Dual-color Correlative Light and Electron Microscopy for the Visualization of Interactions between Mitochondria and Lysosomes
Summary
This protocol describes a mechanism for using correlative light and electron microscopy to visualize the interaction of mitochondria and lysosomes labeled with mEosEM and APEX2, respectively.
Abstract
Cellular organelles, such as mitochondria and lysosomes, display dynamic structures. Despite the higher resolution of transmission electron microscopy for structural analysis, light microscopy is essential for the visualization of dynamic organelles by target-specific labeling. The following protocol describes a method that combines dual-color correlative light and electron microscopy (CLEM) to observe the interactions between mitochondria and lysosomes. In this study, mitochondria were labeled with mEosEM (Mito-mEosEM) and lysosomes with TMEM192-V5-APEX2. The results obtained from CLEM images enable us to observe the changes in the interactions between mitochondria and lysosomes under external stress conditions. Treatment with bafilomycin (BFA), which inhibits lysosomal function, resulted in an increase in contact between mitochondria and lysosomes, leading to the formation of fragmented mitochondria trapped inside lysosomes. Conversely, treatment with U18666A, which inhibits cholesterol export from lysosomes, caused lysosomes to be surrounded by mitochondria, indicating a distinct form of interaction. This study presents an effective method for observing the interactions between mitochondria and lysosomes in fixed cells. Furthermore, CLEM imaging with dual-color probes offers a powerful tool for future investigations of organelle dynamics and their implications for cell function and pathology.
Introduction
Mitochondria and lysosomes are the principal membrane-bound organelles that are essential for the maintenance of cellular homeostasis. Mitochondria are highly dynamic cellular organelles modulated by fission and fusion events1. In general, mitochondria are referred to as the powerhouse of the cell and are known for their important role in oxidative phosphorylation, ATP production, and metabolite storage such as calcium2. In addition to their role in energy metabolism, mitochondria are involved in a number of other cellular signaling processes, including apoptosis3,4, calcium signaling5,6, and lipid metabolism7,8. Similarly, lysosomes are highly dynamic cellular organelles that play multiple roles as cellular recycling centers to maintain cellular cleanliness through protein degradation, autophagy, endocytosis, and phagocytosis9. Apart from their role in degradation, lysosomes also play roles in cellular physiology such as nutrient sensing, homeostasis10, lipid metabolism11, and cellular dedifferentiation12.
Despite the existence of numerous studies on the functions of individual cellular organelles, our recent research has concentrated on the less-understood crosstalk between mitochondria and lysosomes, which has been demonstrated to influence both cellular physiology and pathology. Contacts between mitochondria and lysosomes have been observed in various cell types, including cancer cells, neurons, and induced pluripotent stem cell (iPSC)-derived cells13, and they are mainly observed under cellular stress or in neurodegenerative diseases such as Parkinson's disease13,14 and Charcot-Marie-Tooth disease15. The contact between mitochondria and lysosomes plays an important role in maintaining metabolic homeostasis through the exchange of ions (e.g., Ca2+)16, cholesterol17, and iron18 between the two organelles. Furthermore, the contact between mitochondria and lysosomes plays a crucial role in the quality control of cellular organelles. For example, damaged mitochondria can be targeted for degradation through the process of mitophagy, in which they are engulfed by autophagosomes and delivered to lysosomes for degradation. Consequently, the removal of dysfunctional mitochondria can be facilitated before they cause further damage to the cell or to normal mitochondria. This interaction between mitochondria and lysosomes is crucial for maintaining cellular homeostasis and preventing the accumulation of dysfunctional organelles. Thus, only damaged mitochondria are selectively removed, allowing healthy mitochondria to function optimally within the cell.
Correlative light and electron microscopy (CLEM) is an imaging technique that has recently received attention as a research method by combining the advantages of light microscopy (LM) and electron microscopy (EM) to provide detailed structural information at different scales. In CLEM, a sample is first imaged using LM to identify specific regions of interest, followed by EM sample preparation steps such as fixation, dehydration, and embedding. The same sample is then observed under an electron microscope. Although live-cell CLEM methods have been reported recently19,20,21,22, it remains challenging to accurately combine optical images of highly dynamic biological samples with electron microscopy images of fixed cells into a single plane. For highly dynamic organelles like mitochondria and lysosomes, their precise localization can change as they are observed live under the light microscope, and they continue to move even during the short delay time required for fixation.
