Assessing Cardiac Reprogramming using High Content Imaging Analysis
Summary
We present a protocol to quantify directly reprogrammed induced cardiomyocyte-like cells (iCMs) in vitro using high content imaging analysis. This method allows us to quantify the efficiency of cardiac reprogramming in an automated manner and to directly visualize iCMs.
Abstract
The goal of this protocol is to describe a method for quantifying induced cardiomyocyte-like cells (iCMs), which are directly reprogrammed in vitro by a reprogramming technique. Cardiac reprogramming provides a strategy to generate new cardiomyocytes. By introducing core cardiogenic transcription factors into fibroblasts; fibroblasts can be converted to iCMs without transition through the pluripotent stem cell state. However, the conversion rate of fibroblasts to iCMs still remains low. Accordingly, there have been numerous additional approaches to enhance cardiac reprogramming efficiency. Most of these studies assessed cardiac reprogramming efficiency using flow cytometry, while at the same time performed immunocytochemistry to visualize iCMs. Thus, at least two separate sets of reprogramming experiments are required to demonstrate the success of iCM reprogramming. In contrast, automated high content imaging analysis will provide both quantification and qualification of iCM reprogramming with a relatively small number of cells. With this method, it is possible to directly assess the quantity and quality of iCMs with a single reprogramming experiment. This approach will be able to facilitate future cardiac reprogramming studies that require large-scale reprogramming experiments such as screening genetic or pharmacological factors for enhancing reprogramming efficiency. In addition, the application of high content imaging analysis protocol is not limited to cardiac reprogramming. It can be applied to reprogramming of other cell lineages as well as any immunostaining experiments which need both quantification and visualization of immunostained cells.
Introduction
Cardiac reprogramming has been developed as an alternative approach to stem cell mediated approaches to generate new cardiomyocytes. Given that it does not transition through stem cell state, it has a high potential to bypass some inherited limitations in stem cell mediated approaches. It has been shown that viral infection of at least three or four cardiogenic transcription factors into fibroblasts can convert fibroblasts toward a cardiac fate by eliminating fibroblast gene programs and rebuilding cardiogenic transcriptional networks in fibroblasts1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17.
Since the first landmark study demonstrating cardiac reprogramming in vitro1, the cardiac reprogramming protocol has been optimized by numerous studies3,5,6,7,9,11,12,13,14,15,16,18. Common technical approaches to assess cardiac phenotypes in fibroblasts following cardiac reprogramming have been flow cytometry analysis for quantifying cells expressing specific cardiomyocyte markers and immunocytochemistry for visualizing those cells at a single cell level. Although both experiments (i.e., flow cytometry and immunocytochemistry) are to demonstrate expression of cardiomyocyte markers using the same antibodies, they have to be performed separately. In addition, flow cytometry needs a relatively larger number of cells, thereby increasing the amount of reagents needed for the experiment. Alternatively, cardiomyocyte marker positive cells can be quantified by manual counting following immunocytochemistry. However, it is very labor intensive and tends to be less accurate.
The purpose of this protocol is to describe the method that can quantify and visualize iCMs by a single immunostaining experiment using automated high content imaging analysis. It requires a relatively small number of starting cells because this protocol is performed in a well of 24-well plate. As many as three different markers can be used at the same time. Single, double, and triple positive cells can be automatically quantified. In addition to quantification of immunostained cells, high content imaging analysis provides high quality 2-100x objective images. If necessary, the same immunostained cells used in high content imaging analysis can be re-used for further imaging studies, such as confocal microscopy. The main advantage of this protocol is that it provides not only unbiased quantification of iCMs with a much smaller number of cells, but also visualization of iCMs. Furthermore, this protocol can be utilized for assessing non-cardiac lineage reprogramming (e.g., iPSC, neuron, and hepatocyte reprogramming).
Protocol
All animal procedures were performed with the approval of Vanderbilt University Medical Center Institutional Animal Care and Use Committee.
