -20 °C) to each sample.
- Vortex for 10 sec per tube.
- Centrifuge at 0 °C for 15 min at 18,000 x g.
- Transfer all the supernatant to a new glass culture tube, and then dry under N2.
- If analyzing the protein pellet fraction, proceed to steps 3.8-3.11. If not analyzing the protein pellet, skip to Section 4.
Note: The protein pellet may contain compounds with high hydrophobicity which would be useful when performing drug studies18, food analyses involving hydrophobic flavonoids19 or metabolomic studies related to diseases where very hydrophobic compounds accumulate, such as neuronal ceroid-lipofuscinoses20 and lysosomal lipid storage diseases21. - Add 1 ml of MTBE to the white (or off-white) protein pellet, vortex for 30 sec per tube, then centrifuge at 0 °C for 15 min at 18,000 x g. Decant the MTBE layer to a new glass culture tube.
This is a critical step as errors here will drastically affect results (see Figure 5 in representative results).- Consistently aspirate the same amount of MTBE for ALL samples since the size of the pellet will vary among samples. Therefore, if only 900 µl can be decanted for the sample with the least amount of supernatant, then decant 900 µl for all samples.
- Repeat step 3.9, and combine the organic layer to the same glass culture tube prepared in step 3.7.
- Dry samples by N2 flow and resuspend in 200 µl of 1:1 chloroform:methanol. Vortex briefly.
- Transfer to a centrifuge tube. Centrifuge at 0 °C for 15 min at 18,000 x g, then transfer supernatant to autosampler screw cap vials using glass pipettes.
4. Liquid-liquid Extraction
- Using a glass pipette, add 3 ml MTBE to the dried methanol residual (from section 3, step 6), vortex 30 sec, add 750 µl of water, then vortex 10 sec per tube.
- Spin ~200 x g for 10 min in a centrifuge at RT.
- Aspirate 2.5 ml of the MTBE layer (without getting water) and transfer to a clean glass culture tube.
Note: This is a critical step as differences here will affect results. 2.5 ml of MTBE should be carefully decanted from the top layer because it allows a fixed volume to be decanted without pipetting the aqueous layer below. If the volume of sample at the start of the experiment was less than the indicated 100 μl for this method, scale the volume of MTBE to proportionally reflect this starting sample volume. - Add 3 ml MTBE to the remaining water part of sample, and vortex 10 sec per tube.
- Spin ~200 x g for 10 min in the centrifuge at RT.
- Aspirate 3 ml of MTBE (without getting water) and combine with MTBE tube.
- Concentrate the remaining aqueous layer by drying under N2.
- Re-suspend residue in 100 µl of water.
- Add 400 µl of ice cold MeOH to the glass culture tube, vortex briefly, and then transfer to microcentrifuge tube.
- Leave at -80 °C for 20-30 min. Spin at 0 °C for 15 min at 18,000 x g.
Note: It is recommended to place the entire sample rack in a -80 °C freezer as this will allow any remaining protein to precipitate out in the methanol. If a -80 °C freezer is not available, other options are: storing at -20 °C, placing the samples on dry ice, or keeping them in an ice bucket. It is important to consistently store the samples in a cold environment and at the same temperature to allow precipitation. - Aspirate 450 µl of supernatant and transfer to a clean microcentrifuge tube. Dry completely in a vacuum centrifugal concentrator at no more than 45 °C. (Takes about 1-2 hr).
- Resuspend dried supernatant in 200 µl of 5% acetonitrile/water. Vortex briefly. Freeze at -80 °C.
5. Solid-phase Extraction
- Dry the MTBE fraction under nitrogen at 35 °C with a good flow of nitrogen (takes about 10-15 min).
- When completely dry, stop the flow of nitrogen and quickly resuspend in 1 ml chloroform (CHCl3) using a glass pipette. Vortex briefly.
Note: Solvents such as CHCl3 have low viscosity. During pipetting, low surface tension causes solvent loss from the pipette. It is recommended that the pipette tip be prewet at least twice to allow equilibration between the solvent being pipetted and the space in the pipette. A gas-tight syringe can also be used if available. - Set up an SPE vacuum manifold and NH2 SPE columns for fractionation.
- Warm samples to RT and always keep suspended under a steady flow of N2. Warming to RT will allow lipid resuspension and the nitrogen will prevent oxidation and polymerization of lipids.
- Wash and condition SPE cartridge 2x with 400 µl hexane. Discard waste and replace with new glass collection tube.
- Add sample to SPE column, collect flow through in glass tubes.
- With glass pipette add 1 ml of 2:1 CHCl3:IPA, collect flow through in same glass tubes (this is the Neutral Fraction).
- Dry the neutral fraction under N2 to minimize oxidation (takes about 10-15 min).
- With glass pipette add 1 ml of 5% acetic acid in diethyl ether, collect flow through in new glass tubes (this is the Fatty Acid Fraction).
- Dry the fatty acid fraction under N2 to minimize oxidation (takes about 10-15 min).