Therefore, in this protocol, two different molecular tags that remain active even within fixative solutions were employed to visualize the interaction between mitochondria and lysosomes, which play an important role in maintaining metabolic homeostasis through the exchange of ions (e.g., Ca2+)16, cholesterol17, and iron18 between these organelles. The constructs TMEM192-V5-APEX2 and Mito-mEosEM were used to observe the interactions of mitochondria and lysosomes in cultured cells at both the EM and LM levels. For mitochondria, the mEosEM tag was selected for its ability to preserve the fluorescence signal during EM sample preparation in a fixative solution and OsO423. For lysosomes, the APEX2 tag, a protein of approximately 28 kDa, was used to study the localization of a specific protein at both EM and LM levels24. In the presence of hydrogen peroxide, the APEX2's substrate, Amplex-Red, forms an insoluble polymer that can be observed by confocal microscopy at the tag-targeted location. In this protocol, dual-color CLEM was used to visualize the structure of the interactions between lysosomes and mitochondria in fixed cells.
Protocol
The step-by-step workflow for sample preparation is shown in Figure 1.
1. Cell culture and transfection
- Culture HeLa cells at a density of 1 × 105 cells in 35 mm glass grid-bottomed culture dishes containing DMEM with 10% fetal bovine serum and 100 U/mL of penicillin and streptomycin at 37 °C.
- The day after seeding the cells, co-transfect the cells at 30%-40% confluency with TMEM192-V5-APEX2 and Mito-mEosEM plasmids using the transfection reagent according to the manufacturer's instructions(1 µg each of TMEM192-V5-APEX2 and Mito-mEosEM plasmid DNA with 3 µL of transfection reagent). After transfection, treat the cells with DMSO or U18666A (2 µg/mL), and Bafilomycin A1 (100 nM) for 18 h.
2. Confocal microscopic imaging
- After treating the cells with Bafilomycin and U18666A for 18 h, remove the culture media and immediately add 250 µL of warm (30 °C-37 °C) fixative solution (2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate solution [pH 7.0]) by gentle pipetting. Immediately remove the fixative, replace it with 1.5 mL of fresh fixative, and incubate on ice for 30 min. Then, wash the cells for 3 x 10 min in 1 mL of ice-cold 0.1 M sodium cacodylate buffer.
NOTE: Aldehyde fumes are toxic. All experiments should be conducted in a ventilated fume hood. - Add 1 mL of cold 20 mM glycine solution to quench the aldehyde and incubate on ice for 10 min. Wash 3 x 10 min in cold 0.1 M sodium cacodylate buffer.
- Prepare a fresh solution containing 300 µM Amplex-Red by adding 10 mM H2O2 to a 0.1 M cacodylate solution. Add 500 µL of Amplex-Red solution to the sample dish and incubate for 5 min on ice.
- Remove the Amplex-Red solution and rinse 3 x 10 min with 0.1 M sodium cacodylate buffer.
- After Amplex-Red staining, use a confocal microscope to image the area around the cells of interest in 3 x 3 tile scan mode. Capture both differential interference contrast (DIC) and fluorescence signal to identify and record the grid and letters on the grid dish using a 40x (or 60x) objective.
NOTE: These overview images serve as a map for identifying specific target cells in the plastic-embedded specimens. - Image the target cells at high magnification using a 40x (or 60x) objective and acquire a Z-stack image.
NOTE: This high-magnification image is superimposed on an electron microscope image to create a correlative image.
3. Sample preparation for EM block
- After LM imaging, add 1 mL of 1% reduced osmium tetroxide (prepared by containing 3% potassium ferrocyanide in 0.3 M cacodylate buffer with an equal volume of 2% aqueous osmium tetroxide) for postfixation at 4 °C for 1 h, and then rinse for 3 x 10 min with distilled water at 4 °C.
NOTE: Osmium tetroxide fumes are toxic. All experiments should be performed in a ventilated fume hood. - Rinse 3 x 10 min with distilled water at room temperature, then dehydrate in a graded ethanol series for 15 min each time (50%, 60%, 70%, 80%, 90%, 95%, 100%, and 100%) at room temperature.
