Rapid Genetic Analysis of Epithelial-Mesenchymal Signaling During Hair Regeneration
Summary
Tissue-specific analysis of a hair follicle regeneration model using lentivirus to mediate gain- or loss-of-function.
Abstract
Hair follicle morphogenesis, a complex process requiring interaction between epithelia-derived keratinocytes and the underlying mesenchyme, is an attractive model system to study organ development and tissue-specific signaling. Although hair follicle development is genetically tractable, fast and reproducible analysis of factors essential for this process remains a challenge. Here we describe a procedure to generate targeted overexpression or shRNA-mediated knockdown of factors using lentivirus in a tissue-specific manner. Using a modified version of a hair regeneration model 5, 6, 11, we can achieve robust gain- or loss-of-function analysis in primary mouse keratinocytes or dermal cells to facilitate study of epithelial-mesenchymal signaling pathways that lead to hair follicle morphogenesis. We describe how to isolate fresh primary mouse keratinocytes and dermal cells, which contain dermal papilla cells and their precursors, deliver lentivirus containing either shRNA or cDNA to one of the cell populations, and combine the cells to generate fully formed hair follicles on the backs of nude mice. This approach allows analysis of tissue-specific factors required to generate hair follicles within three weeks and provides a fast and convenient companion to existing genetic models.
Protocol
1. Prepare 0 to 2 days Old Newborn Mice for Skin Dissection
- Euthanize mouse pups using a CO2 chamber for at least 20 min. Leave pups on ice up to an hour until dissection. P0-P2 mice are highly recommended as cells prepared from older mice have reduced progenitor capacity that results in lower graft yields. Prepare one dish of 70% ethanol (for step 1.3) and three dishes of wash solution (for step 3) when ready to perform skin dissection. Thaw Dispase Solution and leave it at 4 °C. Cervical dislocate pups after CO2 overexposure to ensure euthanasia.
- Place euthanized pups in a culture dish on ice and transfer to a sterile flow hood.
- Wash pups by briefly immersing in 70% ethanol and place onto sterile culture dish.
2. Dissect Mouse Skin
- Cut off each limb and tail at the base of the torso with sterile scissors.
- Grasp the body firmly between a pair of curved forceps and make an incision along the dorsal skin from head to tail using a scalpel without penetrating the underlying fascia.
- Carefully peel the skin away from the midline of the mouse.
- Grasp the exposed mouse firmly with the long side of the curved forceps and insert another pair of curved forceps underneath the skin at the posterior end of the mouse and gently pull skin over hips toward the ventral half of the mouse.
- Carefully peel skin completely off the mouse in one smooth motion and discard the carcass.
3. Wash Skin and Incubate with Dispase Solution
- Place the skin in the first dish of wash solution with dermis-side down. Spread skin out and agitate with forceps. Leave skin in the first dish of wash solution while dissecting the next skin.
- After placing the next dissected skin in the first dish of wash solution, transfer the prior skin to the second dish of wash solution. Keep skins in the second wash solution until all skins are collected. Transfer all of the skins to the final wash, agitate briefly.
- Add 10 ml ice cold Dispase to a sterile 10 cm culture dish. Transfer skin to the Dispase Solution and flatten skin dermis-side down.
- Float skin for 8-16 hr at 4 °C (alternative option: 1 hr at 37 °C).
4. Separate Dermis and Epidermis
- Transfer skin to a new sterile culture dish and flatten skin dermis-side down. Carefully hold down the dermis from one corner with a scalpel.
- Grasp the epidermis with forceps and slowly peel away from the dermis. Epidermis should peel away as one piece. The epidermis will be white and thin and the dermis should be brownish, thicker, and gelatinous.
- Separate the two tissues into separate sterile culture dishes.
- To make a dermal specific gene knockdown/overexpression graft, take dermis and mince into very small pieces with two scalpels. Discard epidermis. On day of infection (step 7), dissect another set of mouse skins to prepare fresh, untreated epidermal cells to combine with lentiviral-infected dermal cells.