- Use plastic tips to add 800 µl of methanol to SPE cartridge, and collect flow-through in 15 ml plastic conical tubes (this is the Phospholipid Fraction).
- Transfer phospholipid fraction to 1.5 ml centrifuge tubes. Dry the samples with a vacuum centrifugal concentrator at 45 °C (takes about 1-1.5 hr).
- Resuspend each of the samples from the three fractions in 200 µl of 100% methanol, vortex, and transfer to autosampler vials screw-cap for storage.
6. Sample Storage Conditions
- Store all samples at -80 °C until ready for instrument analysis.
Note: Extracted samples, reconstituted in organic solvent may be stored at -20 °C. However this is not recommended as potential metabolites of interest will degrade at this temperature. Liquid nitrogen storage is also not recommended as investigators have reported contamination issues, special storage vials are needed, and there is lack of homogeneity in the chamber causing wide temperature fluctuations. - If sample volumes are small (<100 µl), use inserts in the vials to prevent solvent evaporation in the headspace during storage.
- Avoid freeze-thaw of the samples. Thaw the samples only once, right before instrument analysis. Repeated freeze-thaws result in sample degradation.
7. Liquid Chromatography Conditions
- Use a C-18 2.1 mm x 50 mm (1.8 μm) analytical column with a C-18 2.1 mm x 12.5 mm (5 μm) guard column to analyze the hydrophobic fraction.
- Set the autosampler temperature to 4 °C, the column temperature to 60 °C, the injection volume to 2 µl and the flow rate to 0.25 ml/min.
- Use 0.1% formic acid in water for mobile phase A, and 0.1% formic acid in isopropanol:acetonitrile:water (60:36:4) for mobile phase B.
- Run the following gradient elution profile: Start at 30% B, and increase to 70% B from 0 to 1 min, then increase to 100% B from 1 to 15 min and hold for 5 min, followed by 5 min 10% B washing and 5 min post run.
- Use a HILIC 2.1 mm x 50 mm (2.6 μm) analytical column with a guard column to analyze the hydrophilic fraction.
- Set the autosampler temperature to 4 °C, the column temperature to 20 °C, the injection volume to 2 µl and the flow rate to 0.5 ml/min.
- Use 50% acetonitrile in 10 mM ammonium acetate, pH 5.8 for mobile phase A, and 90% acetonitrile in 10 mM ammonium acetate pH 5.8 for mobile phase B.
- Run the following gradient elution profile: Start at 100% B from 0 to 2 min, then decrease to 50% B from 2 to 15 min, followed by 5 min 0% B washing and 10 min post run.
Representative Results
The entire sample preparation technique was performed as described above and the most important and/or relevant aspects are presented below. Hydrophilic and hydrophobic internal standards were spiked into pooled plasma samples to perform direct comparisons of the internal standards and endogenous metabolite abundances using various extraction methods. Liquid chromatography-mass spectrometry (LC-MS) data was analyzed using qualitative and quantitative software and resulted in excellent recovery and separation of both the endogenous compounds and internal standards. Figure 1 demonstrates the effectiveness of the MTBE-SPE method in extracting both lipid standards (A) and endogenous compounds (B).
Overall, better extraction and coverage of the metabolites were obtained compared to other methods such as methanol extraction, or ‘MTBE only’ extraction when the number of features was compared using qualitative and quantitative software following LC-MS analysis. For example, using only methanol extraction, the variation for creatinine-D3 was 15.2%. However, with MTBE LLE, this was reduced to 1.04% CV. Using MTBE, the reproducibility of lipids and aqueous compounds were <8% and <5% respectively, compared to a simpler methanol extraction which resulted in larger variation of 29% and 15% respectively for lipids and aqueous compounds. The internal standards used to monitor lipid recoveries – testosterone-D2, C17 ceramide, 15:0 PC, and 17:0 PE increased by 26%, 200%, 100%, 400% respectively compared to using methanol alone. Similar increases were detected for fatty acid internal standards and phosphotidylcholine and phosphotidylethanolamine endogenous metabolites. Other endogenous metabolites such as sphingosines, ceramides, diacylglycerols, triacylglycerols, cholesterol, and sphingomyelin were either not detected using methanol or were detected at negligible levels. However these endogenous lipids were easily detected using MTBE extraction.
In our analysis comparing standard protocols, the following results were obtained: Methanol precipitation alone resulted in 1,851 metabolites, methanol-ethanol precipitation gave 2,073 metabolites, MTBE with liquid-liquid extraction gave 3,125, and MTBE with liquid-liquid and solid-phase extraction recovered 3,806 metabolites. Therefore this approach results in a greater number of metabolites being extracted, most likely due to reduced ion suppression and cleaner samples prior to LC-MS.
Figure 2 demonstrates the efficiency in separating the hydrophobic metabolites into their respective chemical classes for more confident metabolite identification. There is minimal overlap of the compounds identified in the three lipid fractions following SPE. In support, Figure 3 shows the recovery of the internal standards demonstrating that ISTD’s were eluted in the fraction related to their chemical class.
