Structure and Coordination Determination of Peptide-metal Complexes Using 1D and 2D 1H NMR
Summary
The NMR-solution structure of a metallochaperone model peptide with Cu (I) was determined, and a detailed protocol from sample preparation and 1D and 2D data collection to a three-dimensional structure is described.
Abstract
Copper (I) binding by metallochaperone transport proteins prevents copper oxidation and release of the toxic ions that may participate in harmful redox reactions. The Cu (I) complex of the peptide model of a Cu (I) binding metallochaperone protein, which includes the sequence MTCSGCSRPG (underlined is conserved), was determined in solution under inert conditions by NMR spectroscopy.
NMR is a widely accepted technique for the determination of solution structures of proteins and peptides. Due to difficulty in crystallization to provide single crystals suitable for X-ray crystallography, the NMR technique is extremely valuable, especially as it provides information on the solution state rather than the solid state. Herein we describe all steps that are required for full three-dimensional structure determinations by NMR. The protocol includes sample preparation in an NMR tube, 1D and 2D data collection and processing, peak assignment and integration, molecular mechanics calculations, and structure analysis. Importantly, the analysis was first conducted without any preset metal-ligand bonds, to assure a reliable structure determination in an unbiased manner.
Introduction
Peptides are widely used as protein models, potential drug leads and therapeutic agents in their own right. However, their small size and high degree of flexibility often precludes structure determination by X-ray due to difficulties in crystallization.
Nuclear magnetic resonance (NMR) can be used to determine peptide structures and interactions. The method can give information regarding local and overall structure, binding and lower affinity interactions, and is applicable to difficult samples since it can be done in the solution state.
Copper transport in biological systems is achieved by intracellular copper metallochaperone proteins that specifically bind Cu (I) ions and deliver them to their target proteins through a series of protein-protein interactions, to protect the ions from oxidation and prevent the release of toxic copper2-5. The binding site is characterized by the conserved sequence, MXH/TCXanyXanyC, which has been shown both by NMR and crystallography to bind the Cu (I) by the soft thiolato ligands of the two cysteine residues, although an additional external ligand has also been proposed6-8. The structure-function relationship of these proteins has been a subject of intensive research9.
In the study presented here, a peptide model that includes the conserved sequence of copper metallochaperones was synthesized and reacted with Cu (I) under an inert environment. The presented protocol describes the steps of structure determination by NMR, including sample preparation, data collection, data processing, structure generation and structural analysis. The analysis was done in two steps: First structures were generated with no information regarding the mode of binding of the peptide to the copper ion. Once the binding mode was established empirically, these constraints were introduced to provide a high resolution structure. The mode of binding is the essential point in the model and was thus determined in an unbiased manner.
The NMR structural determination of model peptides is an extremely valuable technique that is often used by chemists and biologists. It may be applied relatively easily to different peptides under different conditions, and thus may shed light on relevant mechanisms10. Understanding the structure elucidation process provide a better understanding of the strengths and weaknesses of the proposed structures.
Protocol
1. Sample Preparation
- Apo-sample: Dissolve approximately 1-2 mg of the peptide in 450-500 µl of deuterated NMR grade solvent (the preferable solutions for biological samples are 10% D2O in water or d6-DMSO11-12 if the sample does not dissolve in water or reacts with it).
- Copper-reacted sample: Dissolve approximately 1-2 mg of the peptide with an equimolar amount of metal salt in 450-500 µl of the NMR solvent.
- Filter the solution using a Sinter glass, filtration paper or any other technique that suits the compounds under investigation and does not adsorb them. This is essential to remove any metallic particles, which will affect the homogeneity.
- Transfer the solution to an NMR tube and close it. Make sure that the tube quality fits the magnet strength used (for more details see equipment table).
- If the sample is sensitive to oxygen, these steps must be done under an inert environment and the tube sealed to prevent oxidation.
2. NMR Data Collection and Processing13
- Record 1D 1H spectra of the apo- and copper-reacted samples and compare. The copper-containing peptide spectrum should show a significant change of chemical shift in the amide region and the peaks became resolved since the apo-peptide is flexible and shows an average of conformations, but upon reacting with copper, the bound-peptide amides have a single structure (Figure 2).
- If the spectrum is unchanged, the reaction is assumed to have failed.
- If the sample turns green, the copper will have oxidized in the atmosphere, which may change its binding properties.
- In both these cases, the sample will need to be remade.
- Find optimal temperature conditions for the NMR measurements that give minimal overlap of the amide residues with narrow line-widths.
- Set up COSY14, TOCSY15 and NOESY16, or ROESY17 NMR experiments under identical conditions (temperature, pH, etc.) and run in sequence.
- COSY (Figure 3) and TOCSY (Figure 4) experiments will detect through-bond interactions of adjacent and longer range neighboring proton-proton interactions, respectively. COSY spectrum gives information on HN-Hα correlations while TOCSY spectrum gives, in addition information on HN and other H aliphatic protons of correlated residues.
- NOESY and ROESY (Figure 5) experiments detect through-space proximity with no dependence on bonds to give structural information. The choice between the two is according to the effective size of the compound and the field strength. Some such combinations give theoretical zero signals (Figure 6) and a ROESY experiment will have to be run to detect NOE interactions10,18.
- If the sample is in water, the experiments need to have a component of water suppression. This is most efficiently accomplished using gradients19. In this case, it is advantageous to rerun the sample, once assigned, in pure D2O to obtain interactions to and among the Hα hydrogens.
- Optimize the duration of the TOCSY and NOESY or ROESY mixing times by running a number of short experiments to find maximal signal build-up with minimal loss to spin diffusion. Make sure that this is the linear region of the buildup curve. Values common for peptides at 600 MHz fields include mixing times of around 100 msec for TOCSY and 150 msec for NOESY or ROESY experiments.