- To make an epidermal specific gene knockdown/overexpression graft, slice each epidermis into 6 – 8 smaller pieces. Discard dermis. On day of infection (step 7), dissect another set of mouse skins to prepare fresh, untreated dermal cells to combine with lentiviral-infected epidermal cells.
5. Dissociate Primary Mouse Dermal Cells (mDCs) from Minced Tissue
- Dissociate mDCs by incubating dermal pieces with freshly made Collagenase Solution at 37 °C for 1 hr. Use 10 ml of Collagenase Solution per mouse pup.
- Gently mix solution containing dermis every 10 min. Check the extent of dissociation under a microscope; the digested cell suspension should be mostly single cells. The solution should change from clear to cloudy as dermal cells are dissociated from the dermis.
- Add FBS to Collagenase Solution containing dermis to a 10% final volume to reduce the activity of collagenase.
- Pass the cell suspension through a 70 μm cell strainer into a new 50 ml tube to remove large undigested clumps and whole follicles. Add 10 ml of DMEM/10%FBS per 30 ml of the flow-through to further dilute the collagenase.
- Centrifuge tubes containing digested dermis at 30 x g on a tabletop centrifuge for 5 min to further remove whole follicles.
- Carefully transfer the supernatant to a new 50 ml tube. Pellet cells by centrifuging at 200 x g on a tabletop centrifuge for 5 min.
- Resuspend cells with DMEM/10%FBS (1 ml per pup) and count cells. Plate cells with Amniomax C-100 media for infection (step 7), or to combine with lentivirus-infected KC for grafting (step 9). Typical yield is 20 x 106 cells per pup.
6. Dissociate Primary Keratinocytes (KC) from Minced Tissue
- Dissociate KC by incubating epidermal pieces with freshly thawed 0.05% Trypsin-EDTA at 37 °C for 15 min. Use 7 ml of Trypsin-EDTA per mouse pup.
- Gently swirl solution containing epidermis every 5 min.
- Add equal volume of Neutralizing Solution to Trypsin-EDTA containing epidermis to reduce the activity of trypsin.
- Pellet cells by centrifuging at 200 x g on a tabletop centrifuge for 5 min. Remaining undigested tissue should float on top.
- Carefully pour off most of the supernatant with undigested tissue. Remove remaining supernatant with a 10 ml pipette without disturbing cell pellet. Resuspend cell pellet with PBS. In case KCs are not pelleted completely, pour off most of the supernatant and repeat centrifugation until most of the cells are pelleted.
- Strain cell suspension through 70 μm cell strainer into a new tube to remove remaining tissue or clumps.
- Pellet cells by centrifuging at 200 x g on a tabletop centrifuge for 5 min.
- Resuspend cells in CnT-07 media or PBS (1 ml per pup) and count cells. Use CnT-07 for plating cells for infection (step 7), use PBS to combine with lentivirus-infected mDCs for grafting (step 9). Typical yield is 3-10 x 106 cells per pup.
7. Infect Primary Cells with Lentivirus Containing shRNA or cDNA
On day of infection dissect another set of mouse skins to prepare fresh, untreated primary KCs or mDCs to combine with virus-infected cells on day of grafting (step 9).
-
- For mDCs, plate 4 x 106 mDCs per 10-cm plate with AmnioMAX-C100 media and incubate at 37 °C + 5% CO2 for next day infection at 50% cell density. Usually 3-4 plates of mDCs are used to generate one graft, and a total of 10-12 plates are needed to generate three grafts in one experiment. Alternatively, let mDCs attach and spread for a minimum of 2 hr at 37 °C + 5% CO2 and perform infection the same day of cell plating. mDCs will have fibroblast-like morphology.
- For KC, plate 12 x 106 KC per 10-cm plate with CnT-07 media and incubate at 37 °C + 5% CO2 for next day infection. Alternatively, let KC attach and spread for a minimum of 2 hr at 37 °C + 5% CO2 and perform infection on the same day. Usually 3-4 plates of KC are used to generate one graft. A total of 10-12 plates are needed to generate three grafts for one experiment. KCs will have cobblestone morphology.