Quality control samples are used to evaluate the quality of the sample preparation, to determine any batch effects when multiple days of analysis are required for a large sample set, and to monitor instrument reproducibility. Chromatograms are examined to ensure that spiked-in standards are greater than 90% recovered with mass error of less than ± 3 ppm and retention time window of less than ±5%. If these criteria are not met, the results are discarded and the samples are reanalyzed. In a case of batch effects whereby a shift in retention is observed for one batch, the data analysis software can correct for this. A previously prepared batch of pooled plasma samples underwent sample preparation. The fractions were then sub-aliquoted into autosampler vials and stored at -80 °C for use in monitoring instrument conditions throughout every sample analysis. Table 1 shows the results from these spike-in standards. The fatty acid negative ionization mode fraction (data not shown in table) was not used for analysis because the % CV of the spike-in standards for the QC sample was greater than 10%. The dataset for that fraction was therefore discarded and the instrument inspected and maintained. Table 2 shows the results from endogenous metabolites in the samples following three different days of sample preparation and triplicate instrument injections. The endogenous metabolites in the sample preparation QC samples are all reproducible, signifying the strength of the sample preparation as well as the instrument injection reproducibility.
When sample preparation steps are not properly followed, however, unreliable and inconsistent results are obtained. Figure 4 shows the results when the protein precipitation step of the method is not followed as outlined. Three operators, A, B, and C performed the same sample preparation procedure on pooled plasma samples. Operator A, rather than pipetting the required amount of supernatant per the experimental protocol, instead pipetted >1 ml for both washes with some of the pellet. This not only resulted in a higher number of false positives for that fraction, but increased the variability of the data.
The chromatographic reproducibility of the data can be seen in Figure 7. Pooled plasma samples were prepared in triplicate on separate days using protein precipitation, liquid-liquid extraction, and solid-phase extraction as described in this protocol. Each fraction was analyzed using the chromatographic separation described in section 7 of the protocol. Samples were then injected in triplicate on the LC-MS to evaluate instrument and sample preparation reproducibility. This consistent overlap demonstrates both the strength of the reproducibility of the sample preparation when prepared on three different days, as well as the strength of the chromatographic method in producing reproducible results. An increase in chemical noise is observed for the negative ionization mode of the fatty acid fraction. This may occur due to contaminants in the LC-MS solvents and can result in inconsistent quantitative metabolomic results. Therefore only metabolites which eluted prior to 9 min were analyzed.
When running long worklists, a loss of instrument sensitivity and change in buffer concentrations can occur over time resulting in decreased signal intensity and retention time shift. If the retention time overlap variation is less than 5% and the signal intensity variation is less than 10%, the data is still within standard laboratory limits. Analysis software can be used to align and normalize the data to correct for instrument and retention time drift. However, if the variation is large, then the reason has to be determined. Once this is rectified, the samples can be re-analyzed.
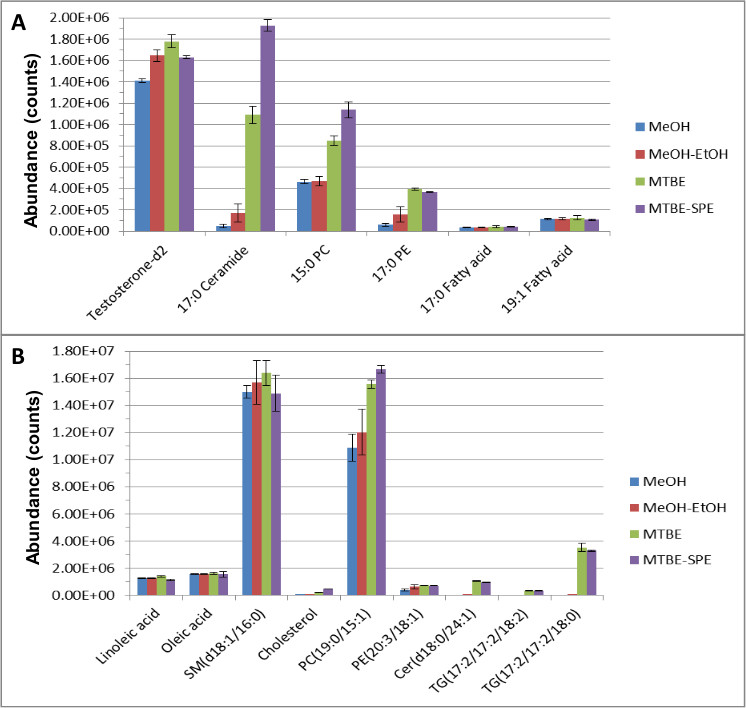
Figure 1. The abundance of lipid ISTDs (A) and endogenous metabolites (B) following extraction and reversed-phase chromatography (RPC)12. Extraction was performed and resulting samples were separated using RPC and analyzed using LC-MS in positive and negative ionization mode. This figure has been modified from Yang et al, Journal of Chromatography A 1300, 217-226 (2013).
