High Throughput Co-culture Assays for the Investigation of Microbial Interactions
Summary
The co-culture interaction assays presented in this protocol are inexpensive, high throughput, and simple. These assays can be used to observe microbial interactions in co-culture, identify interaction patterns, and characterize the inhibitory potential of a microbial strain of interest against human and environmental pathogens.
Abstract
The study of interactions between microorganisms has led to numerous discoveries, from novel antimicrobials to insights in microbial ecology. Many approaches used for the study of microbial interactions require specialized equipment and are expensive and time intensive. This paper presents a protocol for co-culture interaction assays that are inexpensive, scalable to large sample numbers, and easily adaptable to numerous experimental designs. Microorganisms are cultured together, with each well representing one pairwise combination of microorganisms. A test organism is cultured on one side of each well and first incubated in monoculture. Subsequently, target organisms are simultaneously inoculated onto the opposite side of each well using a 3D-printed inoculation stamp. After co-culture, the completed assays are scored for visual phenotypes, such as growth or inhibition. These assays can be used to confirm phenotypes or identify patterns among isolates of interest. Using this simple and effective method, users can analyze combinations of microorganisms rapidly and efficiently. This co-culture approach is applicable to antibiotic discovery as well as culture-based microbiome research and has already been successfully applied to both applications.
Introduction
In nature, microorganisms rarely exist in isolation; consequently, they are constantly interacting with other organisms. Therefore, studying how microorganisms interact with each other is essential to understanding a multitude of microbial behaviors1. Microbial interactions can be mutualistic, commensal, or antagonistic. These in teractions can affect not only the microorganisms themselves but also the environments and hosts that the microorganisms colonize1,2.
Many scientists study microbial interactions to identify new antimicrobial molecules. One of the first clinically important antimicrobial molecules was found through the study of microbial interactions. Sir Alexander Fleming observed a contaminating Penicillium spp. isolate that inhibited the growth of a Staphylococcus strain, which led to the discovery of the commonly used antibiotic penicillin3. Characterization of the mechanisms that microorganisms use to antagonize their competitors remains a fruitful resource for the discovery of antimicrobial molecules. For example, it was recently shown that Streptomyces sp. strain Mg1 produces antibiotic linearmycins, which have a lytic and degradative activity against Bacillus subtilis4.
Further, a non-ribosomally synthesized peptide named lugdunin was recently discovered after the observation that nasal commensal Staphylococcus lugdunensis inhibits Staphylococcus aureus5. Studies have also shown that mutualistic interactions between microorganisms are equally as powerful as antagonistic interactions for the discovery of antimicrobial molecules. For example, many fungus-farming ants in the tribe Attini harbor symbiotic bacteria called Pseudonocardia on their exoskeleton that produces antifungal molecules to inhibit an obligate pathogen of their fungal crop6. As the study of microbial interactions has been beneficial for discovering antimicrobial molecules, the use of high throughput screens may result in the discovery of new antimicrobial molecules.
With respect to the cost and ease of performance, the methodologies used to study microbial interactions range from simple to complex. For instance, an agar plug assay is an inexpensive and simple method that can be used to investigate antagonism between multiple microorganisms7. However, an agar plug assay is not an efficient procedure and can be labor-intensive for many pairwise combinations.To assess the effects of microbially produced products on target isolates of interest in a high throughput manner, many laboratories use disk diffusion assays8. These assays are easy and inexpensive and can be scalable to higher numbers of samples7. However, this assay requires the generation of microbial extracts and may produce misleading results for certain combinations of target organisms and antibiotics, such as Salmonella and cephalosporins9.