- Run 1D experiments in between each experiment to make sure that the sample composition remains constant throughout data acquisition (Figure 7).
- Process the spectra using appropriate apodization functions to obtain maximal resolution with minimal loss of signal strength, adding zero filling in the t1 dimension.
- Further correct the baseline in the F2 then F1 dimension with a quadratic polynomial function.
- Calibrate the chemical shift of the spectra carefully according to the residual water or DMSO peaks in the respective solvents according to their shift from known standards (TMSP or TMS, respectively).
3. Peak Assignment and Integration Using SPARKY20
- Prepare a set of COSY and TOCSY spectra overlaid on the NOESY or ROESY spectrum (Figure 8).
- Assign all NOE peaks in the spectrum according to the sequential assignment methodology developed by Wüthrich21.
- Start with assigning peaks that overlap TOCSY signals in the fingerprint region, as this will facilitate subsequent peak assignment entries with the SPARKY program (Figure 9). Register the assignment chemical shift (Figure 10), and 3JHNHα values (Figure 11).
- Translate peaks into distance constraints and 3JHNHα values into dihedral angles.
- This can be done by integrating the peaks from within the program and translating them into distance constraints using an interaction of known distance (such as the distance between two protons of methylene group or aromatic hydrogen atoms of an aromatic amino acid).
- If the peaks overlap, and integration methods cannot be used, they can be labeled as strong-medium-weak-very weak by visual estimation, and these designations can be translated into distances up to 2.5, 3.5, 4.5, and 5.5 Å, respectively. This to be most effective in work with peptides.
- 3JHNHα values are indicative of dihedral angles according to the Karplus equation where most given coupling values can result from more than one dihedral angle. These can be indicative of secondary structure (helices or sheets), random coil or conformational averages thereof. Therefore, it is important to use the constraints carefully, or to use them as confirmation of conformational results.
4. Molecular Mechanics Calculations to Generate Structure Ensemble using XPLOR22
- Import the distance constraints and dihedral angles with the correct format for XPLOR. (Figure 12, inter-residual constraints only). XPLOR will search conformational space to find structures that adhere to canonical geometric chemical constraints, such as bond lengths, angles, chirality, etc. in addition to the experimentally found distance constraints, to generate an ensemble in which none of the above parameters are violated. This will constitute the starting ensemble.
- The first structure determination run will be without using any constraints to the metal. This will show which residues participate in metal binding without any bias. Skip to step 4.4.
- In addition to the NMR-derived distance constraints, add metal binding constraints to the residues determined.
- Add appropriate parameters to describe the metal ion and its topology.
- The appropriate physical information (mass, bond lengths with other atoms, angles, and nonbonding repulsion parameters) must be put into the parameter file.
- Add the binding information to the topology file: Which bonds are formed and broken and which formal charges are changed as a result of binding.
- Create a small ensemble of usually 10 members.
- Introduce the constraints gradually, starting with backbone-to-backbone interactions, then backbone-to-sidechain, then sidechain-to-sidechain interactions, and going from intra-residual, sequential to increasingly longer range interactions. This facilitates identifying mistakes in assignment.
- Introduce the NOE energy as a square-well potential with a constant force constant of 50 kcal/mol•Å2. Simulated annealing should be done with 1,500 3-fsec steps at 1,500 K and 3,000 1-fsec steps during cooling to 500 K. Finally, minimize the structures using conjugate gradient energy minimization for 4,000 iterations; these are variables that can be changed within XPLOR.
- Create a final ensemble of usually 50 members. Perform steps from step 4.4 on the entire ensemble.
- Report the number of each type of NOE interaction that were found, as shown in Figure 13.
- Create an ensemble of structures that adhere to canonical chemical geometry and the empiric NMR-derived constraints.
- Report the total number of conformations, the number of these that have violations of the NMR-derived constraints, and the RMSD of the entire ensemble, both backbone, and all heavy atom RMSD values.
5. Structure Analysis
- The solved ensemble represents the conformational space adopted by the peptide during the NMR measurement. The local flexibility may change in different regions of the molecule and this can be ascribed to structural or functional reasons (the purpose of structural analysis is to determine the structural basis for the stability and mechanism of action).
- Import all conformations of the structure to the MolMol program23 to create a starting ensemble (Figure 14).
- Examine the ensemble to determine the local stability of the molecule.
- Determine the backbone and sidechain RMSD values by selecting subsequent four-residue regions along the sequence and having the program calculate the RMSD to the lowest energy structure or the mean.
- Determine which regions of the molecule show local stability (Figure 15) by plotting the local RMSD as a function of sequence.
- Overlay the ensemble along this region of the molecule and use this ensemble for further analysis (Figure 16).
- Choose low-energy conformations that adhere to the NMR-derived constraints (nonviolated). These will form the "low energy ensemble".
- Report the number of conformations in the ensemble, the criteria for choosing them and the RMSD values as defined in step 4.6.1.
- If the metal binding mode was already determined, continue to the next step. Otherwise: Analyze the low energy ensemble and determine which residue sidechains are in correct proximity of each other to be able to bind the metal ion. Once these have been determined, go back to step 3.3 and redo the analysis including the copper-binding data (Figure 17 for the ensemble and Figure 18 for the low energy conformer).
- Examine the ensemble and determine local secondary structure within the molecule using the default search parameters of the MolMol program. Secondary structures are held by hydrogen-bonding and indicate stable regions of the molecule.
- Determine the secondary structure of each conformer (Figure 19).
- Create a table of the percentage of conformers of the ensemble that show each secondary structure.
- Determine whether the regions of secondary structure overlap the regions that were determined to be the stable regions by local-RMSD. This is often the case.
- Determine the secondary structure of each conformer (Figure 19).
- Determine distances and hydrogen bonding within the molecule.
- Import the ensemble into Chimera24.
- Use Chimera to determine intramolecular distances between atoms suspected for metal binding. Calculate the average distances in the ensemble.