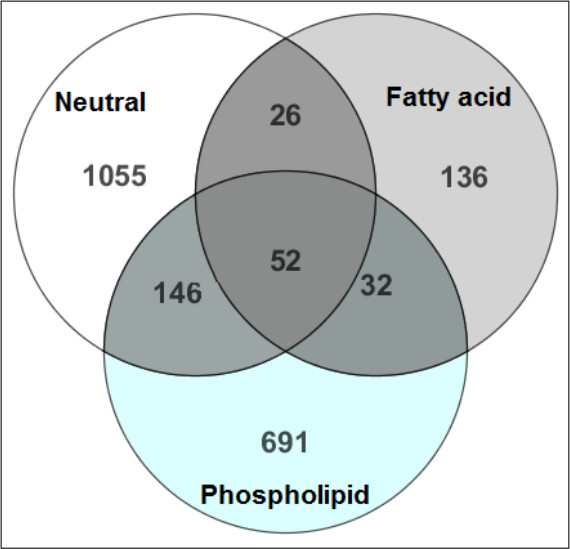
Figure 2. A comparison of MTBE-SPE fractions12. The metabolites identified in each fraction were compared to identify the amount of overlap during the SPE portion of the prep. The numbers in the Venn diagram reflect the number of metabolites detected in each fraction. Here minor overlap is observed among the three fractions, representing successful compound extraction and metabolite class separation during the SPE step. This figure has been modified from Yang et al, Journal of Chromatography A 1300, 217-226 (2013).
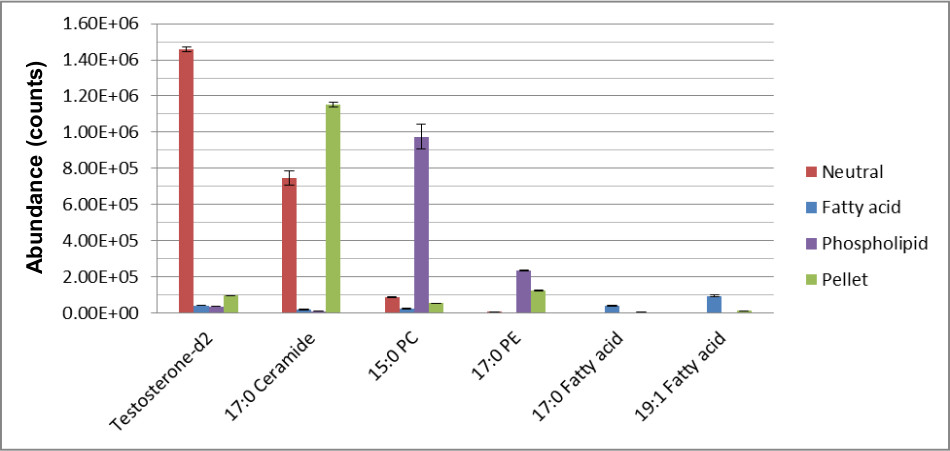
Figure 3. The recovery of ISTDs in fractions using the MTBE-SPE method12. Extraction was performed and resulting samples were separated using RPC and analyzed using LCMS in positive and negative mode as described in text. This figure has been modified from Yang et al, Journal of Chromatography A 1300, 217-226 (2013).
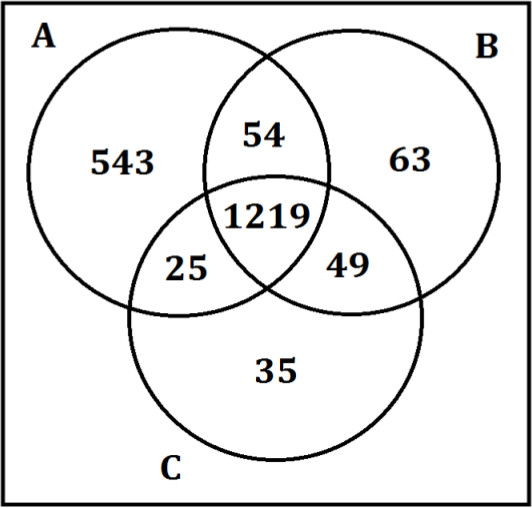
Figure 4. Results from the pellet fraction prepared by three operators. Three sample preparation operators A, B, and C performed the same protein preparation step on pooled plasma samples. The numbers in the Venn diagram reflect the number of metabolites detected by each operator. Operators B and C pipetted the required volume per the sample preparation protocol while operator A pipetted the entire supernatant and some of the pellet, resulting in over 500 more metabolites, the majority being false positives for that specific fraction.