- Mix ethanol with epoxy resin in volume ratios of 3:1, 1:1, and 1:3, preparing a total of 300 µL of each mixture. Pour each epoxy resin into the dish, allowing it to incubate at room temperature for 1 h. Then, use the 100% epoxy resin for overnight impregnation.
- Prepare the embedding capsule or PCR tube by filling it with fresh epoxy resin, place the tube or capsule upside down on the region of interest, and place it in an oven at 60 °C for 48 h to polymerize.
4. Sectioning and TEM imaging
- After the epoxy resin has completely polymerized, separate the bottom glass from the polymer block by immersing it in liquid nitrogen.
- Place the specimen block in the sample holder of the ultramicrotome and set it in the trimming block. Use a razor blade to trim the resin into a trapezoidal or rectangular shape around the object of interest.
- Place the trimmed specimen block in the sample holder and adjust the diamond blade. Ensure that the leading edge of the specimen block is perfectly parallel to the edge of the diamond blade.
- Using an ultramicrotome, cut ultrathin sections approximately 60-70 nm from the flat-embedded cells. Collect the serial sections on a formvar-coated slot grid and dry them on a hot plate.
- Stain the section with the contrast stain for 2 min and lead citrate for 1 min.
- Place the grid in the TEM sample holder for observation. Identify the target cell of interest based on the DIC image from the light microscope. Image the entire cell area using tiled, overlapping images at 1,700x magnification.
5. Image stitching using imageJ Fiji.
- To begin image stitching, start imageJ Fiji25 and click on File | New | TrakEM2 (blank) | Select storage folder to create a new TrakEM226 project (Figure 2A1).
- Open the tile image dataset by dragging and dropping the raw tile image data into the workspace (Figure 2A2).
- Click the right mouse button and select Align | Montage all images in this layer | elastic (non-linear block correspondences), and then click OK to accept the default values remaining parameters (Figure 2A3).
- Click the right mouse button and select Export | Make flat image to create a stitched image, then click File | Save As to save the image.
6. Resizing the fluorescence image
- Open the photo enlargement software (see Table of Materials). Click on File | Open and select the fluorescence image.
- Enter the width and height to be the same size as the electron microscope image, and select the algorithm to be applied in the resize method (Figure 2B1).
- Click on File | Save AS to save the resized image.
7. Landmark-based image registration with the Big Warp plugin
- Open ImageJ Fiji and drag and drop the resized fluorescence micrograph (FM image) and the stitched electron micrograph (EM image).
- Click on Plugins | BigData Viewer | Big Wrap to start the registration , then click on the dropdown menu to select the FM image as the moving image and the EM image as the target image (Figure 2C1).
NOTE: Each FM and EM image will have three new popup windows (BigWrap moving Image, BigWrap fixed image, and Landmarks) to display landmarks. - Compare the FM and EM images using functions such as zoom (mouse wheel), image rotation (moving the mouse while holding down the left button), and move (moving the mouse while holding down the right button), and then press the space bar on the keyboard to switch Landmark mode on, and press the left mouse button to mark at least three landmarks identified in the FM and EM images (Figure 2C2, pink dots and red arrow).
- After marking all the identified landmarks, go to File | Export Moving Image | OK and a new popup window will appear with the transformed FM image (Figure 2C3).
NOTE: The newly created popup window is an FM image transformed based on the basis of the landmark. Review the alignment and make adjustments, if necessary, by adding or modifying landmarks and recalculating the transformation. - Click on File | Save As to save the transformed FM image.
8. Overlay of CLEM images
- In ImageJ, drag and drop the transformed FM image and the stitched EM image.
- Click on Image | Type | 8-Bit Color to set the same image type for both the LM and EM images (Figure 2D1).
NOTE: The EM image must be converted twice in the following order: 8-bit -> RGB Color -> 8-bit Color. - Click on Image | Color >| Merge Channels to combine the images into a composite image by selecting the FM and EM images from the dropdown menu, respectively (Figure 2D2).
- Click on File | Save As to save the CLEM image.
Representative Results
To visualize the interactions between mitochondria and lysosomes using this dual-color CLEM protocol, we employed Mito-mEosEM and TMEM192-V5-APEX2. Fixed cells were first imaged using a light microscope, followed by EM sampling and imaging. The results of the correlated images are shown in Figure 3A,B and Figure 4A. Following the protocol described above, we checked whether we could observe the changes in the interaction between mitochondria and lysosomes under external stress.