- Use Chimera to determine intramolecular hydrogen bonds in each conformer of the ensemble (Figure 20), using the default relaxed hydrogen-bond parameters (by 20%) of the program.
- Create a table of the percentage of conformers of the ensemble that show each hydrogen-bond. Hydrogen bonds that recur in a majority of the conformations indicate a stable bond that adds to the stability of the structure.
- Determine electrostatic interactions within the molecule.
- Identify interactions between charged sidechains.
- Create a table of the percentage of conformers of the ensemble that show each electrostatic interaction.
- Calculate the electrostatic potential distribution using the Amber25 force field within the Delphi26 program.
- Choose a single representative conformation from the ensemble on which to perform the electrostatic potential calculations.
- Derive electrostatic potential using the Poisson-Boltzman equation and a full Coulombic calculation. Required parameters include the ionic strength of the solution, the dielectric values of the solution (80 for water; 40 for DMSO); the internal peptide (usually around 5.0); the size of the grid (usually 65´65´65 points); and the minimal distance between the peptide and the edge of the grid (10 Å). The results are usually quite robust with respect to these values as the main result is governed by the peptide itself (Figure 21).
- Examine the electrostatic potential mapped onto the Van der Waals surface of the peptide (Figure 22).
- Examine the electrostatic potential iso-surfaces, which describe positions having the same potential. Find iso-surfaces that suggest biological relevance, such as extended positive or negative surfaces that might attract or repel other proteins or ligands, or distributions that indicate significance, such as amphipathic distribution or patches of differing properties (Figure 23).
- Sum up all the structural findings to see how they reinforce each other.
- Repeat steps 4.3-5.7.4 where relevant.
Representative Results
As part of an ongoing study of copper-binding protein models, the structure of the conserved sequence MT/HCXXC within the linear peptide MTCSGCSRPG was determined by solution state NMR. The peptide was reacted with CuCl under inert environment and the pH was measured as ~3.0 by a universal stick. The amide region of the peptide showed an expansion upon reaction with copper, from 6.7-8.5 to 6.6-9.0 ppm (Figure 2). Line broadening due to slight copper oxidation is evident in the baseline.
The copper-reacted peptide sample was stable with time (Figure 7) and the spectra were well-resolved (Figure 8) and gave 81 NOE interactions that were acquired by ROESY experiment since the molecule gave near zero NOE interactions in the NOESY spectrum (see theoretical explanation in Figure 6).
The ensemble of the peptide derived for the reacted sample, but using no constraints to the metal gave 47 out of 50 nonviolated structures with an RMSD value of 1.44 and 2.07 Å on the backbone and heavy atoms, respectively. Of these, 13 low-energy conformers were chosen for further analysis, with RMSD values of 0.25 and 0.61 Å on the backbone and heavy atoms, respectively.
The local RMSD plot (Figure 15) showed a region of stability between residues 3 and 7, in addition to the rigid C-terminal region including a proline residue. This region is found in a bend conformation between residues 4 and 7 in all conformations (Figure 19), which are stabilized by hydrogen bonding between backbone donors and acceptors, Gly5 and Thr2; Cys6 and Cys3 (Figure 20). This bend is also evident between C3 and S7 by the reduced 3JHNHα values in this region (Figure 11).
The conformations were superimposed over this region (Figure 16) and analyzed for possible binding residues. When considering Cys3, Cys6 and Met1 as potential binding residues, the shortest S.”.S distance was that between the thiolato groups of Cys3 and Cys6 (7.9±0.1 Å relative to 9.1±1.1 and 9.4±0.9 Å for SMet1…SCys3 and SMet1…SCys6, respectively). Copper-binding was introduced and the calculation was repeated to give the ensemble used for analysis (Figure 17).
The low-energy ensemble of the copper-bound peptide shows that the N-terminal amine is proximate to the bound copper (3.5-5.5 Å).
The electrostatic potential distribution on the surface of the molecule showed that arginine residue extends from the backbone of the peptide, forming a positive lobe of electrostatic potential, whereas the backbone carbonyls are arranged in a line forming a less prominent-1 kT/e electrostatic potential (Figure 23).
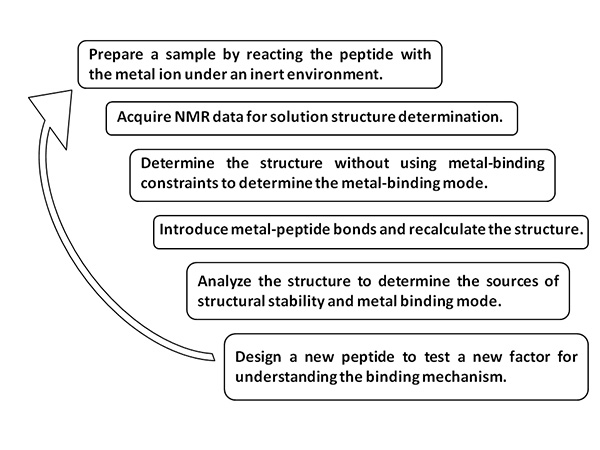
Figure 1. Scheme of the method.Click here to view larger image.
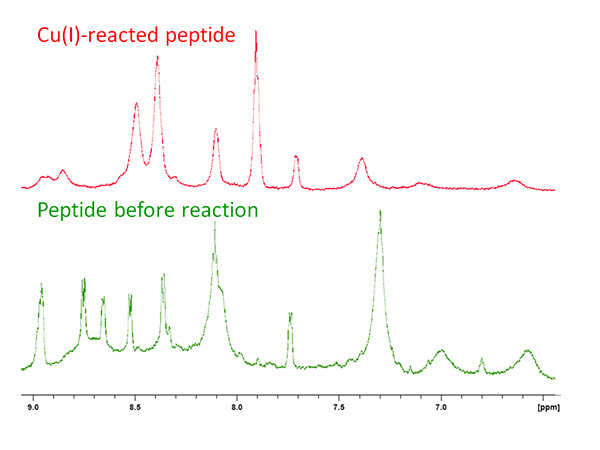
Figure 2. 1D NMR spectrum of apo- (green) and copper-reacted peptide (red) showing the change in distribution of amide resonances.Click here to view larger image.
