Patch-Clamp Recordings from the Dendrite of a Dopaminergic Neuron in a Brain Slice
Abstract
Source: Engel, D. Subcellular Patch-clamp Recordings from the Somatodendritic Domain of Nigral Dopamine Neurons. J. Vis. Exp. (2016).
This video demonstrates a technique for patch-clamp recording from the dendritic membrane surface. This helps to directly examine the electrical properties of dendrites and underlying voltage-gated ion channels in dopaminergic neurons.
Protocol
All procedures involving animal models have been reviewed by the local institutional animal care committee and the JoVE veterinary review board.
1. Preparation of the Solutions
- Standard artificial cerebrospinal fluid (ACSF; Table 1)
- Use high-purity salts and clean glass beakers previously rinsed with double-distilled water to prepare a fresh solution. Use high-quality double-distilled water for the preparation of all extra- and intracellular solutions.
- According to Table 1, take a 2 L beaker to dissolve NaHCO3 in 500 mL water and another to dissolve the other salts to prevent precipitation of divalent ions. Clean the spatula systematically with double-distilled water and dry it before taking a salt. Use a magnetic stirrer to homogenize dissolution. Add the solutions from both beakers to an appropriate volumetric flask and bring them to the appropriate volume.
- Ensure that the final extracellular solution is fully transparent and without any trace of precipitation.
- Apply a gas mixture composed of 95% O2 and 5% CO2 (carbogen gas) with a glass microfilter candle (Ø 13 mm) 20 – 30 min before perfusing the slices. Carefully control the osmolarity (2 – 3 consecutive measurements) of the ACSF with an osmometer (optimal range: 314 – 325 mOsmol/L). Store the remaining solution after preparation at 4 °C and use it within 3 days.
- Cutting solution (sucrose-ACSF; Table 2)
- Prepare 3 L of fresh solution. Use 1 L to prepare brain slices from one animal. Store the remaining solution at 4 °C and use it within 3 days.
NOTE: High Mg2+ and low Ca2+ concentrations are used to decrease synaptic transmission, and some NaCl is substituted with sucrose to preserve the tissue.
- Prepare 3 L of fresh solution. Use 1 L to prepare brain slices from one animal. Store the remaining solution at 4 °C and use it within 3 days.
- Intracellular solutions
- Prepare in advance 100 mL solution for dual whole-cell recordings (Table 3).
- Include methylsulfate to obtain good recovery of cell morphology. Add biocytin (0.1 – 0.4%) to examine the neuron's morphology subsequently. Include a fluorescent dye (e.g., Alexa 594 or Sulforhodamine 101 41) to follow the extension of dendrites during the recording (optional).
- Prepare in advance 100 mL electrode solution for cell-attached recordings (Table 4). In this case, the internal solution is a high-K+ solution designed to isolate the macroscopic hyperpolarization-activated cation current (Ih) and contains blockers for the other voltage-gated ion channels.
- Store intracellular solutions in 2 mL aliquots at -20 °C. Use a new aliquot for every new recording session. Check the osmolarity of every thawed aliquot carefully with an osmometer. Take a new aliquot if the osmolarity does not correspond to the initial value.
- Transfer the solution from the aliquot into a 3 mL syringe on the top of which a 0.22 µm sterile syringe filter is placed. Keep the syringe on ice during the recording session to limit degradation of its compounds (ATP, GTP or phosphocreatine).
2. Fabrication and Filling of Patch Pipette
- Pulling
- Use a horizontal electrode puller and thick-walled borosilicate glass tubing. For whole-cell and cell-attached recordings, use a 2 mm outer diameter/1 mm inner diameter. Ensure that the glass tubing is absolutely clean.
- If this is not the case, immerse glass capillaries in an organic solvent (e.g., ethanol) first and then in double-distilled water. Briefly heat capillary endings with a Bunsen burner. Alternatively, order glass tubing washed and heated directly from the manufacturer (see Materials list).