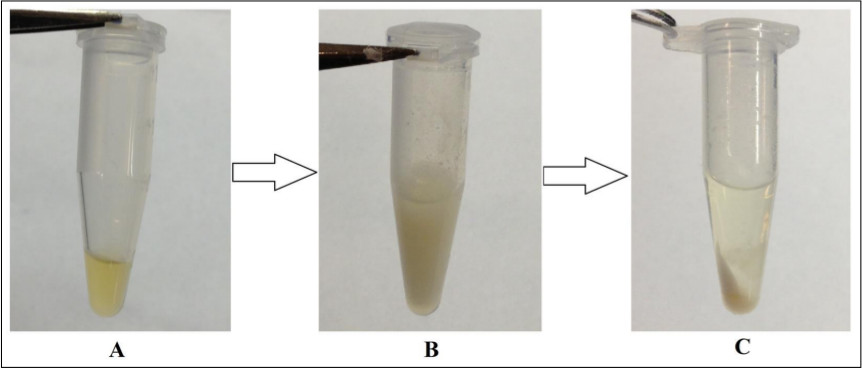
Figure 5. Formation of protein pellet during protein precipitation step. (A) 100 µl of human plasma prior to sample preparation; (B) plasma after addition of ice cold methanol; (C) protein pellet formed on bottom of tube after centrifuging at 0 °C for 15 min at 18,000 x g.
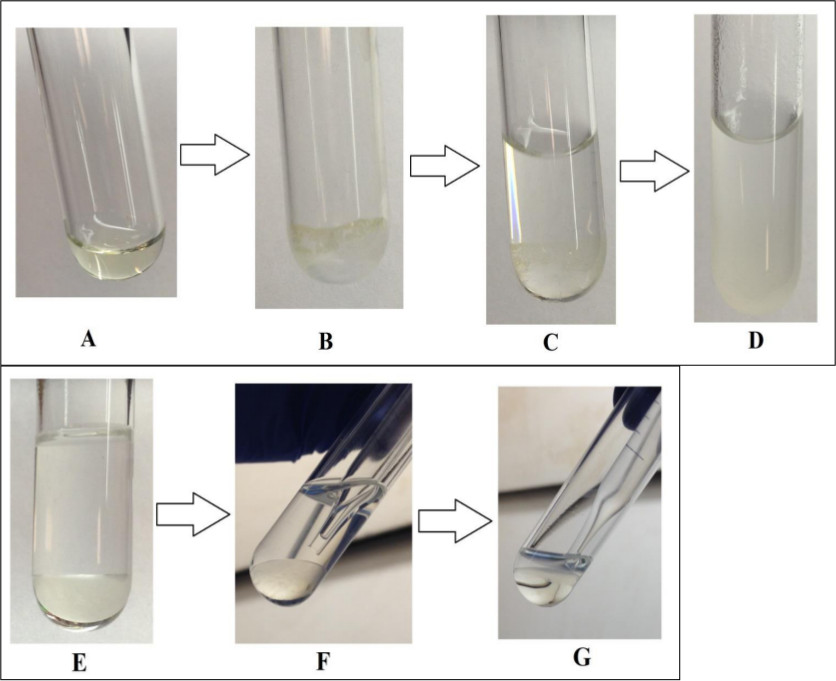
Figure 6. Separation of the hydrophilic and hydrophobic layers during liquid-liquid extraction (LLE) step. The organic solvent methyl tert-butyl ether (MTBE) and water were used to separate the hydrophilic and hydrophobic metabolites. The MTBE layer has dissolved non-polar compounds and the water layer has dissolved polar compounds. (A) plasma supernatant after protein removal; (B) plasma after drying under nitrogen; (C) plasma after addition of MTBE; (D) addition of water to plasma and MTBE; (E) MTBE-water layer formed after centrifuging; (F) removal of top MTBE layer; (G) mainly hydrophilic layer remaining after MTBE removal.
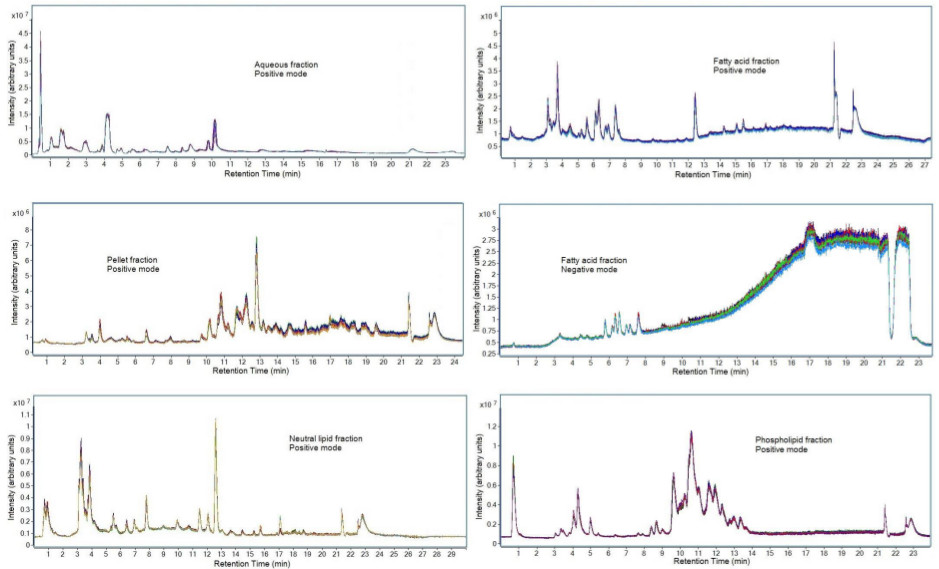
Figure 7. Chromatograms of fractions from a selected dataset. Sample preparation was performed on three separate pooled plasma QC samples and each sample was injected in triplicate on the LC-MS instrument. Represented are total ion chromatograms of the plasma samples acquired using the LC-MS parameters indicated in section 7 of the method protocol. The chromatographic representation of other biological fluids will vary due to differences in metabolite composition. Please click here to view a larger version of this figure.