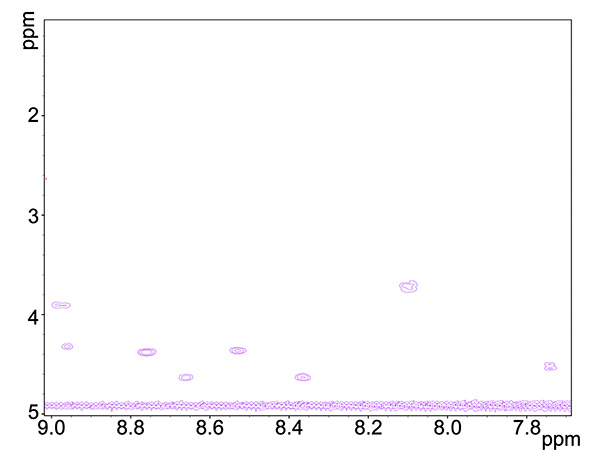
Figure 3. COSY spectrum of copper-reacted peptide, showing HN-Ha neighboring proton group interactions.Click here to view larger image.
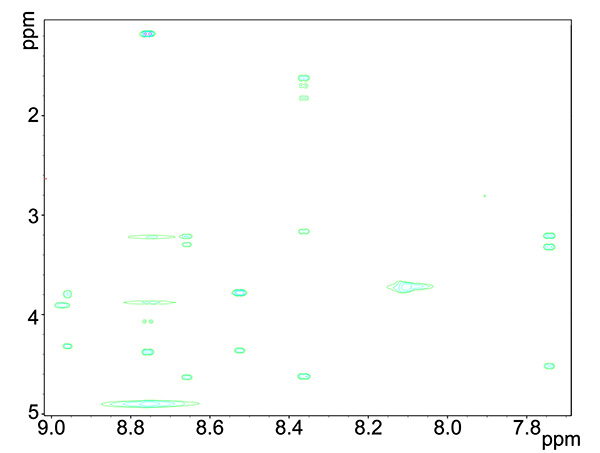
Figure 4. TOCSY spectrum of copper-reacted peptide, showing HN interactions with proton groups within the residue.Click here to view larger image.
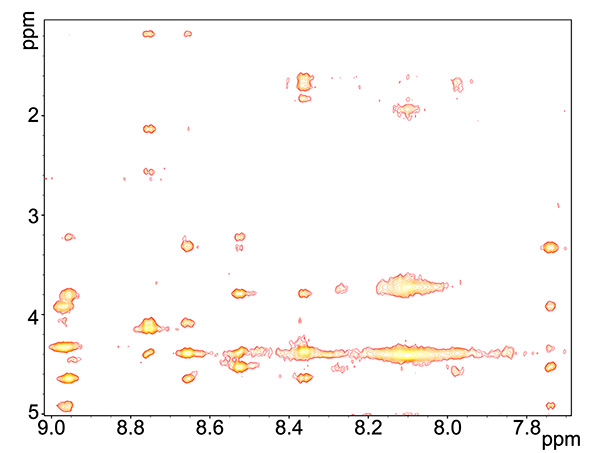
Figure 5. ROESY spectrum of copper-reacted peptide, showing HN interactions with all proximate proton groups, independent of being covalently bound to amide.Click here to view larger image.
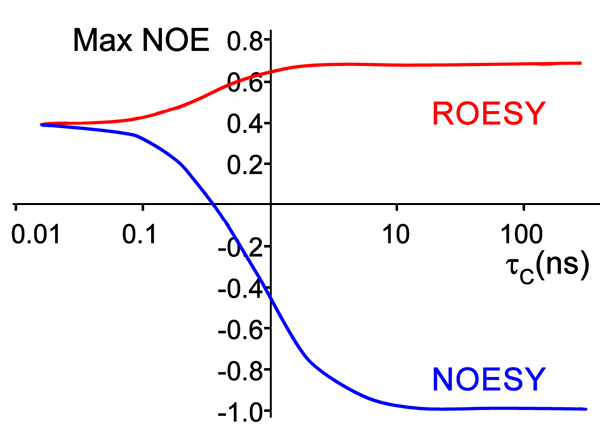
Figure 6. Theoretical calculation of NOE intensity of molecule by NOESY and ROESY experiment versus the correlation time, indicative of the rotation rate, which is dependent on effective size and spectrometer field15.Click here to view larger image.
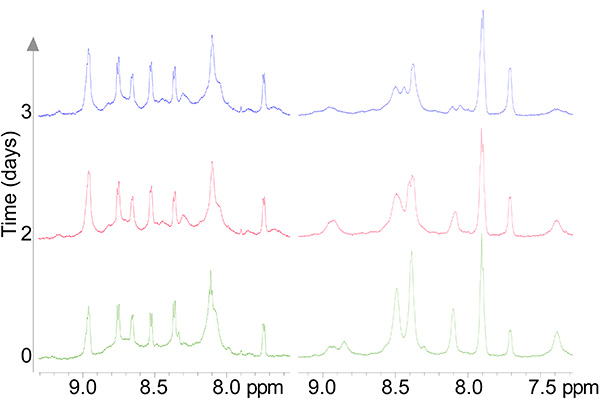
Figure 7. 1D spectra of a stable copper complex (left pane) and the apo peptide (right pane) undergoing oxidation as a function of time.Click here to view larger image.