The preceding approaches rely on isolated components to elicit a response in a target organism, instead of allowing microorganisms to interact with each other. This is of note because interactions between microbes may elicit the production of "cryptic" antimicrobial molecules that are not produced in monoculture. For instance, it was recently shown that the antimicrobial keyicin is only produced by a Micromonospora sp. when co-cultured with a Rhodococcus sp. that is isolated from the same sponge microbiome10. More complex interaction methodologies circumvent this potential monoculture hindrance. For instance, the iChip is useful for isolating rare and difficult to cultivate bacteria from environmental samples and allows for the observation of microbial interactions through growth in situ11. To investigate interactions in detail, matrix assisted laser desorption/ionization time-of-flight imaging mass spectrometry (MALDI-TOF-IMS) can be used. This approach provides detailed information on the composition and distribution of small molecules and peptides produced by interacting microbial colonies with high spatial resolution. MALDI-TOF-IMS has also been used in multiple studies of bacterial interactions to characterize the mechanisms of competition12,13,14,15. However, MALDI-TOF-IMS often requires laborious sample preparation, specialized expertise to operate the equipment, and expensive and specialized mass spectrometers. For these reasons, it is a difficult technique to use for high throughput studies. Thus, a simple, scalable, and high throughput co-culture assay for microbial interactions that overcomes many limitations of the above approaches would be beneficial.
Here, a protocol for high throughput microbial co-culture is presented. This assay is simple and easily incorporated into preexisting studies of microbial interactions. In contrast to many commonly used methods for the study of microbial interactions, our method is simple, inexpensive, and is amenable to investigating large numbers of interactions. These assays are not only easy to perform, but the materials are widely available from most laboratory suppliers or public resources (e.g., libraries and makerspaces). Consequently, this assay is advantageous as a first line of investigation to identify and parse interesting patterns among many pairwise combinations of microorganisms, which may be especially useful for the investigation of microbial ecology.
Protocol
Informed consent was obtained from the donor's parents, and the Human Subjects Committee at the University of Wisconsin-Madison approved the study (Institutional Review Board [IRB] approval number H-2013-1044).
1. Sample Culture
NOTE: This procedure is used here for the study of interactions among bacteria isolates from the human nasal cavity. In principle, the following methods are applicable to any culture condition. Brain-heart-infusion broth (BHI) is used for general propagation of nasal bacteria. All plates are solidified using 1.5% agar. For this study, samples are taken from saline solution flushed into a donor's nose (nasal lavage), transferred into microcentrifuge tubes, and frozen at -80 °C.
- Use standard culture techniques to plate 100 µL of each of the thawed lavage samples onto BHI plates.
- Incubate the plates aerobically at 37 °C for 1 week.
- After incubation, select ≥2 colonies of each distinct morphotype per plate and passage the isolates aerobically on BHI plates by streaking a colony from the initial plate onto a new BHI plate and incubating the plate at 37 °C. Repeat until the bacterial cultures are pure.
NOTE: Bacterial isolates can be identified through colony morphology, gram-staining, 16S rRNA gene sequencing, or another method. However, knowing the isolate's identity is not necessary for continuing the protocol. - Cryopreserve all bacterial isolates at -80 °C after combining 1 mL of 50% glycerol with 1 mL of bacterial overnight culture (see section 3) in cryotubes.
2. 3D Printing Stamps
NOTE: Polycarbonate was selected as the stamping material due to its high glass transition temperature (147 °C) that exceeds standard autoclave temperatures (121 °C), which minimizes the potential for deformation after repeated uses.
- Load the 3D printer with polycarbonate filament.
- Apply white school glue (polyvinyl acetate) to the print bed to aid in the adhesion and minimize warping of the inoculation stamp during the print.
- Load the .STL model file (Supplementary Data File) for the inoculation stamp (Figure 1) into the 3D printer software.
- Print the inoculation stamp at a 290 °C nozzle temperature, 60 °C bed temperature, and layer height of 0.38 mm.
- Wrap the stamp in the aluminum foil and sterilize by autoclaving for 1.5 h on a gravity cycle with 15 min of drying.
NOTE: Though polycarbonate is hygroscopic, the stamps only retain approximately 0.5% water weight after autoclaving.
3. Preparation of Overnight Cultures
- Using a serological pipette, pipette 3 mL of sterile BHI broth into 14 mL culture tubes.
- Using a sterile 1 µL inoculating loop, inoculate a bacterial colony into the broth. Swirl the loop to ensure the clump disperses into the broth. Vortex the culture tubes briefly before incubation.
- Incubate the culture tubes at 37 °C overnight (~16 h) on a shaker at 250 rpm.