| Fraction | Ionization Mode | Internal Standard | n | Average Peak Area | Peak Area % CV |
| Aqueous | Positive | Creatinine-D3 | 31 | 2217311 | 3.8% |
| Neutral lipid | Positive | Triglyceride-D5 | 31 | 4837032 | 9.9% |
| C17 Ceramide | 31 | 12736707 | 7.9% | ||
| Phospholipid | Positive | 15:0 PC | 32 | 1248929 | 9.3% |
| 17:0 PE | 32 | 517234 | 7.9% |
Table 1. Quality control results from spiked internal standards. Pooled plasma samples from an emphysema mouse model dataset were analyzed to monitor instrument conditions on a daily basis for this multi-week study. Quantitative analysis software was used to determine the peak areas of the internal standards, (n = number of instrument QC injections).
| Fraction | Ionization Mode | Endogenous metabolites | Average Peak Area | Peak Area % CV |
| Aqueous | Positive | Creatinine | 2554574 | 2.3% |
| Valine | 3712151 | 3.3% | ||
| Glucose | 2669190 | 6.9% | ||
| Neutral lipid | Positive | 3-Dehydrosphinganine | 226644 | 3.9% |
| DG(16:0/16:1/0:0) | 11301 | 8.2% | ||
| DG(P-14:0/18:1) | 364119 | 1.9% | ||
| Phospholipid | Positive | PC(24:0/0:0) | 27599 | 0.9% |
| PC16:0/22:6) | 2873326 | 4.5% | ||
| PI(16:0/18:1) | 112998 | 4.4% | ||
| Fatty acid | Positive | 10-oxo-5,8-decadienoic acid | 1363284 | 2.3% |
| 16-oxo-heptadecanoic acid | 83700 | 2.9% | ||
| 2-methyl valeric acid | 285782 | 5.7% | ||
| Fatty acid | Negative | 10-hydroxy-8-octadecenoic acid | 10042 | 4.9% |
| (R)-laballenic acid | 173929 | 6.5% | ||
| 2-keto valeric acid | 35488 | 6.0% |
Table 2. Quality control results from endogenous metabolites. Pooled plasma samples from a human disease dataset were analyzed to monitor sample preparation reproducibility on separate days. Samples were prepared in triplicate over three days, (n = 9 prep QC injections).
Discussion
One goal of clinical metabolomic studies is to identify changes in the metabolome related to disease or treatments. Therefore sample preparation techniques need to be robust, consistent, and transferable from technician to technician and from laboratory to laboratory22. The resulting data needs to be representative of the sample, and identified changes need to reflect the sample set rather than sample preparation errors. Therefore accurate pipetting, correct temperature, efficient decanting of immiscible layers, drying under nitrogen, and use of the same brands and sizes of glassware and tips are necessary.
During the protein precipitation step, it is crucial that the same amount of solution is decanted from each pellet. This reduces variation in volume and as such reduces variation in sample data. This protein precipitation step is necessary for metabolomic studies and cannot be skipped since it removes protein from the samples prior to small molecule profiling analysis on the mass spectrometer. It eliminates pathogens and large macromolecules, and releases bound metabolites from proteins7. Lack of protein accumulation in the samples expands the HPLC column lifetime and increases the accuracy and quality of results. Figure 5 is a depiction of the protein pellet formed when performing this technique on plasma samples. This allows the detection of the small molecules, enhances ion abundance, and reduces matrix effects from proteins in the sample. In addition, since it is assumed that all proteins are removed in this step, the amino acids which are detected during LC-MS analysis would originate from metabolic changes rather than from protein breakdown.
The liquid-liquid extraction step is critical since it separates the hydrophilic and hydrophobic metabolites into two immiscible layers. Figure 6 shows the LLE procedure and a representation of the LLE layer. An improper separation of the two layers results in metabolites either being lost or being eluted in both fractions. Careful application of this step reduces the number of hydrophilic compounds which appear in the hydrophobic fraction. The results for these compounds become unreliable since it cannot be determined which fraction contains the representative results. When done correctly, metabolite overlap is reduced.
To prevent oxidative degradation, particularly in lipids but also in small molecules which may contain thiol groups for example, exposure to oxygen has to be kept to a minimum. Therefore, this procedure is always performed under nitrogen to reduce/prevent oxidation of lipid or thiol containing compounds. In addition, transfer of sample and/or solution is rapid (within the first minute) to reduce oxygen exposure, then samples are quickly placed under a steady stream of nitrogen to dry down. Once dried, they are immediately resuspended in 100% methanol for the above discussed reasons.