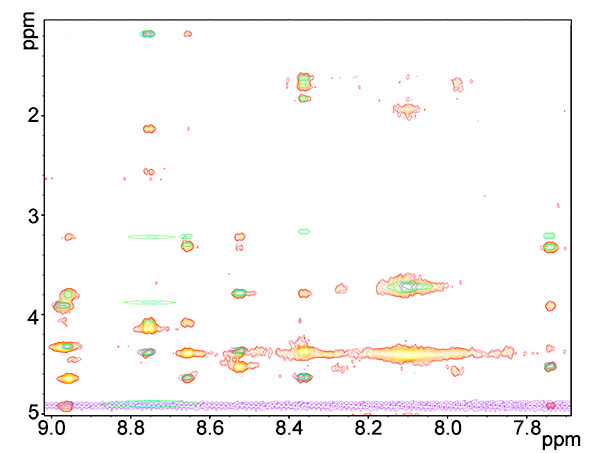
Figure 8. Overlay of fingerprint regions of ROESY (red-yellows), TOCSY (blue-greens) and COSY (purple) spectra of copper-bound peptide.Click here to view larger image.
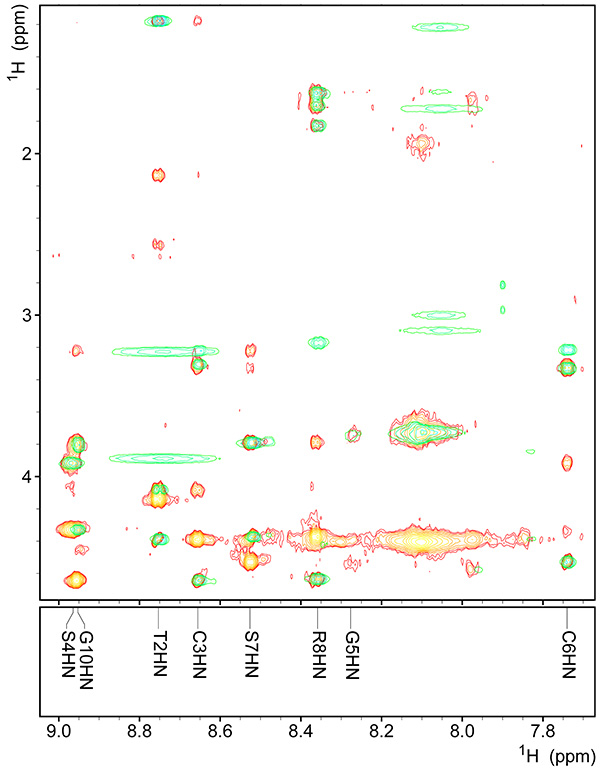
Figure 9. Assigned fingerprint region of spectrum of copper-bound peptide.Click here to view larger image.
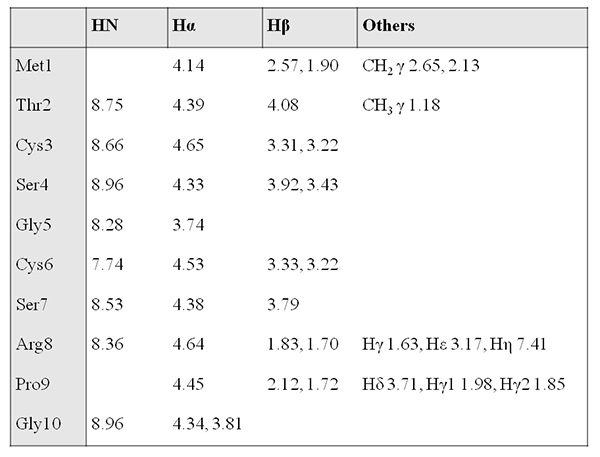
Figure 10. 1H chemical shift assignment table (ppm).Click here to view larger image.
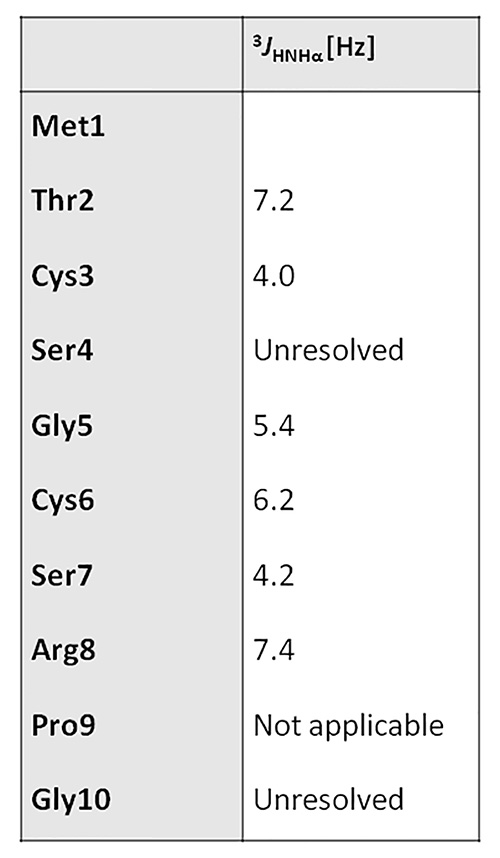
Figure 11. 3JHNHα values table.Click here to view larger image.

Figure 12 . Inter-residual constrains list.Click here to view larger image.
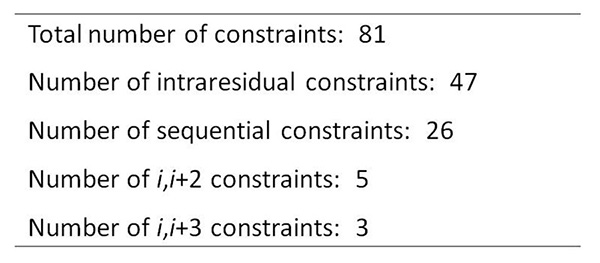
Figure 13. Statistics on NMR-derived NOE constraint file used to generate ensemble of peptide.Click here to view larger image.