1. Retrovirus generation and in vitro cardiac reprogramming
- Culture Platinum E cells in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 1 µg/mL puromycin, and 10 µg/mL blasticidin until Platinum E cell confluency reaches 70%–80%.
- On Day 1, seed ~0.55 x 106 cells (first well) and ~0.18 x 106 cells (second well) into two separate wells of 12-well plate with 1 mL of DMEM supplemented with 10% FBS and 1% penicillin/streptomycin a day prior to the first transfection.
NOTE: Different sizes of culture dishes can be used depending on the amount of viral media needed for the experiments. Refer to Table 1 for other scales of experiments. - On Day 2, transfect the quad-cistronic M-G-T-H retroviral construct encoding Mef2c, Gata4, Tbx5, and Hand2 described in the previous study9 or empty vector into Platinum E cells of the first well. Add 3 µL of transfection reagent to 30 µL of reduced serum media. Five minutes later, add 1 µg retroviral construct or the empty vector into the mixture of transfection reagent and reduced serum media. After 20 min incubation at room temperature, add the mixture to Platinum E cells.
- On Day 3, 16–20 h after transfection, remove the media and replenish fresh DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
- On Day 3, 24 h after the first transfection, perform the second transfection into the second well in which Platinum E cells were plated on Day 1 as described in step 1.3).
- On Day 3, seed ~5 x 104 frozen mouse embryonic fibroblasts (MEFs) isolated from Titin-GFP reporter knock-in mice19 into a well of 24-well plate. A total of two wells of 24-well plate are needed (i.e., uninfected control and M-G-T-H infection).
NOTE: The number of MEFs plated may need to be adjusted, because the recovery rate of frozen MEFs can vary depending on cell freezing conditions. About 10% confluency a day after seeding cells is adequate for reprogramming. Reprogramming efficiency could be enhanced using fresh unfrozen MEFs. - On Day 4, 16–20 h after the second transfection, remove the media and replenish fresh DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
- On Day 4, 48 h after the first transfection, collect the viral media in the first well of Platinum E cells using a 5 mL syringe and filter them through a 0.45 µm polyethersulfone (PES) membrane filter. Remove the fibroblast growth media on MEFs, and replace them with the viral media supplemented with polybrene at 6 µg/mL (first infection).
- On Day 5, 48 h after the second transfection, perform the second infection using the viral media in the second well as described in step 1.8).
- On Day 6, 24 h after the second infection, the viral media are replaced with cardiac induction media composed of DMEM/199 (4:1), 10% FBS, 5% horse serum, 1% penicillin/streptomycin, 1% non-essential amino acids, 1% essential amino acids, 1% B-27, 1% insulin-selenium-transferrin, 1% vitamin mixture, and 1% sodium pyruvate, 1 µM SB431542, and 0.5 µm A83-01. Change cardiac induction media every three days until the cells are harvested.
2. Immunostaining
- At 14–15 days post infection, fix the cells on a 24-well plate with 2% paraformaldehyde for 15 min.
- Permeabilize the fixed cells with permeabilization buffer (0.05% Triton-X in PBS) by washing the cells three times with permeabilization buffer every 5 min.
- Incubate the cells with universal blocking buffer for 45 min.
- Incubate the cells with mouse α-actinin (1:400 dilution) and chicken GFP antibodies (1:400 dilution) for 1.5 h at room temperature.
NOTE: About 150 µL of antibody solution can cover the whole area of a well of 24-well plate. - Wash the cells with permeabilization buffer for 5 min three times.
- Incubate the cells with anti-mouse Alexa-555 and anti-chicken Alexa-488 secondary antibodies (1:400 dilution) for 1 h at room temperature
- Wash the cells with permeabilization buffer for 5 min three times.
- Add 2.5 µL of DAPI solution into 250 µL of permeabilization buffer.
3. High content imaging
- Turn on the imaging system.