Treatment of cells with BFA, a lysosomal V-ATPase inhibitor, inhibits autophagy, resulting in reduced mitochondrial quality and increased interactions between mitochondria and lysosomes27. In BFA-treated cells, we observed mitochondria trapped within lysosomes (Figure 3B), compared to DMSO-treated control cells (Figure 3A). Although it is difficult to distinguish fragmented electron-dense mitochondria and cellular compartments within lysosomes in electron micrographs alone, they can be easily distinguished in correlated images (Figure 3B1,B2). The mitochondrial disruption structure (Figure 3A1,A2; blue arrow) observed in the mitochondria of DMSO-treated cells is not related to lysosomes, and the structure in the fluorescence image that appears to be a lysosome trapped inside a large mitochondrion is a structure where the mitochondrion surrounds the lysosome (Figure 3A2; red arrow).
Next, we tested the effects of U18666A (2 µg/mL for 18 h), a well-known inhibitor of NPC1 that blocks the movement of cholesterol out of lysosomes28. Cells treated with U18666A showed increased direct interaction between mitochondria and lysosomes (Figure 4). In addition, the contacts between mitochondria and lysosomes in U18666A-treated samples were different from those in BFA-treated samples. In HeLa cells treated with BFA, the fragmented mitochondrion is completely trapped in the lysosome, a previously reported mitophagy morphology12,29. However, in the U18666A-treated group, some of the TMEM192-positive lysosomes appear engulfed by mitochondrion (Figure 4D,E)30,31.
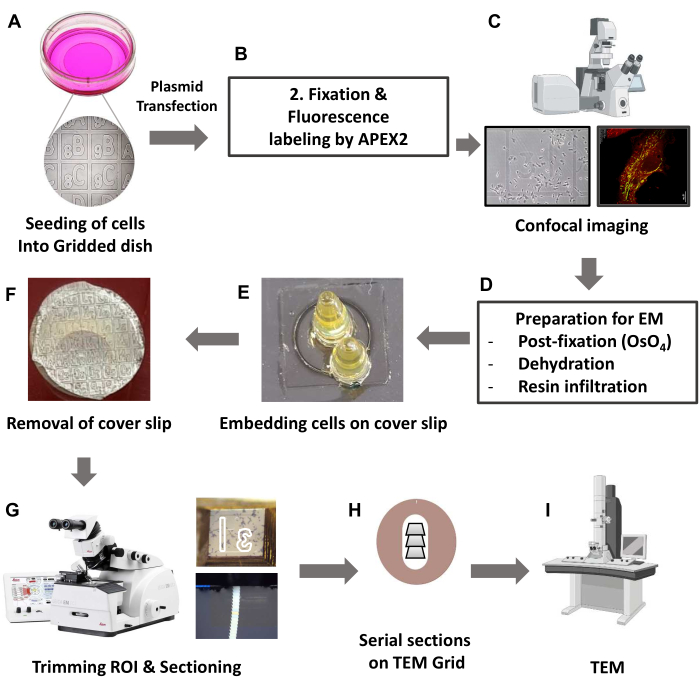
Figure 1: Schematic overview of TEM-based CLEM by using a genetically tagged fluorescence probe. (A) HeLa cells were grown in 35 mm glass grid-bottom culture dishes. (B) Fixed cells were labeled with Amplex-Red by APEX2 protein. (C) Fluorescence images were acquired with a confocal microscope. A motorized stage and tile-scan function were used to capture a large overview image and a high-resolution image of the target cell. The large image serves as a navigation map to identify the location of the target cell in plastic-embedded specimens. (D) Proceed with post fixation and dehydration embedding. (E) Embedding was done using a PCR tube or BEEM capsule filled with resin over the target cell on the coverslip. (F) After resin polymerization, removing the coverslip left an imprinted grid pattern on the bottom surface of the block. (G) The target cell was identified by the imprinted pattern on the resin surface, and the target cell was sectioned at approximately 60 nm thickness using an ultramicrotome. (H) Serial sections were collected on a formvar-coated copper slot grid. (I) The grids were imaged using transmission electron microscopy. Abbreviations: TEM = transmission electron microscopy; CLEM = correlative light and electron microscopy; ROI = region of interest. Please click here to view a larger version of this figure.