- Vortex to break up the clumps of cells once the bacterial cultures reach sufficient turbidity (OD600 ≥ 1).
4. Preparation of Bioassay Plates
NOTE: Bioassay plates are prepared in a laminar flow hood to maintain sterility.
- Prepare BHI media with 1.5% agar and sterilize by autoclaving according to manufacturer instructions.
- After autoclaving, cool the BHI media to 55 °C in a temperature-controlled water bath.
- Using a serological pipette, pipette 3 mL of molten BHI media into each of the 12 wells on a 12 well plate. Ensure that the wells are as exact and even as possible. One liter of media will yield ~27 bioassay plates.
- Allow the agar to set overnight.
5. Inoculating Bioassay Plates with the Test Organism
NOTE: A test organism refers to the organism for which the production of inhibitory activity (e.g., antibiotic production) is determined using the co-culture interaction assay. For this experiment, the test organisms are Actinobacteria isolated from nasal lavages samples.
- Inoculate the test organism on a bioassay plate by inserting a sterile 10 µL inoculating loop into the overnight culture and streaking a culture droplet over the left third of a plate well.
NOTE: It is recommended to only streak one-third of the well, as overgrowth of the well prohibits later inoculation of target organisms. - Repeat until all 12 wells on the plate have been inoculated with the test organism.
- Incubate plates upside down at the appropriate temperature for 7 days. At higher temperatures (≥37 °C) or in drier climates, store the plates in a humid container to prevent the plates from drying out.
6. Preparation of Target Organisms
NOTE: A target organism refers to the organism whose inhibition status is determined using the co-culture interaction assay. For this experiment, the target organisms are Staphylococcus spp. isolated from nasal lavages samples.
- After incubating the bioassay plates for 6 days, prepare overnight cultures of the specified target organisms, as done above (see section 3).
7. Target Organism Inoculation
NOTE: After overnight incubation, ensure that the cultures are turbid (OD600 ≥ 1). Some bacterial cultures may flocculate at the bottom of the culture tube. Vortex the culture tubes to disperse clumps and assess the culture turbidity.
- Prepare the target plate by filling each well of an empty 12 well plate with 1.8 mL of BHI and 200 µL of the target overnight culture.
- Unwrap and place a sterile inoculation stamp into the target plate. Gently swirl the cultures around in the wells, taking care to ensure that the cultures do not cross contaminate neighboring wells.
- Lift the inoculation stamp and ensure that there is a droplet of diluted target culture on each stamp tip.
- Prepare a monoculture control plate by placing the inoculation stamp on an uninoculated bioassay plate and gently rock the stamp so that a culture drop inoculates each well. Once the inoculation stamp is removed, a droplet of culture should be visible in the wells of the bioassay plate.
- If any wells are not inoculated with the inoculation stamp due to uneven levels of media, spot 3 µL of diluted overnight culture onto the right side of the wells using a pipette.
- Inoculate the bioassay plates as done above (see step 7.4.1), but align the stamp so that the tips align with the right side of the 12 well plate. Ensure that the stamp does not contact the existing bacterial colony when inoculating the wells.
- Carefully remove the stamp and place back into the target plate.
- Repeat the inoculation for each bioassay plate until all the plates are inoculated.
- Incubate bioassay plates upside down at the appropriate temperature for 7 days.
8. Scoring
- After co-culturing the test and target organisms for 1 week, score the interactions based on the following visual assessment:
- Score the wells with target organism growth that exhibits growth that is indistinguishable from the monoculture control as "0" (no inhibition) (Figure 2A,B).
- Score the wells with target organism that exhibits diminished growth compared to the control as "1" (weak inhibition) (Figure 2C).
- Score the wells where the target organism did not grow as "2" (strong inhibition) (Figure 2D).
Representative Results
Co-culture interaction assays can be used to understand microbial interactions, identify patterns of interest, and uncover microbial isolates with intriguing activities. In these assays, a test organism is monocultured on one side of a 12 well agar plate and incubated for 7 days. Subsequently, a target organism is spotted next to the test organism and the two microbes were co-cultured for 7 days before scoring for the growth phenotype of the target organism. The assays are scored based on a visual analysis of the growth or inhibition of the target organism.