- Open the associated software.
- Log into the system.
- On the taskbar, select Run a Plate.
- Click on Open Door-Eject Plate to open the door on the machine and put the plate in. Make sure the plate is in the right direction. Click on Close Door-Load Plate to close the door.
- Click on Load Plate Settings. Select a template protocol, and then click on Load From DB.
- Click on Acquisition Setup.
- In the Plate Acquisition Setup dialog, click on Configure.
- In the Objective and Camera tab, select 10x objective.
- In the Plate tab, select a 24-well plate.
- In the Sites to Visit tab on Site Options, choose "Fixed number of sites" to determine the number of imaging sites by choosing the number in columns and rows. A total of 36 imaging sites are used by selecting 6 in columns and 6 in rows. Select the Spacing between each imaging site (i.e., 500 μm).
- In the Acquisition tab, set the number of wavelength as 3.
- In the Wavelengths tab on Illumination, select DAPI, FITC, or Texas Red for each wavelength separately.
- Click on the Run tab and then set up the "Folder Name" and "Plate Name" for the plate. Click the wells on the plate diagram for imaging and then on Calculate to set up the focus offset for each wavelength. Set up the foucs offset for DAPI first. Click on Auto Expose to set up the exposure time automatically for each wavelength. Then, click on Acquire Plate to start imaging the selected sites.
4. Analysis of high content imaging
- Once high content imaging is completed, click on the Screening menu, and select Review Plate Data to select a plate for analysis.
- Click on Select Plate. In the Select Plate for Review dialog, open the folder and select the plate saved in the database and then click on Select.
NOTE: To display the images, select DAPI, FITC, and Texas Red in the Wavelengths field. In the Sites field, select All Sites. Click on a site among the selected 36 sites to display it in each wavelength image window, and then click on Look-Up Table to select a color for the wavelength. Use the Print Screen key on the keyboard and paste the image in a photo editing software (e.g., Paint and Photoshop) to save it. - Click on Run Analysis, and select a template setting.
- Click on Configure Settings and then on Number of Wavelengths. Select three wavelengths (i.e., DAPI for nuclei staining, Teaxs Red for α-actinin, and FITC for Titin-GFP).
- Using the Line tool in the tool bar, measure the width across the short axis of a cell. Based on the measured widths of cells, set the “Approximate min width” and “Approximate max width” to include most of cells in the selected imaging site.
- Click on Preview to check whether nearly all nuclei are selected. The selected nuclei exhibit white color, while the unselected nuclei remain blue (DAPI stained). If necessary, make adjustments for “Approximate min width” and “Approximate max width”.
- To set Intensity Above Local Background, place a mouse cursor inside and outside of a cell. The intensity value appears at the bottom of the window. Slightly reduce the intensity of a dim cell to evenly exhibit the intensity throughout the whole area of each cell. Define this intensity value as Intensity Above Local Background value. This value needs to be set separately for each channel.
- Click on the Screening menu and select Plate Data Utilities. In the Plate Data Utilities dialog, click on Run Analysis to select the plate.
- In the Settings field, select the saved setting for analysis; select Add to Auto Run List, and then click on OK to run the analysis.
- After the analysis is completed, click on Screening menu and select Plate Data Utilities. Then, in the Plate Data Utilities dialog, click on Export Measurements to export the analysis results.
- Click on Cell and Image Measurements and then on OK.
- On the Export Measurements Wizard – Step 1 page, select the plate and click on Next.
- On the Export Measurements Wizard – Step 2 page, click on Finish.
- On the Configure Data Export page, select the data types (i.e., well name, total cell number, subtotal cell number for DAPI+, subtotal cell number for Texas Red+, subtotal cell number for FITC+, subtotal cell number for DAPI+Texas Red+, subtotal cell number for DAPI+FITC+, subtotal cell number for DAPI+FITC+Texas Red+, percentage of DAPI+ cells, percentage of DAPI+Texas Red+ cells, percentage of DAPI+FITC+ cells, and percentage of DAPI+Texas Red+FITC+ cells) and then click on OK.