- For dual somatodendritic recordings, ensure that the somatic pipette resistance is between 6 and 10 MΩ and between 8 and 19 MΩ for dendritic pipettes when filled with intracellular solution (Table 3). Standardize pipette resistance for cell-attached recordings to a resistance of 10 MΩ to achieve comparable results along the somatodendritic domain of the neuron.
- Prepare fresh pipettes before every recording session (every day) or immediately before patching and use them 5 – 8 h after their fabrication. Store pipettes in a covered glass container to protect them from dust and obstruction of the tip.
- Polishing
- Inspect and heat-polish every pipette tip with a microforge to obtain better seals with the membrane. Before using the microforge, melt a tiny piece of glass on the platinum heating filament. Replace this glass coating on the platinum filament daily for constancy (Peter Jonas, personal communication).
NOTE: Pipettes used for cell-attached recordings can be coated before the heat-polishing step to reduce background noise. An insulating agent such as molten dental wax or a silicone elastomer can be used to coat the pipettes.
- Inspect and heat-polish every pipette tip with a microforge to obtain better seals with the membrane. Before using the microforge, melt a tiny piece of glass on the platinum heating filament. Replace this glass coating on the platinum filament daily for constancy (Peter Jonas, personal communication).
3. Preparation of Brain Slices
- Use healthy Wistar rats aged between 16 – 19 days old. Do not use unhealthy animals or animals suffering from hypothermia or dehydration. Before starting the preparation of slices keep the animal in a safe and silent place. Avoid having any other animal in the room while preparing an animal. Manipulate animals gently.
- Pour sucrose-ACSF into two 400 mL polypropylene (PP) beakers and place them in the -80 °C freezer for 45 min. Mix the solution to get a homogenous ice-cold liquid/frozen solution and place the beakers on ice. Supply the sucrose-ACSF solution with carbogen gas using a microfilter candle (Ø 13 mm) in each PP beaker.
- Prepare the slicer and the reserve chamber
- Use a high-quality tissue slicer with extremely low vertical vibrations to minimize damage to the superficial layers of slices as much as possible. Fix a fresh and entire razor blade on the slicer.
- Use a new razor blade for each cutting session. Avoid bending of the razor blade. Do not remove the fatty film at the surface of the razor blade (Josef Bischofberger, personal communication).
- If available, check the vertical vibration with a vibroprobe provided by the manufacturer. Alternatively, use a custom-made vibroprobe. Reduce the vertical vibrations of the blade so as to be as close as possible to 0 µm.
- Prepare a 150 mL reserve chamber for slices. Pour standard ACSF into the reserve chamber and place it in a water bath. Supply the reserve chamber solution with carbogen using a microfilter candle (Ø 6 mm). Ensure that the carbogen bubbles are as tiny as possible.
NOTE: Build a new reserve chamber every 2 – 3 months to keep the slice quality constant.
- Cut slices
- In a silent room in which the experimenter is not disturbed or stressed, decapitate the animal with surgery scissors (length 150 mm) at the level of the cervical medulla.
- Cut the skin at the top of the animal's head in the nasal to caudal direction with a scalpel and remove it laterally. Cut the top of the skull from the caudal to the nasal part of the head with fine scissors and remove both parts of the skull laterally. Remove the brain with a thin spatula and drop it carefully into a PP beaker containing ice-cold (0 – 4 °C) sucrose-ACSF equilibrated with carbogen.
NOTE: This sequence of steps needs to be done as quickly as possible, but also meticulously (<<1 min, roughly 40 s). - Keep the brain in the solution of the PP beaker for ~2 to 5 min. Adjust the carbogen pressure to prevent brain movements. If the carbogen bubbles are too large, replace the microfilter candle with a fresh one.
- Place the brain on a 9 cm Petri dish whose inside bottom is covered with a ~0.5 cm thick Sylgard layer. Prepare this petri dish in advance.
- Surround the brain with ice-cold sucrose-ACSF. Ensure that some liquid phase of ACSF-sucrose solution submerges the brain. For coronal slices, cut off the frontal part of the brain in the coronal plane with a scalpel or a razor blade. Remove the cerebellum with a coronal cut.