Laboratories can benefit from this comprehensive method in a number of ways; Researchers looking to isolate one class of compounds can choose the part of the method which best suits their needs. Those seeking to only perform a protein precipitation to obtain a pool of metabolites may do so. If hydrophilic metabolites are desired, such as many pharmaceutical drugs, amino acids, and sugars, or if only hydrophobic metabolites are desired, such as triglycerides, epoxides, fat soluble vitamins, and phospholipids for example, then researchers can perform the liquid-liquid extraction step following protein precipitation and discard the undesired fraction. Investigators who require further sub-classification of the hydrophobic compounds (neutral lipids, fatty acids, and phospholipids) may proceed to the fractionation step.
Storage considerations are important in maintaining the viability of samples for later analysis. If samples are stored incorrectly, degradation or decomposition can occur. Ideally, samples should be stored in screw cap amber vials away from light to prevent degradation of light sensitive species. Samples should also be kept frozen at -80°C to prevent metabolite degradation23-25. Although not discussed in detail here, samples are always kept at 4°C in the autosampler tray during LC-MS analysis. This ensures that all samples are kept at a constant temperature and that changes in ambient temperature do not affect the viscosity, solubility, or stability of the samples. It is recommended that the manual aspects of this procedure, such as LLE and SPE, be practiced in order to gain confidence and comfort with the steps involved.
A few limitations exist for this technique. Discreet separation of the hydrophobic and hydrophilic metabolites is not guaranteed as certain compounds will inherently partition into both fractions due to their chemical composition and charge state. In addition as shown in Figure 4, improper technique during the protein pellet extraction step can result in poor metabolite reproducibility in both the samples and quality controls. This affects the statistics, especially in small datasets because the statistical power is not available. Therefore it is crucial that this step be performed exactly the same every time for each sample. Another limitation is time. Although there are stop points throughout this protocol where samples can be frozen and the prep continued the following day, an entire day should be set aside to perform this procedure. Thirdly, not every compound within a biological sample can be evaluated for ion suppression. Since it is not possible to identify how the matrix is affecting each individual metabolite, the current option is to evaluate the internal standards which theoretically mimic some classes of endogenous metabolites. Lastly, absolute identifications cannot be performed solely with this method. Tandem MS in collaboration with database searches and standards are required for absolute metabolite identification.
An important part of metabolomics is the identification of compounds. Although not discussed in detail here, quality control samples were analyzed using LC-MS. Multiple sample preparation blanks and instrument blanks were prepared for use as background subtraction to reduce the rate of false positives from contaminants, thereby resulting in more reliable metabolite hits. Following this step, the number of “molecular features” were grouped together based on m/z, retention time, isotope ratio, and adducts to produce a list of actual compounds. Although the list of compounds was greatly reduced, the results were more reliable as they were not based on multiple adducts from the same compound. The full method is comprehensive and allows isolation of hydrophobic metabolites such as neutral lipids, phospholipids, fatty acids, triglycerides, and steroids, while also isolating hydrophilic classes in the aqueous fraction, of which eicosanoids, sugars, flavonoids, and amino acids have been identified12,26.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The presented tutorial was performed and developed within the Mass Spectrometry Core Facility at National Jewish Health. The NJH MS facility is supported in part by CCSTI UL1 TR000154. Funding from NIH grants P20 HL-113445 and R01 HL-095432 also supported this work.
Materials
| Acetonitrile | Fisher Scientific | A955-4 | – |
| Methanol | Fisher Scientific | L-6815 | – |
| Chloroform | Fisher Scientific | C606-1 | – |
| Hexane | Sigma Aldrich | 34859 | – |
| Acetic acid | Sigma Aldrich | 49199-50ML-F | – |
| Methyl tert-butyl ether | J.T. Baker | 9042-03 | – |
| Isopropyl alcohol | Sigma Aldrich | 34965-2.5L | – |
| Water | Honeywell Burdick & Jackson | 365-4 | – |
| OA-SYS heating system | Organomation Associates, Inc | – | Used to keep samples under a constant flow of nitrogen while at 35oC |
| 12-position vacuum manifold | Phenomenex | – | – |
| Strata NH2 (55µM, 70Å) 100mg/mL SPE cartridges | Phenomenex | 8B-S009-EAK | – |
| Glass pipette tips | Fisher Scientific | 13-678-20C | Used to transfer sample to SPE column |
| Plastic pipette tips | USA Scientific | 1182-1830 | Used when glass tips are not necessary |
| 1182-8810 | |||
| Microcentrifuge tubes | Fisher Scientific | 02-681-320 | – |
| Graduated glass pipets | Fisher Scientific | 13-678-27B | Used to transfer organic solvents during sample prep |
| 13-678-27E | |||
| Pyrex glass culture tubes | Corning Incorporated | 99499-16X | Used to store aqueous and lipid fractions until the next step |
| Autosampler vials | Agilent Technologies | 5182-0545 | – |
| Snap cap vials for autosampler vials | Agilent Technologies | 5182-0541 | – |
| Glass inserts | Agilent Technologies | 5183-2085 | Used for small sample volumes |
| Mass Hunter Qualitative Analysis software | Agilent Technologies | Version B.06.00 | Used to monitor retention times and pressure curves |
| Mass Hunter Quantitative Analysis software | Agilent Technologies | Version B.05.02 | Used to analyze quality control and sample data |
| Mass Profiler Professional software | Agilent Technologies | Version B.12.50 | Used to determine statistics, fold changes, and perform metabolite identification |
Referenzen
- Beckonert, O., et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols. 2, 2692-2703 (2007).