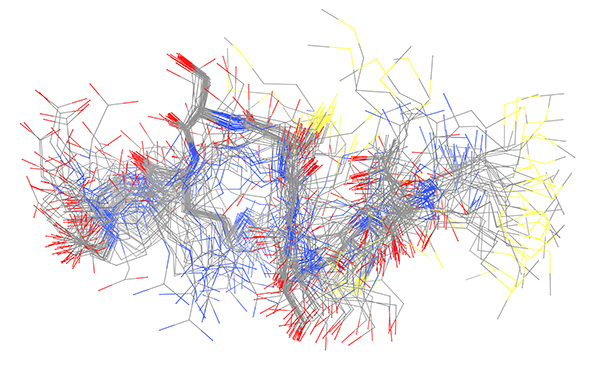
Figure 14. Ensemble derived from NMR data of copper-bound peptide. Complete 50-member ensemble representing all samples conformations superimposed on backbone.
Click here to view larger image.
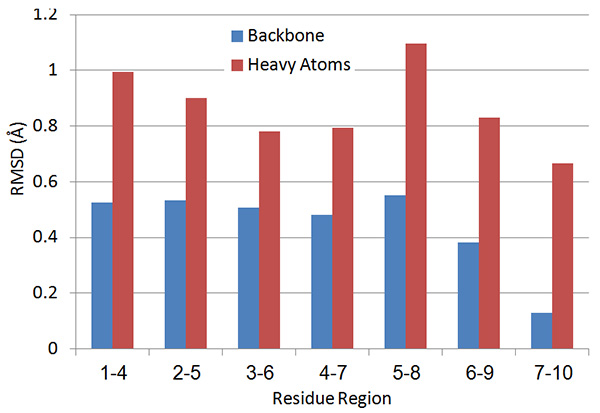
Figure 15. Local four-residue RMSD values along the peptide sequence.Click here to view larger image.
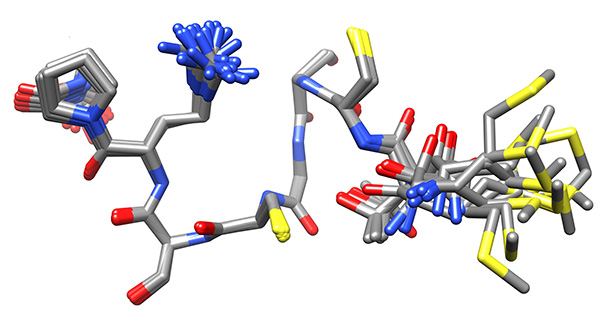
Figure 16. Low-energy ensemble of copper-bound peptide without any restraints to the metal, representing the low-energy conformations that have no violations of experimental NMR-derived constraints, superimposed on core region of larger stability. This ensemble will be used to determine the residues that interact with the copper ion.Click here to view larger image.
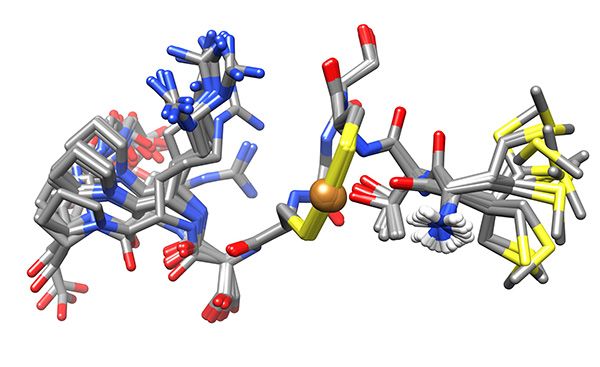
Figure 17. Low-energy ensemble of copper-bound peptide, representing the low-energy conformations that have no violations of experimental NMR-derived constraints.Click here to view larger image.
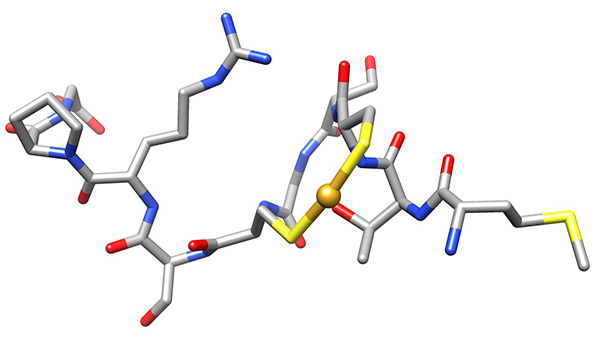
Figure 18. Low-energy conformer of copper-bound peptide, representing the low-energy conformations that have no violations of experimental NMR-derived constraints.Click here to view larger image.
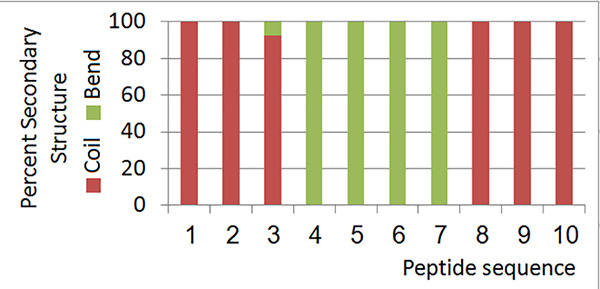
Figure 19. Secondary structure information for all members of the low-energy ensemble.Click here to view larger image.
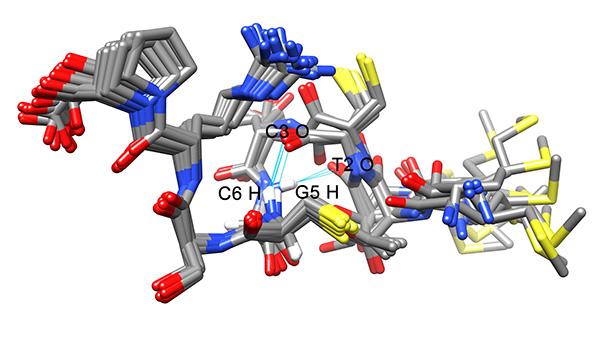
Figure 20. Intramolecular hydrogen bounds found in the conformers of the ensemble.Click here to view larger image.