- Preincubate cells for 5-10 min with 8 μg/ml Polybrene Solution at 37 °C + 5% CO2.
- Exchange media and add lentivirus + 8 μg/ml Polybrene Solution. Lentiviral titer will vary depending on the construct used and should be determined before infection.
- Centrifuge the plates at 200 x g on a tabletop centrifuge for 1 hr at 32 °C to infect cells with lentivirus.
- Remove lentivirus-containing media and add fresh media. For KC, wash cells once with PBS before adding back fresh media. Usually infected cells recover overnight at 37 °C + 5% CO2. Alternatively, allow at least two hours of recovery at 37 °C + 5% CO2 before trypsinizing the cells for grafts. Continue to culture a small portion of infected cells to check for infection efficiency as described in the Representative Results section.
8. Prepare Infected Cells for Grafting
- Isolate fresh primary mDCs or KCs from skin using steps 4-6. Resuspend cells in ice cold sterile PBS. Count cells.
-
- For dermal specific gene knockdown/overexpression graft, dissociate infected mDCs from plates using 0.05% Trypsin-EDTA at 37 °C for 8 min.
- For epidermal specific gene knockdown/overexpression graft, dissociate infected KC from plates using 0.125% Trypsin-EDTA at 37 °C for 15 min.
- Neutralize trypsin activity using DMEM + 10%FBS (mDCs) or Neutralizing Solution (KC). Transfer the trypsinized cells to a 50 ml tube.
- Pellet cells by centrifuging at 200 x g on a tabletop centrifuge for 5 min.
- Resuspend cells in ice cold sterile PBS. Count cells.
- Combine the virus-infected cells with the freshly prepared untreated counterpart cell type. For one graft, combine 7-10 x 106 mDCs with 7-10 x 106 KCs in ice cold sterile PBS and pellet the cell slurry in a tabletop centrifuge at 4 °C. Resuspend cells in 50 -100 μl ice cold sterile PBS. Keep cells on ice until nu/nu mouse chambers are ready.
9. Create Wound Bed and Fix Chamber onto Back of nu/nu Mouse
A day before grafting, sterilize surgical tools with autoclave and sterile silicon chambers by soaking in 70% ethanol. Replace the 70% ethanol with sterile PBS before using the chambers.
- Anesthetize nu/nu mouse (7-12 week old females) using ketamine (8 mg/ml)/xylazine (1.6 mg/ml) via intraperitoneal injection (100 μl/10 g body weight) or other preferred method. Apply eye lubricant. Place heating pad under mouse.
- Disinfect nude mouse back skin with iodine and clean with alcohol swabs.
- Add 1-3 drops 0.25% Bupivicaine to incision site topically. Pinch the nude mouse back skin at base of neck with forceps.
- Cut a small circular hole (~1 cm in diameter) in pinched skin with sterile scissors.
- Place sterile chamber onto wound bed and cover chamber edges with surrounding skin.
- Secure chamber into place with sutures along chamber edges (6 stitches per chamber) as shown in Figure 2A.
- When chambers are ready, gently swirl cell slurry and draw up into 23 gauge needle attached to 1 ml syringe. If cell slurry has condensed at the bottom, gently resuspend with 1 ml pipette tip before drawing into the needle.
- Puncture chamber with needle and slowly drip cells onto wound.
- Place mice into clean, autoclaved cages. House 2 mice per cage and add antibiotics and Tylenol (3 mg/ml) to drinking water. In our experience, two female mice per cage have not resulted in adverse trauma to the chamber, graft, or animals.
- Place warming pad under half the cage and observe mice until anesthesia wears off.
10. Remove Chamber After 7-9 Days
- Anesthetize chambered nu/nu mouse as in step 9.1. Apply eye lubricant.
- Disinfect skin around chamber with iodine and clean with alcohol swabs.
- Carefully cut stitches and remove from chamber.