These co-culture assays were recently used to assess the inhibitory activity of Actinobacteria (test organisms) toward Staphylococcus spp. (target organisms) isolated from the human nasal cavity (Figure 2)16. The co-culture assays were used to identify specific inhibition patterns between Actinobacteria (n = 21) and Staphylococcus isolates (n = 39) and showed that Actinobacteria isolates showed variation in their ability to inhibit coagulase-negative staphylococci (CoNS). A total of 812 pairwise combinations were tested. In particular, Corynebacterium propinquum strongly inhibited CoNS (Figure 2D), especially compared to other Corynebacterium that weakly inhibited CoNS (Figure 2C) or had no effect on CoNS (Figure 2B), and compared to the monoculture control (Figure 2A)16.
Using comparative genomics, a biosynthetic gene cluster for siderophore production was identified in C. propinquum genomes that was absent in the genomes of other Corynebacterium isolates16. Siderophores are chelators produced by microorganisms to scavenge iron from the environment17. Siderophore production by C. propinquum was confirmed and the siderophore was identified as dehydroxynocardamine16. This result led to the hypothesis that inhibition of CoNS was due to siderophore-mediated iron depletion. Subsequently, by performing co-culture interaction assays between C. propinquum and CoNS on both standard and iron-supplemented BHI medium, it was determined that the inhibition phenotype was iron-dependent (Figure 2E). Together, these results suggested that siderophore-mediated iron depletion was responsible for the strong inhibition of CoNS by C. propinquum16.
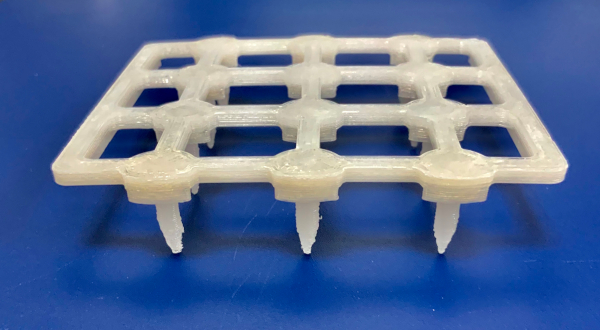
Figure 1: Photograph of the bioassay inoculation stamp. Please click here to view a larger version of this figure.
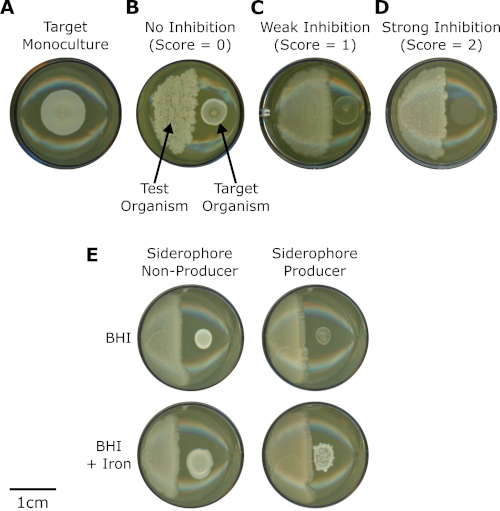
Figure 2: Co-culture interaction assays uncover siderophore-mediated inhibition of CoNS by Corynebacterium propinquum. (A) Monoculture of CoNS (target organism) inoculated on a BHI bioassay plate. (B,C,D) Co-cultures between different strains of Corynebacterium spp. (test organisms, left) and the same strain of CoNS (target organism, right) inoculated on BHI bioassay plates. Each panel is a representative image showing interactions with (B) no inhibition (score = 0), (C) weak inhibition (score = 1), or (D) strong inhibition (score = 2). (E) Comparison of interactions between Corynebacterium pseudodiphtheriticum (siderophore non-producer) or Corynebacterium propinquum (siderophore producer) with the same strain of CoNS on BHI media (BHI) and BHI media supplemented with 200 µM FeCl3 (BHI + iron). Please click here to view a larger version of this figure.