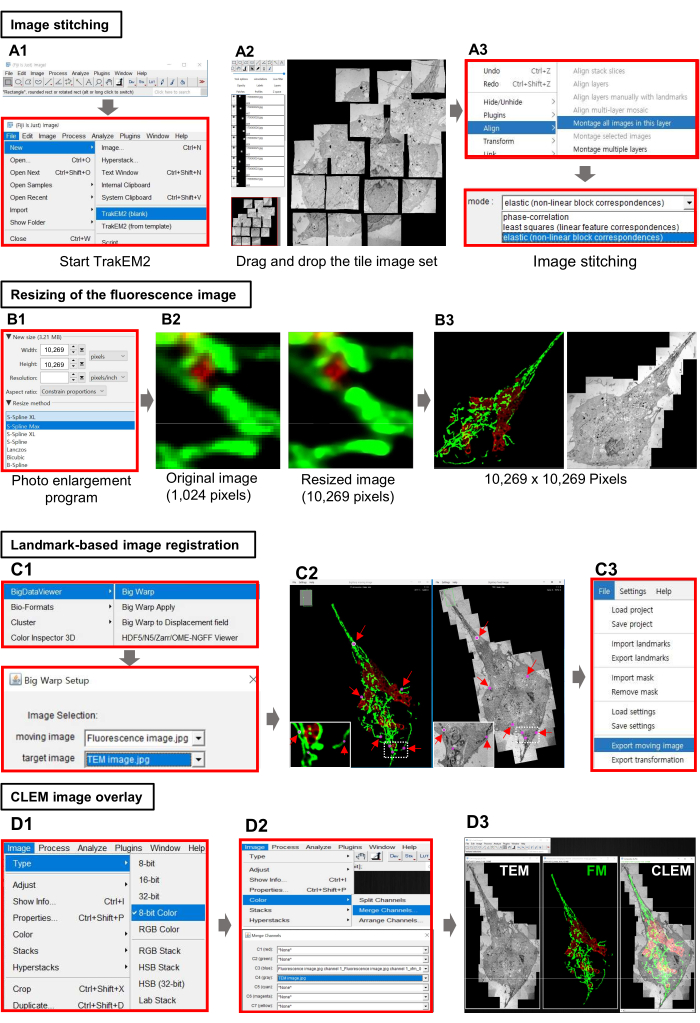
Figure 2: Image processing and generation of the CLEM image using imageJ Fiji. (A1) Main GUI of Fiji and TrakEM2. (A2) Import of the tile image dataset acquired with TEM at 1,700x magnification. (A3) Alignment and example of a stitched image using TrakeEM2's image alignment method. (B1–B3) Conversion of small light micrograph into large high-quality images using photo enlargement software. (C1–C3) Workflow for registration between light microscopy images and electron microscopy images using the Big Wrap plug-in software. (D1–D3) The process of overlaying a transformed FM image and an EM image to synthesize a correlated microscope image. Abbreviations: TEM = transmission electron microscopy; CLEM = correlative light and electron microscopy; FM = fluorescence microscopy. Please click here to view a larger version of this figure.

Figure 3: CLEM images of the mitochondrial and lysosomal contact sites treated with or without bafilomycin A1. HeLa cells were co-transfected with Mito-mEosEM (mitochondria; green) and TMEM192-APEX2 (lysosomes; red) and then stained with Amplex-Red dye. (A) Untreated HeLa cells. Molecular information from the fluorescence signal (left panel) and ultrastructural information obtained by TEM (middle panel) were superimposed (right panel) based on mitochondrial signal and morphology. (A1,A2) In untreated HeLa cells, the mitochondrial inner membrane is disrupted (blue arrow), or the mitochondria and lysosomes are very close to each other (red arrow). (B1,B2) In HeLa cells treated with bafilomycin A1 for 18 h, mitophagy, characterized by the trapping of undegraded mitochondria in lysosomes, was observed (green arrow). Scale bars = 1 µm (A1,A2,B1,B2). Please click here to view a larger version of this figure.