- Apply cyanoacrylate glue on the specimen tray (area ~1 cm2). Paste the brain block such that the frontal part faces the slicing stage. Carefully drip sucrose-ACSF on top of the pasted brain using the large opening of a glass Pasteur pipette and subsequently slowly submerge the cutting chamber of the slicer. Maneuver the specimen tray so that the cutting surface (i.e., the first tissue to hit the blade) is the ventral surface of the brain.
- Apply carbogen with a bended glass microfilter candle (Ø 6 mm) to the cutting chamber (optional).
- Adjust the razor blade at an angle of ~15° with reference to the horizontal plane. Cut coronal brain slices of ~500 µm in the caudal to nasal direction before reaching the substantia nigra. Decrease the thickness to cut 300 – 350 µm thick slices containing the region of interest.
- Separate slices from the tissue block with a very thin perfusion needle attached to a syringe parallel to the edge of the razor blade. Bend the needle so as to have an angle of 90° between the needle and the syringe. Ensure that no pressure is applied to the razor blade while removing the slice from the tissue block.
- Store slices
- Transfer the slices using the largest opening of a glass Pasteur pipette (attached to a bulb) into the reserve chamber. Once all the slices are transferred into the reserve chamber (Christoph Schmidt-Hieber, personal communication) bring the temperature of the water bath to 34 °C for 0.5 – 1 h.
- After this period switch off the water bath and keep the slices at room temperature. Adjust the carbogen pressure in order to avoid movements of the slices in the reserve chamber. Keep the slices for another 10 to 20 min before starting recordings.
- Clean the equipment and the microfilter candles carefully yet thoroughly with double-distilled water at the end of the slicing procedure.
4. Dual Somatic and Somatodendric Recordings in Nigral Neurons and Biocytin Filing
- Follow descriptions for the assembly of a patch-clamp setup for slices given in The Axon Guide.
- Prepare recording chamber
- Place 1 – 2 mL of double-distilled water in the recording chamber. Verify that it is not leaky. Place the recording chamber on the shifting x-y table.
- Feed standard ACSF through an oxygen-impermeable perfusion tubing system (polytetrafluoroethylene) to the recording chamber. Stabilize the perfusion flow in the chamber at a rate of 4 – 5 mL min-1. Keep the length of the tubing joining the beaker containing ACSF and the recording chamber as short as possible (1 m).
- Select a brain slice from the reserve chamber and transfer it into the recording chamber using the largest opening of a glass Pasteur pipette. Cover the slice with a platinum ring to anchor it at the bottom of the recording chamber. Use a flat platinum ring (Ø 1.5 cm) and glue parallel nylon threads (spacing >2 mm) on it.
- Adjust and optimize the optics
- Visualize the slices using infrared (IR) video microscopy. Adjust the Köhler illumination 51 and optimize differential interference contrast (DIC) optics or the oblique illumination (Dodt Gradient contrast – DGC).
- Check the quality of the slice. Only keep slices with smooth and even surfaces that are not too strongly contrasted and have only small, dispersed craters.
- Fill 2 fresh and unused patch pipettes with intracellular solution (Table 3). Use a 0.5 mL tube containing the pipette solution to fill the tip of the pipette by capillarity action. To accelerate the tip filling of pipettes without filament, apply negative pressure to the end of the pipette using tubing attached to a syringe.
- Position the pipettes above the surface of the slice
- Check the presence of chloride coating on the silver wires.
- Insert the pipettes sequentially into the pipette holders and apply positive pressure (~70 mbar) using a tubing circuit connecting the pipette holder, a three-way tap and a manometer. Lower the pipette into the bath of the recording chamber.
- Check that the pressure value on the manometer is constant for ~1 – 2 min. If the pressure is decreasing, check the tubing or the sealing O-rings inside the pipette holder.
- Place the pipette tip in the middle of the video monitor and ensure that it is not obstructed. Ensure that the pipette tip does not move when applying or releasing pressure to the pipette.
- Check for drift and vibrations of the pipette tip by drawing a small cross at the center of the monitor exactly at the position of the pipette tip and observe for 5 min possible movements of the tip.