Supplementary Data File. Please click here to download this file.
Discussion
Antibiotics and other secondary metabolites that mediate microbial interactions are useful for a multitude of applications, including drug discovery. Herein, a protocol for co-culture assays to assess large numbers of microbial interactions is presented. These co-culture interaction assays are a simple, affordable, scalable, and high throughput means to investigate many pairwise combinations of microorganisms in tandem. Target organisms are spotted next to test organisms in a well of a 12 well plate using an inoculation stamp, and inhibition of the target organisms is scored based on visual inspection of the target organism's phenotype. The majority of materials used in these assays are readily available through most laboratory suppliers or through public resources. Therefore, the assays can be easily tailored to many laboratory environments. In the laboratory, these co-culture assays have been successful in investigating the interactions of microorganisms associated with many hosts from a variety of environments.
While there are many benefits to this technique, there are also a few limitations. The first notable limitation is that patterns from the co-culture assays are difficult to interpret without other metadata. In two recent papers that employed these co-culture assays16,18, inhibition patterns of interest were only identified in conjunction with other metadata, such as the taxonomic identity of the test organisms. Nevertheless, even without accompanying metadata, the methods outlined in this paper are amenable to identifying bacterial isolates with inhibitory activity toward specific pathogens or target microbes of interest.
An additional limitation is that these assays are not commercially available, and the bioassay plates must be hand-prepared, which can limit the assay's efficiency. Plate preparation is critical to the experiment, and care should be taken while preparing the plates. If the wells are uneven, then the inoculation stamp may not be able to inoculate all wells. However, missed wells can simply be inoculated by directly pipetting the target organism culture onto the right side of each uninoculated well. Indeed, even if a few wells on each plate are missed by the stamp, the procedure is still more efficient than directly pipetting the target organism into every single well. Alternatively, users can forgo the inoculation stamp and pipette the target organism into each well, but this process is more time-consuming, especially compared to stamping 12 wells simultaneously.
Finally, the test organism may consume the available nutrients in the well during monoculture before the target organism is inoculated. Though nutrient depletion may affect the observed inhibition patterns, it appears to be uncommon among the pairwise combinations that have been tested thus far. Notably, depletion of iron in the wells by C. propinquum allowed for the discovery of siderophore-mediated competition from members of the human nasal microbiota16.
These co-culture interaction assays are customizable by modifying the media composition, timing, or even including multiple organisms or microbial consortia as the test organism. Furthermore, different scoring systems can be used depending on the desired level of detail required to describe the interaction phenotype. Examples scoring scales include those from 0-2 used to describe competitive interactions among bacteria isolated from the human nasal cavity16 (Figure 2) and from 0-3 used to assess the antimicrobial potential of Streptomyces isolated from insect microbiomes18. However, with more nuanced scales, scoring becomes increasingly difficult. Thus, inhibition is most easily recognized using a binary scoring system (e.g., 0 is defined as no inhibition and 1 as inhibition), which can eliminate any confusion and standardize scoring across multiple individuals. Moreover, in addition to scoring for inhibition phenotypes, these assays can also be scored for other phenotypes, including pigment production, sporulation, or any other phenotype that can be assessed visually. As these assays are highly scalable with analysis based on visual inspection of the target organism, training sets for machine learning algorithms can be generated to facilitate phenotype scoring and further increase assay throughput.