- Gently remove chamber from mouse. If new skin graft sticks to the chamber, lift part of the chamber and gently separate graft from chamber using forceps. Hold graft down during removal of the chamber. Graft should look dull and opaque as in Figure 2B.
- Apply a non-adherent pad with a thin film of antibiotics on the open wound and suture into place (2 stitches).
- Wrap bandage around mouse to secure the pad and protect the wound. Suture bandage into place.
- The silicon chambers are cleaned by scrubbing with mild soap and thoroughly rinsed with deionized water. Soak the chambers in 70% ethanol overnight, air dry, and store.
11. Examine Mouse for Hair Growth
- Remove bandages and pad after 3 days of chamber removal. Graft should be dry and opaque to brown in color.
- Check mice periodically for hair growth. Hair should begin to show at 16 days after graft.
Procedure Outline
Day 1 (2-3 hr): Sacrifice newborn pups, remove skin, and leave on dispase at 4 °C overnight.
Day 2 (2-3 hr): Isolate primary cells from skin and plate at recommended cell density.
Day 3 (3-4 hr): Infect cells with lentivirus. Remove skin from another set of newborn pups and leave on dispase at 4 °C overnight.
Day 4 (6-8 hr): Isolate primary cells from skin, count, and leave on ice. Recover lentivirus-infected cells by trypsinization and centrifugation, count, and combine 7-10 x 106 dermal cells and keratinocytes into 50-100 μl of PBS per graft. Create wound bed on mouse, attach chamber, and apply cell mixture to wound bed.
Alternative Procedure Outlines
Alternative procedure outlines will shorten the procedure from four days to three.
- Combine Day 1 and Day 2 by treating mouse skins in dispase at 37 °C for one hour (estimated time 5-7 hr).
- Combine Day 2 and Day 3 by infecting cells with lentivirus after plating cells for two hours (estimated time 5-7 hr).
Representative Results
In Figure 1 we show the result of dermal-specific lentiviral shRNA knockdown of Smoothened (Smo), a critical Shh signaling component, in a 3-week old regenerative hair graft. Control grafts using lentiviral vector alone show robust hair growth (Figure 1A). By contrast, dermal-specific knockdown of Smo results in loss of hair growth (Figure 1B). Hematoxylin and eosin stain depicts the stage of hair follicles in control and experimental conditions (Figure 1C, D). The hair growth phenotype from similar treatments can be variable among experiments, including the positive control with lentiviral vector alone infection. Therefore positive control grafts should always be included in every experiment as a reference as 30% of grafts can fail as a result of scarring or incomplete attachment of grafts. In the case of testing an interaction between two factors such as overexpressing cDNA of one factor to rescue shRNA-mediated knockdown of another factor, one should always include knockdown alone grafts to manifest the loss-of-function result in each experiment. This will provide a necessary range to determine how a particular gene perturbs the hair follicle regeneration pathway and how two genes interact.
Removal of the chamber (Figure 2A) must be done with care. The newly formed graft should be slightly opaque with a dull surface (Figure 2B). The graft may stick to the chamber, so gently lifting one side is important to make sure the graft remains attached to the wound bed. The graft is quite stable after three days and should present little difficulty when removing the dressing. Robust hair growth should be observed after three weeks (Figure 2C). Omitting either mDCs or KCs will result in a recovered wound site without any hair (Figure 2D)11. Cell death or cell loss in one of the cell types also results in no hair, in contrast to reduced hair regeneration in dermal-Smo knockdown as shown in Figure 1B.
To ensure a successful assay, it is critical to achieve greater than 90% viral infection of cells with appropriate overexpression or knockdown efficiency before grafting. Due to the time constraints of the grafting procedure, we recommend trouble shooting viral infection efficiencies before starting a grafting experiment, and always save a small portion of cells on day of grafting procedure to verify loss- or gain-of-expression. Lentiviral titer will vary depending on the construct used and should be determined before infection to achieve 90-100% efficiency. We monitor viral infection efficiency with GFP coexpressed from the lentiviral vector. We routinely perform quantitative PCR and/or Western blot to determine the extent of knockdown or overexpression. Utilizing dual-expression vectors with fluorescent tags to label virally infected cells provides a way to report perdurance of signal and number of cells expressing our constructs in mature grafts. We use GFP driven by a CMV promoter from the lentiviral vector pSicoR-CMV-GFP 10 in combination with Smo shRNA. In Figure 3, we show a 3-week old dermal-specific graft where GFP is lentivirally-expressed in dermal papilla of a representative hair follicle.