- Apply a voltage step (5 mV) to monitor the pipette resistance on an oscilloscope. Set the holding potential of the patch-clamp amplifier to 0 mV. Cancel pipette offset potential such that the DC pipette current reads zero at the amplifier meter.
- Move the pipettes down to the slice and maintain them above the surface.
- Select and patch a neuron with long dendrites
- Within the substantia nigra select a healthy neuron with long dendrites that can be followed over a long distance in approximately the same plane (Figure 1A and 1B) using IR-Dodt Gradient Contrast (IR-DGC). Choose a smooth and homogeneous cell body. Avoid the two strongly contrasted cells at the surface of the slice (Figure 1C and 1D), because it is difficult to break in and to preserve stable recording conditions over time (stable series resistance). Select neurons that have their soma 10 – 30 µm underneath the slice surface.
- Move the somatic electrode close to the soma (40 – 50 µm away) and the dendritic electrode close to the dendrite at the same distance. Use a 2X magnification (fourfold changer) to select and patch a portion of a dendrite. Obtain on the monitor an absolute magnification of 2,100X without magnification (1X) and of 5,500X with a 2X magnification.
- Slightly release the pressure of the somatic pipette by 10 mbar. If necessary, regulate dendritic and somatic pipette pressure to avoid displacement of the cell structure (soma and dendrite) within the slice.
- Position the dendritic pipette close to the membrane in order to see a small dimpling. Release the pressure on the pipette tip and patch the dendrite while controlling the pipette resistance. In ideal conditions, only a very slight suction is sufficient to obtain a good seal.
- Reduce the amplitude of the pipette capacitance transients as much as possible on top of the current step using a voltage pulse with high gain and little filtering 51 and the oscilloscope. Enter the whole-cell mode with the dendritic pipette.
Representative Results
Table 1: Artificial cerebrospinal fluid (ACSF)
| Substance | g/mol | Concentration | for 1 L |
| NaCl | 58.443 | 125 mM | 7.305 g |
| NaHCO3 | 84.007 | 25 mM | 2.100 g |
| KCl | 74.551 | 2.5 mM | 0.186 g |
| NaH2PO4 | 137.99 | 1.25 mM | 0.172 g |
| glucose | 198.17 | 25 mM | 4.95 g |
| MgCl2 | 1 M (solution) | 1 mM | 1 mL |
| CaCl2 | 1 M (solution) | 2 mM | 2 mL |
| Osmolarity : ~ 310 mOsmol/L (optimal range: 314 – 325 mOsmol/L) , pH = 7.4 | |||
Table 2: Sucrose-ACSF to prepare slices
| Substance | g/mol | Concentration | for 1 L |
| NaCl | 58.443 | 87 mM | 5.084 g |
| NaHCO3 | 84.007 | 25 mM | 2.1001 g |
| KCl | 7.551 | 2.5 mM | 0.18637 g |
| NaH2PO4 | 137.99 | 1.25 mM | 0.17248 g |
| MgCl2 | 1 M (solution) | 7 mM | 7 mL |