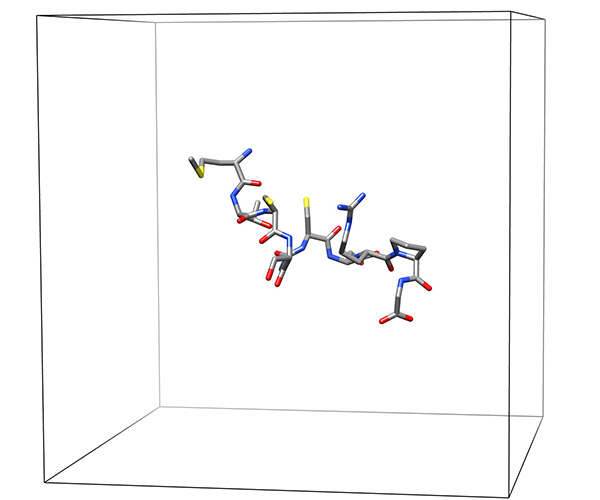
Figure 21. Low-energy representative conformation in grid used to calculate electrostatic potential distribution.Click here to view larger image.
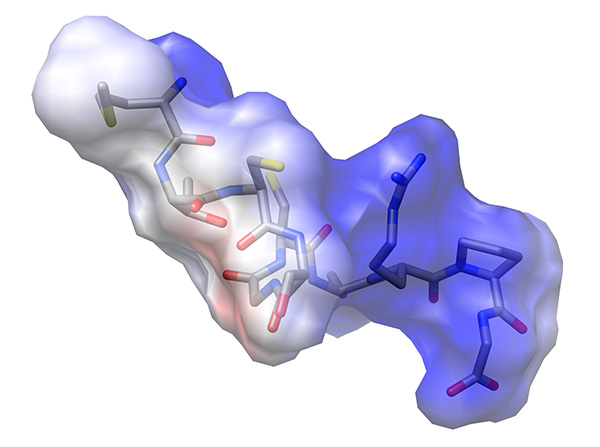
Figure 22. Electrostatic potential mapped onto Van der Waals surface of low-energy representative conformation of peptide.Click here to view larger image.
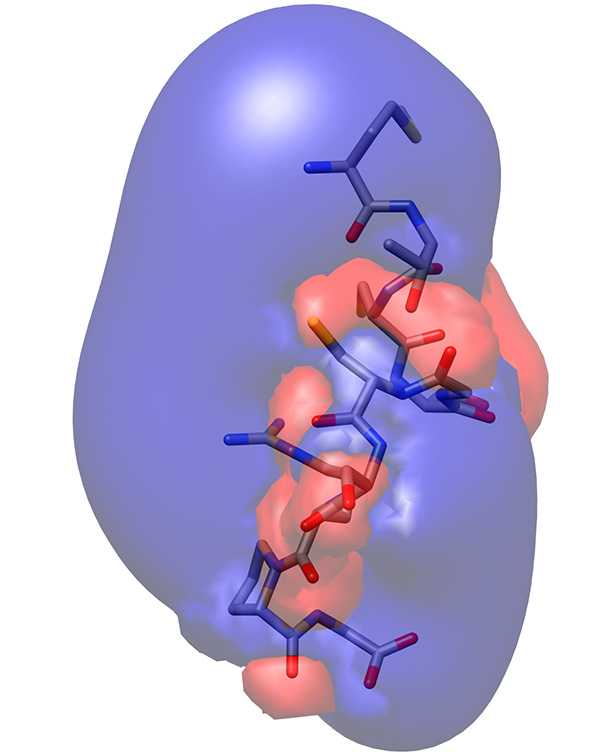
Figure 23. Electrostatic potential iso-surface of low-energy representative conformation of peptide showing +/- 1 kT/e in blue and red, respectively.Click here to view larger image.
Discussion
The contribution of structural information to understand binding mechanisms is well-accepted. Peptides are useful as models for protein binding and interactions; however they are not amenable to the main method for structure determination, X-ray crystallography. NMR is particularly useful for these systems, since the structures can be readily solved in solution. This is especially for the case of metallochaperone-mimetics that additionally require structure determination under an inert environment to prevent oxidation of the metal ion.
The MTCSGCSRPG peptide, containing the conserved MT/HCXXC motif, bound Cu (I) as was evident by the significant change of spectrum from the apo-form to the peptide reacted with copper. The need for a ROESY experiment at the field of 600 MHz, due to a spectrum with null interactions in the NOESY spectrum, indicates a compact peptide, since our experience shows that smaller peptides of 6-7 residues fall in the null signal of the NOESY regime, but peptides of this size usually give adequate signal. In the ROESY spectrum 81 cross-peaks were observed, N of these were inter-residue cross-peaks and (81-N) were intra-residue cross-peaks. This is a small number of peaks compared to proteins, but is expected in small peptides; Particularly cyclic peptides, which tend to give a small number of interactions since all the sidechains point outward and undergo little interaction with one another.
As the metal itself cannot be detected directly by the 1H NMR measurements, one must conclude on the metal binding residues from the distances obtained between suspected donor atoms. To assure a reliable structure, no metal-ligand binding constraints should be added to the initial calculations. Previous studies have shown that forcing metal binding in an incorrect form may still lead to reasonable structural factors even if the structure is incorrect10.