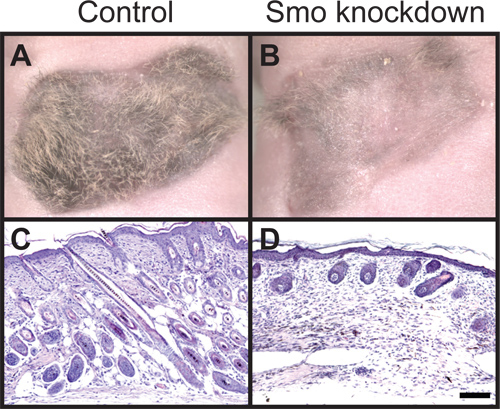
Figure 1. Histological analysis of hair follicle growth. (A) View of control hair regeneration graft with dermal cells infected with empty lentiviral vector and untreated keratinocytes. (B) Smoothened knockdown using lentiviral shRNA in dermal cells combined with untreated keratinocytes results in loss of hair follicles. (C) Hematoxylin and eosin staining shows anagen hair follicles in control grafts extend down into the dermis and (D) Smoothened knockdown hair follicles remain stunted and largely absent. All images are three weeks after grafting. Scale bar denotes 100 μm.
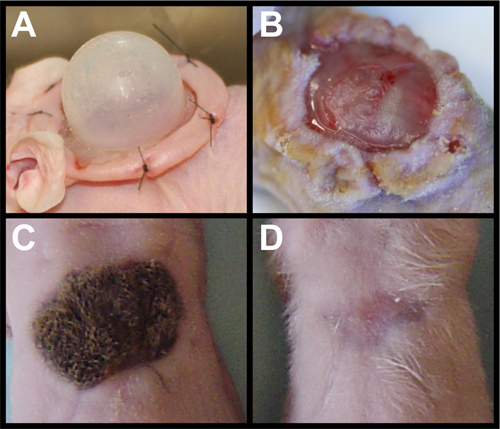
Figure 2. Grafting primary cells. (A) nu/nu mouse with sutured chamber housing primary dermal cells and keratinocytes. (B) Removal of chamber exposes newly formed skin that is dull and opaque in character. (C) Fully grown hairs on graft using wild-type primary dermal cells and keratinocytes at three weeks after grafting. (D) Addition of primary dermal cells without keratinocytes leads to no hair after three weeks. Similar results are seen with addition of primary keratinocytes without dermal cells.
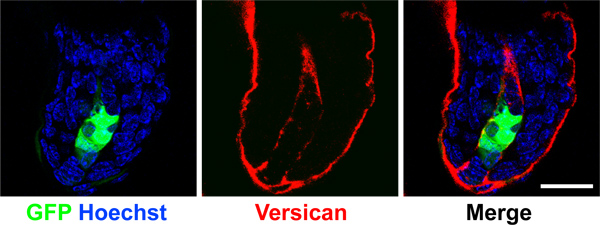
Figure 3. Maintenance of GFP expression in dermal papilla. Confocal images of dermal papilla from a 3-week old regenerated graft. GFP was expressed in the dermal cell population from a lentiviral vector and detected in the dermal papilla (green). Versican (red) demarcates the dermal papilla and nuclei were label with Hoechst (blue). Note GFP is expressed specifically in dermal cells but not in the surrounding keratinocytes. Scale bar denotes 20 μm.