| glucose | 198.17 | 10 mM | 1.9817 g |
| sucrose | 342.29 | 75 mM | 25.672 g |
| CaCl2 | 1 M (solution) | 0.5 mM | 0.5 mL |
| Osmolarity : ~ 326 mOsmol/L, pH = 7.4 | |||
Table 3: Intracellular solution for dual recordings
| Substance | g/mol | Concentration | for 100 mL |
| KMeSO4 | 150.2 | 120 mM | 1.8024 g |
| KCl | 74.56 | 20 mM | 0.14912 g |
| MgCl2 | 1 M (solution) | 2 mM | 200 µl |
| Na2ATP | 551.1 | 2 mM | 0.1102 g |
| Na2GTP | 523.2 | 0.5 mM | 0.02661 g |
| Na2-Phosphocreatine | 255.1 | 5 mM | 0.1275 g |
| EGTA | 380.4 | 0.1 mM | 3.803 mg |
| Hepes | 238.31 | 10 mM | 0.23831 g |
| Biocytin | 1 mg/mL | 0.1 g | |
| Osmolarity : ~ 302 mOsmol/L , pH = 7.2 adjusted with KOH | |||
Table 4: Electrode solution for cell-attached recordings
| Substance | g/mol | Concentration | for 100 mL |
| KCl | 74.56 | 120 mM | 0.8947 g |
| CaCl2 | 1 M (solution) | 2 mM | 200 µl |
| MgCl2 | 1 M (solution) | 1 mM | 100 µl |
| Hepes | 238.31 | 10 mM | 0.23831 |
| TEA-Cl | 165.7 | 20 mM | 0.3314 g |
| 4-AP | 94.11 | 5 mM | 0.04705 g |
| BaCl2 | 244.26 | 1 mM | 0.02443 g |
| CdCl2 | 183.32 | 0.02 mM | 0.3666 mg |
| TTX | 1 mM | 200 nM | 20 µl |
| Osmolarity: ~ 290 mOsmol/L, adjusted with D-glucose (Ref. 16); pH = 7.4 | |||
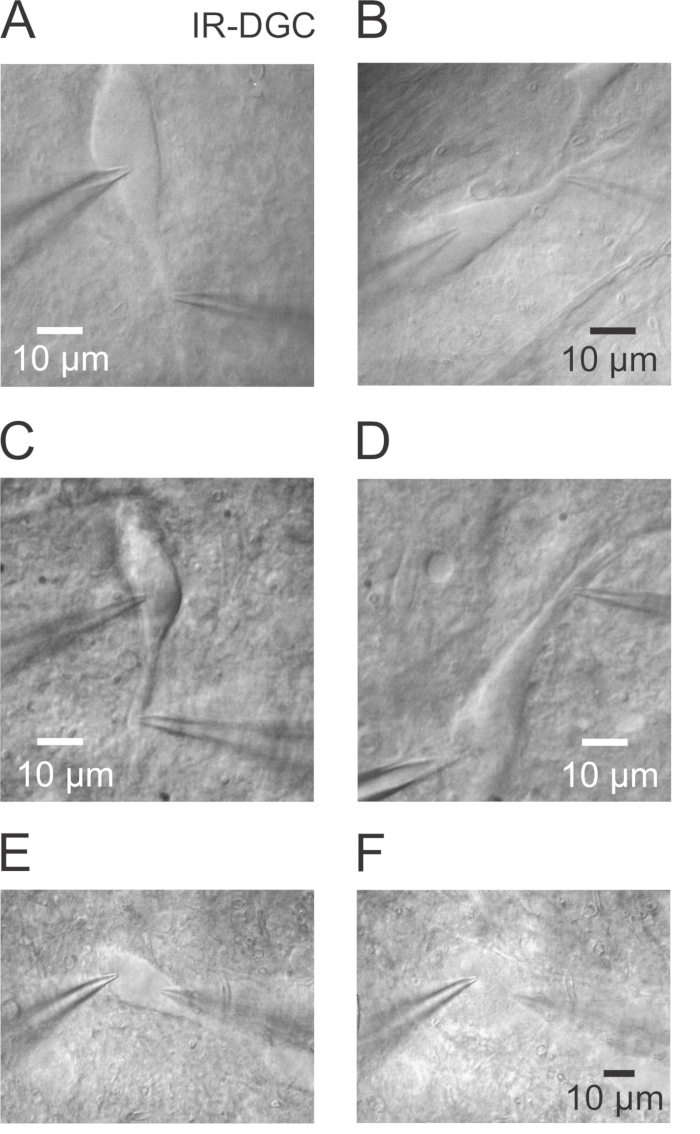
Figure 1. Visualizing the Soma and Proximal Dendrites of Nigral Dopaminergic Neurons Using Infrared Videomicroscopy. (A) IR-DGC image of a nigral DA neuron showing the soma and proximal dendrite and both somatic and dendritic pipettes. A dual somatodendritic recording was successfully performed on this healthy neuron. Observe the low contrast and the smooth surface all over the somatodendritic domain of the cell. (B) Another example of a healthy DA neuron. (C) DA neuron with a rougher and uneven surface and a stronger contrast. While a dual recording has been obtained, the stability was suboptimal, and the recording duration was short (~ 15 min) due to a gradual increase of the access resistance at both pipettes. (D) Second example of a DA neuron showing a strongly contrasted soma and proximal dendrite. In this case, a strong increase of the access resistance was observed shortly after the break in the whole-cell mode. An attempt to decrease the access resistance by negative pressure was unsuccessful. Neurons in panels C and D might have been damaged during the slicing procedure and should be avoided for experiments. (E) Simultaneous double somatic recording in a healthy DA neuron at the beginning of the recording. (F) Same neuron as in panel E with a swollen cell body after ~17 min. Note the full absence of contrast, the ball-like appearance of the soma and the presence of the large nucleus which is not apparent in healthy neurons.