The experiments gave highly nonviolated conformations in an ensemble of low RMSD. The low RMSD of a potentially flexible peptide lends further support for copper binding, which would reduce the conformational flexibility of the molecule. The RMSD values of the binding region were reduced to values around 0.05 Å, which shows tremendous stabilization as expected by the ring closure. The secondary bend and hydrogen-bonding found in the 3-7 region, also indicated binding in this region.
The negative charge obtained when two thiols bind the copper (I) peptide is offset by the N-terminal amine that was held proximate to the bound copper.
When inspecting the resulting distances between potential donor atoms, including the two cysteine residues and the methionine group, the ones located at positions most probable to bind metal were the sidechains of Cys3 and Cys6. Therefore, binding constraints were added between these residues and the metal center, and the resulting structure was evaluated. To further support the resulting structure, various additional control measurements that include preset bonds to other residues may be performed and the structural factors compared. This is especially important where the result of the model is unexpected. In previous studies using similar measurements using protein-mimetic peptides, unusual binding modes were observed, including methionine instead of cysteine7.
Excess copper is toxic to biological systems and copper transport is very tightly controlled. Therefore, it is interesting and mechanistically important to understand how copper is transferred from one protein to another. The transport cannot depend on simple release and acquire mechanism, but must somehow include both stronger and weaker modes of binding, much like how one would transfer an object carefully from the fingers of one hand to another. This type of study provides much information regarding the mechanism of copper binding in biological systems and can be used to further investigate many different aspects of metallochaperone activity in nature. The systems may be easily mutated and manipulated to mimic many different aspects of copper-binding in nature, and may be analyzed without using prior assumptions of the binding mode.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank the Human Frontier Science Program (Young investigator grant (RGY)0068-2006) for financial support.
Materials
| Avance DMX 600 MHz Spectrometer | Bruker | ||
| NMR sample tubes | Wilmad | 535-PP | |
| Glove box | MBraun | LM05-019 | |
| Lyophilizer | VirTis | benchtopK | |
| Peptide | BioChemia | Custom made | >95% purity |
| Copper (1) chloride | Aldrich | 224332 | |
| Hydrochloric acid | BioLab | 231-595-7 | |
| Sodium hydroxide | Gadot | 1310-73-2 | |
| d6-Dimethylsulfoxide | Aldrich | 236926 | |
| Deuterium oxide | Aldrich | 151882 |
Referenzen
- Robinson, J. A. Protein epitope mimetics as anti-infectives. Curr. Opin. Chem. Biol. 15, 379-386 (2011).
- Huffman, D. L., Function O’Halloran, T. V. structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 70, 677-701 (2001).
- Tapiero, H., Townsend, D. M., Tew, K. D. Trace elements in human physiology and pathology. Copper. Biomed., & Pharmacotherapy. 57, 386-398 (2003).
- Boal, A. K., Rosenzweig, A. C. Structural Biology of Copper Trafficking. Chem. Rev. 109, 4760-4779 (2009).
- Rubino, J. T., Franz, K. J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129-143 (2012).
- Wernimont, A. K., Huffman, D. L., Lamb, A. L., O’Halloran, T. V., Rosenzweig, A. C. Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Biol. 7, 766-771 (2000).
- Singleton, C., Hearnshaw, S., Zhou, L., Le Brun, N. E., Hemmings, A. M. Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem. J. 424, 347-356 (2009).
- Hearnshaw, S., et al. A Tetranuclear Cu(I) Cluster in the Metallochaperone Protein CopZ. Biochem. 48, 9324-9326 (2009).
- Shoshan, M. S., Tshuva, E. Y. The MXCXXC class of metallochaperone proteins: model studies. Chem. Soc. Rev. 40, 5282-5292 (2011).
- Shoshan, M. S., et al. NMR characterization of a Cu(I)-bound peptide model of copper metallochaperones: Insights on the role of methionine. Chem. Comm. 47, 6407-6409 (2011).
- Schmitt, W., Zanotti, G., Wieland, T., Kessler, H. Conformation of different S-deoxo-Xaa(3)-amaninamide analogues in DMSO solution as determined by NMR spectroscopy. Strong CD effects induced by beta I, beta II conformational change. J. Am. Chem. Soc. 118 (3), 4380-4387 (1996).
- Behrens, S., Matha, B., Bitan, G., Gilon, C., Kessler, H. Structure-activity relationship of the ring portion in backbone-cyclic C-terminal hexapeptide analogs of substance P – NMR and molecular dynamics. Int. J. Peptide and Protein Res. 48, 569-579 (1996).
- Aue, W. P., Bartholdi, E., Ernst, R. R. 2-Dimensionsl spectroscopy- application to nuclear magnetic resonance. J. Chem. Phys. 64, 2229-2246 (1976).
- Bax, A., Davis, D. G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65, 355-360 (1985).
- Kumar, A., Ernst, R. R., Wüthrich, K. A two-dimensional nuclear overhauser enhancement (2D NO) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Comm. 95, 1-6 (1980).
- Bax, A., Davis, D. G. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 63, 207-213 (1985).
- Williamson, M. P. Chapter 3 Applications of the NOE in Molecular Biology. Annu. Reports on NMR Spectroscopy. 65, 77-109 (2009).
- Piotto, M., Saudek, V., Sklenar, V. Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous solutions. J. Biomol. NMR. 2, 661-665 (1992).
- Wüthrich, K. . NMR of proteins and nucleic acids. , (1986).
- Nilges, M., Kuszewski, J., Brünger, A. T. . Comp. aspects study biol. macromol. by NMR. , (1991).
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Molecular Graphics. 14, 51 (1996).
- Pettersen, E. F., et al. UCSF chimera – A visualization system for exploratory research and analysis. J. Computational Chem. 25, 1605-1612 (2004).
- Pearlmen, D. A., et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. 91, 1-41 (1995).
- Honig, B., Sharp, K., Yang, A. S. Macroscopic models of aqueous solutions- biological and chemical applications. J. Phys. Chem. 97, 1101-1109 (1993).