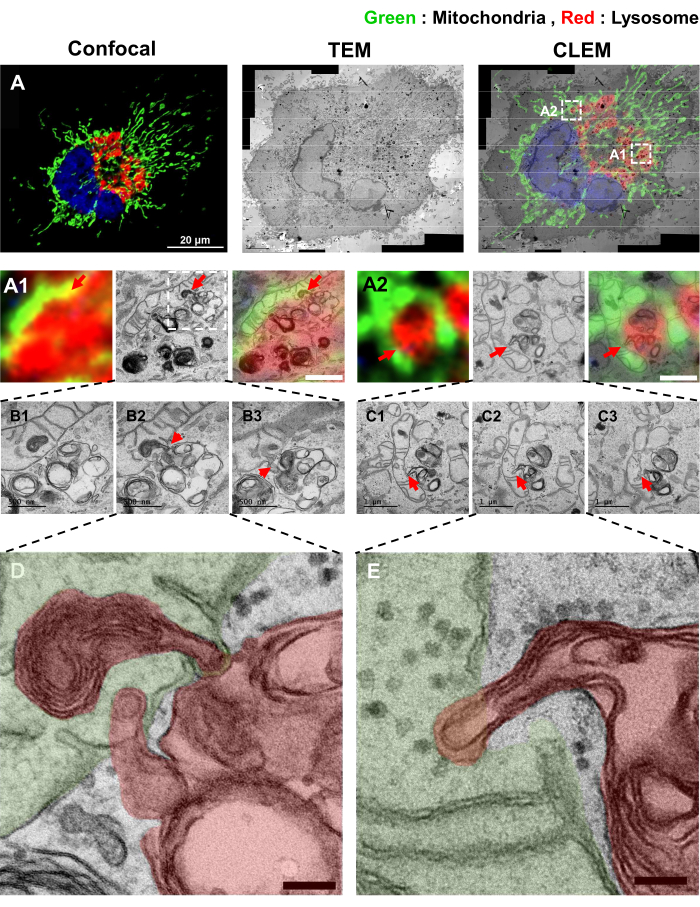
Figure 4: CLEM images of the mitochondrial and lysosomal contact sites treated with U18666A (A) Two-color CLEM images of Mito-mEosEM and TMEM192-V5-APEX2 (Amplex-Red-labeled) expressed HeLa cells under U18666A treatment (2 µg/mL for 18 h). (A1,A2) Red arrows indicate contact sites between mitochondria and lysosomes. Scale bars = 1 µm. (B1–B3,C1–C3) Red arrow indicates the appearance of inter-organelle fusion between mitochondria and lysosomes by serial images. Scale bars = 500 nm (B1–B3). Scale bars = 1 µm (C1–C3). (D,E) High-magnification TEM images of the fusion sites between mitochondria and lysosomes. Pseudocolors are used to distinguish between mitochondrion (green) and lysosome (red). Scale bars = 100 nm. Please click here to view a larger version of this figure.
Discussion
This protocol describes dual-color CLEM to observe the interaction of mitochondria and lysosomes in fixed cells using genetically encoded mEosEM and APEX2 tags. Over the past few decades, advances in microscopy have greatly improved the ability to observe changes in organelle networks32,33, and the study of organelle interactions has become increasingly interesting. Super-resolution microscopy has been used to observe changes in highly dynamic contacts between mitochondria and lysosomes34,35,36. However, despite the use of super-resolution microscopy, it is difficult to distinguish changes in the thickness of the inner and outer mitochondrial membranes (approximately 22 nm) and cristae junctions (approximately 28 nm)37. Transmission electron microscopy has the advantage of being able to observe the structure of biological samples at very high resolution but has the disadvantage that the images can only be viewed in black and white, making it difficult to distinguish organelles in complex images. In our protocol, we used a CLEM method that combines the advantages of light and electron microscopy to monitor the interaction of two different organelles by staining them with different colors. In particular, dual-color CLEM is an invaluable tool for observing and analyzing mitophagy, the process of mitochondrial degradation within lysosomes, as shown in Figure 3.
Live cell imaging is typically performed under physiological conditions, enabling researchers to observe the dynamic cellular processes in real time. However, electron microscopy techniques often require time-consuming sample preparation procedures, including fixation and staining. The time delay between live cell imaging and electron microscopy sample preparation limits the temporal resolution of the technique and can result in potentially missed dynamic cellular events. To overcome the limitation of live cell CLEM, we have developed a dual-color CLEM method in which the mEosEM probe23 and chemical dye (Amplex-Red) are stained by the APEX2 proximity labeling enzyme38 without losing fluorescence signal even in harsh fixative environments. In a previous live cell CLEM study39, discrepancies were found between fluorescence signals and EM images due to temporal resolution issues in TEM sampling after live cell imaging and technical limitations in sample preparation. However, by acquiring fluorescence images of fixed cells for a sufficient time (z-stack imaging of whole cells at a high magnification), we can resolve the temporal resolution issue and improve the correlation between LM and EM images (Figure 3 and Figure 4).