A major strength of these co-culture assays is their ability to facilitate screening many combinations of pairwise interactions inexpensively and rapidly to uncover activities or inhibition patterns of interest. Subsequently, more complex and intensive methods (i.e., genomic characterization, MALDI-TOF-IMS, or natural product isolation and characterization) may be used for deeper characterization of the microbes and interactions of interest identified by co-culture assays. As a recent example, these co-culture inhibition assays were used to show that insect-derived Streptomyces can inhibit Gram-negative bacteria and fungi better than their soil-derived counterparts. The inhibition assays allowed for quick and efficient visualization of inhibition patterns among 2,003 Streptomyces isolates and led to the discovery of a new antifungal called cyphomycin, which is active against drug-resistant fungal pathogens18. Thus, these co-culture interaction assays are a powerful tool for microbiome research, antimicrobial discovery, and gaining deeper insights into patterns of microbial interactions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Daniel May, Marc Chevrette, and Don Hoang for critical reading of the manuscript. This work, including the efforts of Cameron R. Currie, was supported by the University of Wisconsin-Madison, Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation, funding also provided by the National Institutes of Health Centers for Excellence for Translational Research (U19-AI109673-01). Reed M. Stubbendieck was supported by a National Library of Medicine training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM 5T15LM007359). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Materials
| 1 μL disposable polystyrene inoculating loops, blue | VWR | 12000-806 | |
| 10 μL disposable polystyrene inoculating loops, yellow | VWR | 12000-810 | |
| 12-well cell culture plate, sterile with lid | Greiner bio-one | 665 180 | |
| 14 mL polystyrene round bottom tube, 17 x 100mm style, nonpyrogenic, sterile | Falcon | 352057 | |
| 2.0 self standing screw cap tubes with caps, sterile | USA scientific | 1420-9710 | |
| 25 mL serological pipet | Cell Treat | 229225B | |
| Agar, bacteriological | VWR | J637 | |
| Brain Heart Infusion Broth | Dot Scientific | DSB11000-5000 | |
| Polycarbonate filament, white, 3mm diameter | Keene Village Plastics | 12.1-3MM-WH-581.2-1KG-R | |
| School Glue | Elmer's | EPIE304 | |
| Taz 6 3D printer | Lulzbot |
References
- Stubbendieck, R. M., Vargas-Bautista, C., Straight, P. D. Bacterial Communities: Interactions to Scale. Frontiers in Microbiology. 7 (August), 1234 (2016).
- Green, J., Bohannan, B. J. M. Spatial scaling of microbial biodiversity. Trends in Ecology & Evolution. 21 (9), 501-507 (2006).
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. British Journal of Experimental Pathology. 10 (3), 226 (1929).
- Stubbendieck, R. M., Straight, P. D. Escape from Lethal Bacterial Competition through Coupled Activation of Antibiotic Resistance and a Mobilized Subpopulation. PLoS Genetics. 11 (12), e1005722 (2015).
- Zipperer, A., et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 535 (7613), 511-516 (2016).
- Currie, C. R., Scott, J. A., Summerbell, R. C., Malloch, D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 398 (6729), 701-704 (1999).
- Balouiri, M., Sadiki, M., Ibnsouda, S. K. Methods for in vitro evaluating antimicrobial activity: A review. Journal of Pharmaceutical Analysis. 6 (2), 71-79 (2016).
- Heatley, N. G. A method for the assay of penicillin. The Biochemical Journal. 38 (1), 61-65 (1944).
- Clinical and Laboratory Standards Institute. . Performance standards for antimicrobial disk susceptibility tests; approved standard -11th ed. CLSI document M02-A11. , (2012).
- Adnani, N., et al. Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chemical Biology. 12 (12), 3093-3102 (2017).
- Nichols, D., et al. Use of iChip for high throughput in situ cultivation of "uncultivable" microbial species. Applied and Environmental Microbiology. 76 (8), 2445-2450 (2010).
- Gonzalez, D. J., et al. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology (Reading, England). 157 (Pt 9), 2485-2492 (2011).
- Hoefler, B. C., et al. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proceedings of the National Academy of Sciences of the United States of America. 109 (32), 13082-13087 (2012).
- Hoefler, B. C., Straight, P. D. Imaging Mass Spectrometry, Metabolism, and New Views of the Microbial World. Natural Products Analysis. , 349-396 (2014).
- Yang, Y. L., Xu, Y., Straight, P., Dorrestein, P. C. Translating metabolic exchange with imaging mass spectrometry. Nature Chemical Biology. 5 (12), 885-887 (2009).
- Stubbendieck, R. M., et al. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Applied and Environmental Microbiology. 85 (10), 1-17 (2019).
- Winkelmann, G. Microbial siderophore-mediated transport. Biochemical Society transactions. 30 (4), 691-696 (2002).
- Chevrette, M. G., et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nature Communications. 10 (1), 516 (2019).