- On the Export as text file page, select the destination to save the file and click on OK.
- Open the file in Microsoft Excel. The results will be presented by each site (Table 2). There are 36 sites per well. Using the analysis tool, Pivot Table, summarize the data of the 36 sites (i.e., the sums of cell numbers and the averages of the indicated cell percentage in all the 36 sites) (Table 3).
Representative Results
Following reprogramming experiments, we quantified iCMs using high content imaging analysis as described above. Composite images of 36 imaging sites that were used for high content imaging analysis were shown in Figure 1. iCMs are defined as double-positive cells (α-actinin+Titin-eGFP+) in these experiments. High content imaging analysis shows that ~26% of cells exhibited both cardiac markers following M-G-T-H transduction, while ~1% of empty vector transduced control cells show double-positive cells. Individual iCMs were visualized in 10x images taken by high content imaging system (Figure 2).
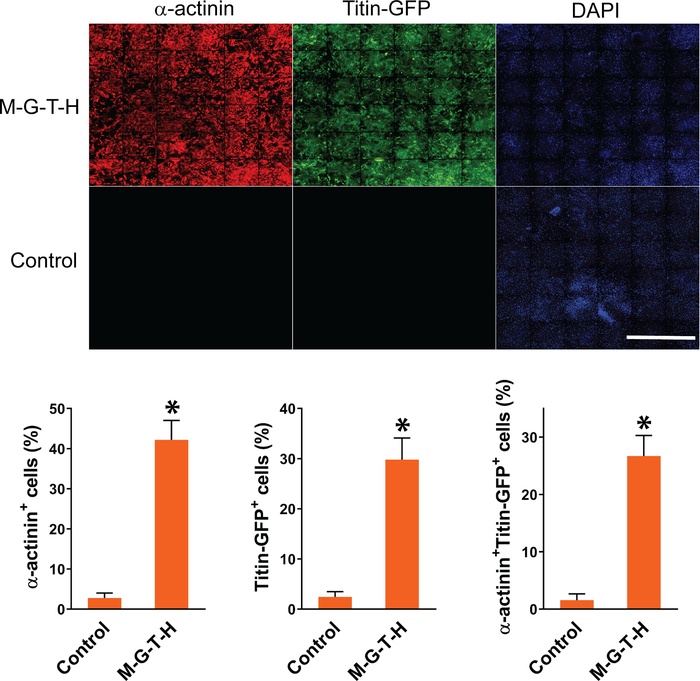
Figure 1: Representative composite images of automated high content imaging analysis. MEFs isolated from Titin-GFP knock-in mice were transduced with M-G-T-H construct or empty vector. Empty vector transduced cells were used as control. Fifteen days after transduction, the MEFs were immunostained for GFP and α-actinin. The immunostained cells were analyzed by automated high content imaging system to quantify DAPI+GFP+, DAPI+α-actinin+, or DAPI+GFP+α-actinin+ cells. The graph shows summary of high content imaging analyses. Three independent experiments are presented as mean ±S.D. *p < 0.005. Scale bar = 2.5 mm. Please click here to view a larger version of this figure.
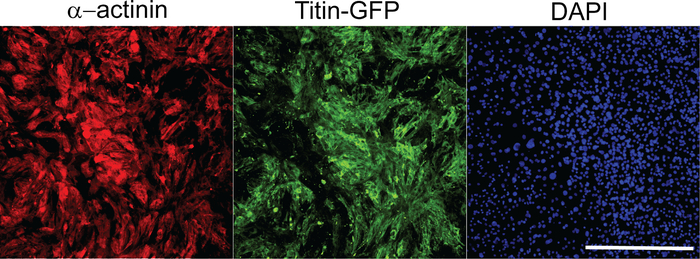
Figure 2: Representative images of 10x objective pictures used for high content imaging analysis. These images were chosen from the composite images of 36 imaging sites as shown in Figure 1. Each image represents an individual imaging site per each channel. Scale bar = 400 µm. Please click here to view a larger version of this figure.