Another key factor for successful CLEM results is how to accurately register the LM image with the EM image. To do this, we first used a combination of DIC and fluorescence imaging modes during light microscopy imaging to create a navigation map around the target cells. Using this navigation map, we could easily identify the cells of interest on the ultramicrotome and TEM. Second, by acquiring TEM tile images of whole cells at high magnification, we were able to obtain many landmarks that could be used for image registration. In our experience, the higher the number of landmarks, the better the alignment accuracy of LM and EM images in the Big Warp plugin.
In this study, we showed the interaction domains of lysosomes and mitochondria with high-resolution CLEM results. In addition, the dual-color probe allowed for precise distinction between mitochondria and lysosomes. However, this method has some limitations. First, if the reaction time during the proximal labeling of Amplex-Red by the APEX2 enzyme is too long, the Amplex-Red will diffuse and stain wider than the target protein location38. Second, the thickness (z-axis volume) of the area where the lysosome is engulfed in the mitochondria is quite small, so the fusion of the two organelle membranes cannot be determined in a 50 nm thick serial section. Furthermore, it is a significant challenge to accurately align the z-axis of an optical section by light microscope and a mechanical section by ultramicrotome. To overcome these limitations, dual-color CLEM can be used in combination with conventional electron-tomography40 or serial section volume-CLEM41.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by the KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (24-BR-01-03). TMEM192-V5-APEX2 plasmids were kindly provided by Hyun-Woo Rhee (Seoul National University). TEM data were acquired at Brain Research Core Facilities in KBRI.
Materials
| Chemicals and reagents | |||
| 25% Glutaraldehyde | Electron Microscopy Sciences | 16200 | Aldehyde fumes are extremely toxic. Use only in fume hood. |
| 30% Hydrogen peroxide solution | Merck | 107210 | |
| 4% Paraformaldehyde solution | Biosesang | PC2031-100-00 | ldehyde fumes are extremely toxic. Use only in fume hood. |
| Amplex Red | ThermoFisher | A12222 | |
| Epon 812 | Electron Microscopy Sciences | 14120 | |
| Ethanol | Merck | 100983 | |
| Fugene HD | Promega | E2311 | Plasmid transfection |
| Glycine | SIGMA | G8898 | |
| Lead Citrate 3% | Electron Microscopy Sciences | 22410 | |
| Osmium tetroxide 4 % aqueous solution | Electron Microscopy Sciences | 16320 | Very toxic. Use only in fume hood. |
| Potassium hexacyanoferrate(II) trihydrate | SIGMA | P3289 | |
| Sodium cacodylate trihydrate | Sigma-Aldrich | C0250-500G | |
| Uranyl acetate | Electron Microscopy Sciences | 22400 | |
| UranyLess EM stain | Electron Microscopy Sciences | 22409 | |
| Plasmid construction | |||
| Mito-mEosEM | addgene | 132706 | DOI: 10.1016/j.chembiol.2024.02.007 |
| TMEM192-V5-APEX2 | Sharma N. et al. | N/A | DOI: 10.1016/j.chembiol.2024.02.007 |
| Tools | |||
| 0.22 um syringe filter | Sartorius | 16534 | |
| 35 mm Gridded coverslip dish | Mattek | P35G-1.5-14-CGRD | |
| BEEM Embedding Capsules | Ted Pella | 130 | |
| Formvar Supported Single slot copper grid | Ted Pella | 01705 | |
| Diamond knife | DiATOME | DU4530 | |
| Ultra-microtome | Leica | ARTOS 3D | |
| Microscopes | |||
| Inverted confocal microscopy | Nikon | A1 Rsi/Ti-E | |
| Transmission electron microscopy | FEI | Tecnai G2 | |
| Software and Algorithms | |||
| Fiji/Image J | NIH / open source | https://imagej.nih.gov/ij/ | |
| ImageJ BigWarp software package | NIH / open source | https://imagej.net/plugins/bigwarp | |
| Photozoom Pro 8 | BenVista | https://www.benvista.com/photozoompro Alternative software Image Resizer : https://imageresizer.com/ Gigapixel AI : https://www.topazlabs.com/gigapixel ON1 resize : https://www.on1.com/products/resize-ai/ |
References
- Giacomello, M., Pyakurel, A., Glytsou, C., Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 21 (4), 204-224 (2020).