Discussion
Hair reconstitution assays provide a unique organ regeneration model for examining the mechanism of de novo hair follicle formation. Here, we describe a modified chamber hair assay useful for determining gene function in hair follicle formation. Our assay is based on the reported chamber hair reconstitution assay 5, 6,11, which we modified to introduce a lentiviral expression technique to achieve gene overexpression or knockdown in either the dermal or epidermal cell types. Our assay provides a rapid alternative to generating tissue-specific genetic knockout. This assay allows us to demonstrate gene function in hair follicle formation in as little as three weeks from infecting primary cells with lentivirus to graft collection. Our assay can be extended to address genetic interactions using combination shRNA/cDNA delivery by lentivirus into one cell type 12, as well as allows examination of signaling between the dermal and epidermal cell types.
Our hair assay is best suited for detecting strong gain- or loss-of-function phenotypes in hair follicle regeneration, while subtle defects are harder to observe. We have successfully demonstrated dermal specific gene function of a few genes using our hair regeneration assay 1,12. Based on the GFP marker co-expressed from lentivirus vector expressing shRNA 10, we demonstrated that the shRNA expressing donor dermal cells persisted in the regenerated hair grafts. GFP-positive cells were present in the dermal papilla (DP) of the regenerated hair follicles (Figure 3). The DP cells likely consisted of different levels of gene knockdown as cells expressed varying levels of GFP, thus the observed hair follicle phenotype was a range from genetic hypomorph to null.
One improvement for this assay will be to establish a quick and efficient selection for the high shRNA expressing cells before grafting such as using FACS to purify strong GFP-expressing cells. An alternative is to substitute mutant or conditional knockout cells in this assay 9. On the other hand, a culture system that can preserve DP cell function is very useful as DP potency is gradually lost once the primary dermal cells are passaged. Potential culture systems include the use of biomaterials derived from artificial niche or aggregation cultures2,7,8,13. Another improvement will be to generate a reliable epidermal cell freezing method with a high cell recovery rate as this will reduce the use of the second set of mouse pups (step 7) to generate fresh keratinocytes in dermal-specific loss- or gain- of-function studies.
This chamber hair assay produces hair follicle density and quality similar to normal mouse skin, although hair growth is slightly less synchronized and follicle orientation is more random. A few limitations of the hair regeneration genetic assay include the requirement of large amount of lentivirus and cell numbers, and it is very labor intensive in terms of surgical methods when compared to other forms of hair reconstitution assays such as the patch and flap assays 3,4,14. However, the quality of hair follicles and timeline to detect hair growth is superior to other assays 4. Our targeted hair regeneration assay provides a fast and convenient companion to existing genetic models.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH NRSA 1F32CA14208701 (S.X.A.) and NIH grants AR052785 and AR046786 (A.E.O. and W.-M.W.).
Materials
| Name | Company | Catalog # | Comments |
| PBS | Invitrogen | 14190-144 | |
| HBSS | Invitrogen | 14170-122 | |
| DMEM | Invitrogen | 11995-065 | |
| 0.25% Trypsin-EDTA | Invitrogen | 25200-056 | |
| Cnt-07 | Cell N Tec | CnT-07 | |
| AminoMAX-C100 | Invitrogen | 17001-074 | |
| AminoMAX-C100 Supplement | Invitrogen | 12556-015 | |
| FBS | Invitrogen | 26140-079 | |
| Penicillin/Streptomycin | Invitrogen | 15140-122 | |
| Fungizone | Invitrogen | 15290-018 | |
| Gentamycin | Invitrogen | 15710-064 | |
| Dispase | Roche | 4942078001 | |
| Collagenase, Type 1 | Sigma | C-0130 | |
| Polybrene | Sigma | 107689-10G | |
| Tissue culture dish | Fisher Scientific | 08-772E | |
| Dissecting Scissors | Fisher Scientific | 11-999 | |
| Forceps | Fisher Scientific | 10-275 | curved |
| Scalpal | Medex Supply | GRF-2975#10 | Size no. 10 |
| 70 μm cell stainer | VWR | 21008-952 | |
| 15 ml conical centrifuge tube | Fisher Scientific | 14-959-49D | |
| Alcohol swabs | Fisher Scientific | 1368063 | |
| 23 gauge needle | Fisher Scientific | 14-821-13F | |
| Eye lubricant | CVS | 8883660 | |
| Providone-lodine swabs | Fisher Scientific | NC0116841 | |
| Silicon Chambers | Renner GmbH | F2U, 30268 | |
| Sutures | Acuderm | SUP3524 | |
| Flexible bandage | Fisher Scientific | 22-363-100 | |
| Non-adherent dressing | TELFA | 1050 | |
| Materials | |||
|
Wash solution PBS Dispase solution HBSS Neutralizing media HBSS Collagenase solution 0.25% (w/v) Collagenase, Type 1 100 μg/ml Penicillin/Streptomycin 2.5 μg/ml Fungizone 50 μg/ml Gentamycin Polybrene solution HBSS |
References
- Bershteyn, M., Atwood, S. X., Woo, W. M., Li, M., Oro, A. E. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell. 19, 270-283 (2010).