Offenlegungen
The authors have nothing to disclose.
Materials
| Double-distillated water | Millipore | Super Q | Resistivity >14 MΩ cm, ideal 18.2 MΩ cm at 25°C, filtered (0.22 µm), https://www.merckmillipore.com/BE/en/product/Super-Q-Plus-Water-Purification-System,MM_NF-C1760 |
| Microfilter candles | Robu | Porosity 3, 6 mm or 13 mm diameter | |
| Tissue slicer with vibroprobe | Leica | VT1200 | cutting parameters: speed 0.07, amplitude 1.40 |
| Tissue slicer | Dosaka | DTK-1000 | cutting parameters: speed 4, frequency 7 |
| Razor blades | Gilette | Super Silver | |
| Dissection tools | Braun, Aesculap | ||
| Reserve chamber | Custom-made | ||
| PP beckers | VWR | 213-1725 | 400 ml |
| Syringe filter | Merk Millipore | Millex – GV 0,22 µm | |
| Cyanoacrylate glue | UHU | "Sekunden Kleber" | liquid glue |
| Recording chamber | Luigs & Neumann | Slice mini chamber | |
| Horizontal pipette puller | Sutter Instruments | Brown-Flaming P-97 | |
| Pipettes | Hilgenberg | 1807524 | 2 mm o.d./1 mm i.d. glass capilaries, ends firepolished, washed |
| Dental wax | Coltène/Whaledent | orthodontic tray wax strips | |
| Platinum grid | to anchor slices in the recording chamber, custom made with a platinum disk | ||
| Microforge | Narishige | MF-830 | to fire-polish pipette tips |
| Potassium methyl sulfate | MP Biomedicals | 215481 | |
| Biocytin | Molecular probes | B1592 | |
| Manometer | GREISINGER electronic | GDH 13 AN | |
| Microscope | Zeiss | FS | |
| Dodt Gradient Contrast | Luigs & Neumann | ||
| Manipulators | Luigs & Neumann | LN Mini 25 | |
| Frame grabber | The Imaging Source | DFG/USB2pro | |
| Camera | DAGE-MTI | NC-70 | |
| Condenser | Zeiss | high numerical aperture condenser working with oil | |
| Objective | Zeiss | W Plan Apochromat 441470-9900 VIS-IR | 63x magnification, long-distance, high numerical aperture 1.0 water-immersion objective |
| Fourfold-changer | Luigs & Neumann | between the camera and the microscope | |
| Black & white video monitor | Sony | SSM-175CE | 16-inch, 850 TV lines, analog |
| Sample Bottles, Amber Glass, with Cap | VWR | 215-2409 | to transfer slices from the setup to histology room |
| The Axon Guide | Molecular Devices | Book | |
| Paraformaldehyde | Sigma | 441244 | see SAFETY DATA SHEET and Dangerous Substances Directive (EU) before use |
| Well cell culture plate | Greiner-Bio-One | 662160 | for the staining of fixed slices |
| Triton X-100 | Merk | 108643 | see SAFETY DATA SHEET and Dangerous Substances Directive (EU) before use |
| ProLong Diamond Antifade | Molecular Probes – Thermo Fisher Scientific | P36961 |