- Clarke, C. J., Haselden, J. N. Metabolic Profiling as a Tool for Understanding Mechanisms of Toxicity. Toxicologic Pathology. 36, 140-147 (2008).
- Wang, X., Zhang, A., Sun, H. Power of Metabolomics in Diagnosis and Biomarker Discovery of Hepatocellular Carcinoma. Hepatology. 57, 2072-2077 (2013).
- Collino, S., Martin, F. -. P. J., Kochhar, S., Rezzi, S. Monitoring Healthy Metabolic Trajectories with Nutritional Metabonomics. Nutrients. 1, 101-110 (2009).
- Cevallos-Cevallos, J. M., Reyes-De-Corcuera, J. I., Etxeberria, E., Danyluk, M. D., Rodrick, G. E. Metabolomic analysis in food science: a review. Trends in Food Scienc., & Technology. 20, 557-566 (2009).
- Nicholson, J. K., Connelly, J., Lindon, J. C., Holmes, E. Metabonomics: a platform for studying drug toxicity and gene function. Nature reviews. Drug Discovery. 1, 153-161 (2002).
- Wishart, D., et al. . Metabolome Analysis: An Introduction. , 253-288 (2006).
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–mass spectrometry. Analytical and Bioanalytical Chemistry. 403, 1523-1548 (2012).
- Bollard, M. E., Stanley, E. G., Lindon, J. C., Nicholson, J. K., Holmes, E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR in Biomedicine. 18, 143-162 (2005).
- Dong, M. W. . Modern HPLC for Practicing Scientists. , (2006).
- Want, E. J., et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Analytical Chemistry. 78, 743-752 (2006).
- Yang, Y., et al. New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. Journal of Chromatography A. 1300, 217-226 (2013).
- Folch, J., Lees, M., Stanley, G. H. S. A simple method for the isolation and purification of total lipides from animal tissues. The Journal of Biological Chemistry. 226, 497-509 (1957).
- Matyash, V., Liebisch, G., Kurzchalia, T. V., Shevchenko, A., Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. Journal of Lipid Research. 49, 1137-1146 (2008).
- Bligh, E. G., Dyer, W. J. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 37, 911-917 (1957).
- Ferreiro-Vera, C., Priego-Capote, F., Luque de Castro, M. D. Comparison of sample preparation approaches for phospholipids profiling in human serum by liquid chromatography–tandem mass spectrometry. Journal of Chromatography A. 1240, 21-28 (2012).
- Michopoulos, F., Lai, L., Gika, H., Theodoridis, G., Wilson, I. UPLC-MS-Based Analysis of Human Plasma for Metabonomics Using Solvent Precipitation or Solid Phase Extraction. Journal of Proteome Research. 8, 2114-2121 (2009).
- Kerns, E. H., Di, L. . Drug-like Properties: Concepts, Structure Design and Methods: from ADME to toxicity optimization. First edn. , (2008).
- Manach, C., Scalbert, A., Morand, C., Rémésy, C., Jiménez, L. Polyphenols: food sources and bioavailability. American Journal of Clinical Nutrition. 79, 727-747 (2004).
- Weimer, J. M., Kriscenski-Perry, E., Elshatory, Y., Pearce, D. A. The neuronal ceroid lipofuscinoses. Mutations in different proteins result in similar disease. NeuroMolecular Medicine. 1, 111-124 (2002).
- Schulze, H., Sandhoff, K. Lysosomal lipid storage diseases. Cold Spring Harbor Perspectives in Biology. 3, (2011).
- Kuhn, E., et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Molecular and Cellular Proteomics. 11, (2012).
- Rist, M. J., et al. Influence of Freezing and Storage Procedure on Human Urine Samples in NMR-Based Metabolomics. Metabolites. 3, 243-258 (2013).
- Boomsma, F., Alberts, G., van Eijk, L., Manin‘t Veld, A. J., Schalekamp, M. A. Optimal Collection and Storage Conditions for Catecholamine Measurements in Human Plasma and Urine. Clinical Chemistry. 39, 2503-2508 (1993).
- Wood, J. T., et al. Comprehensive profiling of the human circulating endocannabinoid metabolome: clinical sampling and sample storage parameters. Clinical Chemistry and Laboratory. 46, 1289-1295 (2008).
- Bahr, T. M., et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. American Journal of Respiratory Cell and Molecular Biology. 49, 316-323 (2013).