- Driskell, R. R., Juneja, V. R., Connelly, J. T., Kretzschmar, K., Tan, D. W., Watt, F. M. Clonal growth of dermal papilla cells in hydrogels reveals intrinsic differences between Sox2-positive and -negative cells in vitro and in vivo. J. Invest. Dermatol. 132, 1084-1093 (2012).
- Lee, L. F., Jiang, T. X., Garner, W., Chuong, C. M. A simplified procedure to reconstitute hair-producing skin. Tissue Eng. Part C Methods. 17, 391-400 (2011).
- Liang, Y., Silva, K. A., Kennedy, V., Sundberg, J. P. Comparisons of mouse models for hair follicle reconstitution. Exp. Dermatol. 20, 1011-1015 (2011).
- Lichti, U., Anders, J., Yuspa, S. H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3, 799-810 (2008).
- Lichti, U., Weinberg, W. C., Goodman, L., Ledbetter, S., Dooley, T., Morgan, D., Yuspa, S. H. In vivo regulation of murine hair growth: insights from grafting defined cell populations onto nude mice. J. Invest. Dermatol. 101, 124S-129S (1993).
- Lutolf, M. P., Gilbert, P. M., Blau, H. M. Designing materials to direct stem-cell fate. Nature. 462, 433-441 (2009).
- Osada, A., Iwabuchi, T., Kishimoto, J., Hamazaki, T. S., Okochi, H. Long-term culture of mouse vibrissal dermal papilla cells and de novo hair follicle induction. Tissue Eng. 13, 975-982 (2007).
- Rendl, M., Polak, L., Fuchs, E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543-557 (2008).
- Ventura, A., Meissner, A., Dillon, C. P., McManus, M., Sharp, P. A., Van Parijs, L., Jaenisch, R., Jacks, T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. U.S.A. 101, 10380-10385 (2004).
- Weinberg, W. C., Goodman, L. V., George, C., Morgan, D. L., Ledbetter, S., Yuspa, S. H., Lichti, U. Reconstitution of hair follicle development in vivo: determination of follicle formation, hair growth, and hair quality by dermal cells. J. Invest. Dermatol. 100, 229-236 (1993).
- Woo, W. -. M., Zhen, H. H., Oro, A. E. Shh maintains dermal papilla identity and hair morphogenesis via a Noggin-Shh regulatory loop. Genes Dev. 26, 1235-1246 (2012).
- Young, T. H., Lee, C. Y., Chiu, H. C., Hsu, C. J., Lin, S. J. Self-assembly of dermal papilla cells into inductive spheroidal microtissues on poly(ethylene-co-vinyl alcohol) membranes for hair follicle regeneration. Biomaterials. 29, 3521-3530 (2008).
- Zheng, Y., Du, X., Wang, W., Boucher, M., Parimoo, S., Stenn, K. Organogenesis from dissociated cells: generation of mature cycling hair follicles from skin-derived cells. J. Invest. Dermatol. 124, 867-876 (2005).