| Cell culture dish | Platinum E cells (miilion/plate) | Media (ml) | Fugene 6 (µl) | Opti-MEM (µl) | DNA (µg) |
| 15 cm | 13.75 | 25 | 75 | 750 | 25 |
| 10 cm | 5.5 | 10 | 30 | 300 | 10 |
| 6 cm | 2.2 | 4 | 12 | 120 | 4 |
| 35 mm (6 well) | 1.1 | 2 | 6 | 60 | 2 |
| 12 well | 0.55 | 1 | 3 | 30 | 1 |
| 24 well | 0.275 | 0.5 | 1.5 | 15 | 0.5 |
Table 1: Transfection scale depending on the size of a cell plate.
Table 2: Raw data set analyzed by high content imaging system. Please click here to download this table.
Table 3: Summarized data set. Please click here to download this table.
Discussion
The previous reprogramming studies assessed reprogramming efficiency using flow cytometry and demonstrated the structural quality of iCMs using immunocytochemistry in two separate experiments. Flow cytometry analysis requires a much larger number of starting cells, thereby increasing the scale of experiments. In contrast, high content imaging analysis can evaluate both quality and quantity of iCM reprogramming by a single experiment with a relatively small number of cells. Therefore, this new method can provide an efficient new technical platform for future reprogramming studies. In particular, this new method will be useful for the reprogramming experiments for screening new genetic or pharmacological factors. For testing a large number of factors, the format of analysis can be down-scaled to a 384-well format.
Transduction efficiency is directly correlated to reprogramming efficiency. In previous studies, we have shown that ensuring the expression of all reprogramming factors into fibroblasts is important to achieve high reprogramming efficiency8,9. Since a relatively small number of fibroblasts expressed all reprogramming factors by transducing individual viral vectors, using a single polycistronic vector harboring all reprogramming factors is beneficial for enhancing reprogramming efficiency7,9. Additionally, to increase transduction efficiency, we performed two sequential transductions using two separate transfections of reprogramming vectors into Platinum E cells. We found that this method is superior to the previous sequential transduction with a single transfection by re-using transfected Platinum E cells for the second transduction.
Success of cardiac reprogramming can be defined in different ways. The most commonly used method was to quantify the percentage of cells expressing a cardiac structural protein, which is absent in starting fibroblasts. In this study, we used cardiac muscle specific α-actinin as well as striated muscle specific Titin as markers for cardiac structural protein induction during cardiac reprogramming. The reason for using Titin was that we were able to more clearly demonstrate sarcomere formation using fibroblasts isolated from Titin-GFP knock-in mouse line8,9. Regardless, we define iCMs as a double-positive cell (actinin+Titin+) to exclude Titin expressing non-cardiac muscle cells, if any. Importantly, any combination of cardiac structural proteins can be used for high content imaging analysis as long as there are specific antibodies available against those proteins. Although the method we described here is solely based on structural protein expression, it is important to recognize that the cells expressing a cardiac structural protein are not necessarily functional cardiomyocytes. In our previous studies, we have shown that sarcomere assembly is required for iCMs to be functional4,8,9. Importantly, the structural development of iCMs can be directly examined through high content imaging analysis. In addition, further structural evaluation (e.g., confocal microscopy) can be subsequently performed using the same immunostained cells for high content imaging analysis. However, the true functionality of iCMs should be determined by recording spontaneous contraction and electrophysiological properties (i.e., calcium transients and action potentials) of iCMs. In addition, demonstrating whole transcriptomic changes from fibroblast to cardiomyocyte would be necessary for defining successful iCM reprogramming (e.g., single cell RNA-seq). Therefore, the major goal of high content imaging analysis for assessing cardiac reprogramming is not to quantify functional iCMs, but to evaluate overall structural progresses of iCM reprogramming as an initial assessment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
High content imaging analysis was performed in the Vanderbilt High-Throughput Screening (HTS) Core Facility with assistance provided by David Westover and Joshua Bauer. The HTS Core receives support from the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center (P30 CA68485). This work was supported by AHA Innovative Project Award 18IPA34110341 and NIH R01 HL146524 (Y-.J. N.), and AHA post-doctoral fellowship award 20POST35210170 (Z.Z).
Materials
| A83-01 | Tocris | 2939 | |
| anti-chicken Alexa 488 | Thermofisher | A11039 | |
| anti-GFP antibody | Invitrogen | A10262 | |
| anti-mouse Alexa 555 | Thermofisher | A21422 | |
| anti-α-actinin antibody | Sigma | A7811 | |
| DAPI solution | Vector labs | H1200 | |
| Fugene 6 | Promega | E2691 | |
| Insulin-Transferrin-SeleniumG supplement | Invitrogen | 41400-045 | |
| Medium 199 | Invitrogen | 11150059 | |
| MEM vitamin solution | Invitrogen | 11120-052 | |
| MetaXpress software | Molecular device | ||
| Micro XL automated cell imagining system | Molecular device | ||
| Minimal essential amino acid solution | Sigma | M7145 | |
| Opti-MEM | Gibco | 31905-070 | |
| PES filter (0.45 µm) | Thomas scientific | 1159T84 | |
| Platninum E cells | Cell Biolabs | RV-101 | |
| Polybrene | Sigma | H9268 | |
| SB431542 | Sigma | S4317 | |
| Universal blocking buffer | BiogeneX | HK083-50K |
References
- Ieda, M., et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 142 (3), 375-386 (2010).
- Song, K., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 485 (7400), 599-604 (2012).
- Ifkovits, J. L., Addis, R. C., Epstein, J. A., Gearhart, J. D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One. 9 (2), 89678 (2014).
- Nam, Y. J., et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 141 (22), 4267-4278 (2014).
- Muraoka, N., et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. The EMBO Journal. 33 (14), 1565-1581 (2014).
- Umei, T. C., et al. Single-construct polycistronic doxycycline-inducible vectors improve direct cardiac reprogramming and can be used to identify the critical timing of transgene expression. International Journal of Molecular Sciences. 18 (8), 1805 (2017).
- Wang, L., et al. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circulation Research. 116 (2), 237-244 (2015).
- Zhang, Z., Zhang, A. D., Kim, L. J., Nam, Y. J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Science Reports. 9 (1), 6362 (2019).
- Zhang, Z., Zhang, W., Nam, Y. J. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Science Reports. 9 (1), 14970 (2019).
- Zhao, Y., et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nature Communications. 6, 8243 (2015).
- Zhou, H., Dickson, M. E., Kim, M. S., Bassel-Duby, R., Olson, E. N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America. 112 (38), 11864-11869 (2015).
- Zhou, H., et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes & Development. 31 (17), 1770-1783 (2017).
- Zhou, Y., et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 18 (3), 382-395 (2016).
- Muraoka, N., et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nature Communications. 10 (1), 674 (2019).
- Yamakawa, H., et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports. 5 (6), 1128-1142 (2015).
- Abad, M., et al. Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports. 8 (3), 548-560 (2017).
- Protze, S., et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. Journal of Molecular and Cell Cardiology. 53 (3), 323-332 (2012).
- Addis, R. C., et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. Journal of Molecular and Cell Cardiology. 60, 97-106 (2013).
- da Silva Lopes, K., Pietas, A., Radke, M. H., Gotthardt, M. Titin visualization in real time reveals an unexpected level of mobility within and between sarcomeres. The Journal of Cell Biology. 193 (4), 785-798 (2011).

.