- Spinelli, J. B., Haigis, M. C. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 20 (7), 745-754 (2018).
- Lopez, J., Tait, S. W. G. Mitochondrial apoptosis: Killing cancer using the enemy within. Br J Cancer. 112 (6), 957-962 (2015).
- Wang, C., Youle, R. J. The role of mitochondria in apoptosis*. Annu Rev Genet. 43, 95-118 (2009).
- Rossi, A., Pizzo, P., Filadi, R. Calcium, mitochondria and cell metabolism: A functional triangle in bioenergetics. Biochim Biophys Acta Mol Cell Res. 1866 (7), 1068-1078 (2019).
- Giorgi, C., Marchi, S., Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 19 (11), 713-730 (2018).
- Talari, N. K., et al. Lipid-droplet associated mitochondria promote fatty-acid oxidation through a distinct bioenergetic pattern in male wistar rats. Nat Commun. 14 (1), 766 (2023).
- Enkler, L., et al. Arf1 coordinates fatty acid metabolism and mitochondrial homeostasis. Nat Cell Biol. 25 (8), 1157-1172 (2023).
- Trivedi, P. C., Bartlett, J. J., Pulinilkunnil, T. Lysosomal biology and function: Modern view of cellular debris bin. Cells. 9 (5), 1131 (2020).
- Mony, V. K., Benjamin, S., O’Rourke, E. J. A lysosome-centered view of nutrient homeostasis. Autophagy. 12 (4), 619-631 (2016).
- Thelen, A. M., Zoncu, R. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 27 (11), 833-850 (2017).
- Ma, X., et al. Mitochondria-lysosome-related organelles mediate mitochondrial clearance during cellular dedifferentiation. Cell Rep. 42 (10), 113291 (2023).
- Kim, S., Wong, Y. C., Gao, F., Krainc, D. Dysregulation of mitochondria-lysosome contacts by gba1 dysfunction in dopaminergic neuronal models of Parkinson’s disease. Nat Commun. 12 (1), 1807 (2021).
- Pickrell, A. M., Youle, R. J. The roles of pink1, parkin, and mitochondrial fidelity in parkinson’s disease. Neuron. 85 (2), 257-273 (2015).
- Wong, Y. C., Peng, W., Krainc, D. Lysosomal regulation of inter-mitochondrial contact fate and motility in charcot-marie-tooth type 2. Dev Cell. 50 (3), 339-354.e4 (2019).
- Peng, W., Wong, Y. C., Krainc, D. Mitochondria-lysosome contacts regulate mitochondrial Ca(2+) dynamics via lysosomal trpml1. Proc Natl Acad Sci U S A. 117 (32), 19266-19275 (2020).
- Juhl, A. D., et al. Quantitative imaging of membrane contact sites for sterol transfer between endo-lysosomes and mitochondria in living cells. Sci Rep. 11 (1), 8927 (2021).
- Khalil, S., et al. A specialized pathway for erythroid iron delivery through lysosomal trafficking of transferrin receptor 2. Blood Adv. 1 (15), 1181-1194 (2017).
- Russell, M. R., et al. 3d correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy. J Cell Sci. 130 (1), 278-291 (2017).
- Van Rijnsoever, C., Oorschot, V., Klumperman, J. Correlative light-electron microscopy (clem) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Methods. 5 (11), 973-980 (2008).
- Loginov, S. V., et al. Correlative organelle microscopy: Fluorescence guided volume electron microscopy of intracellular processes. Front Cell Dev Biol. 10, 829545 (2022).
- De Beer, M., et al. Visualizing biological tissues: A multiscale workflow from live imaging to 3D cryo-CLEM. Microscopy and Microanalysis. 27 (S2), 11-12 (2021).
- Fu, Z., et al. Meosem withstands osmium staining and epon embedding for super-resolution clem. Nat Methods. 17 (1), 55-58 (2020).
- Lam, S. S., et al. Directed evolution of apex2 for electron microscopy and proximity labeling. Nat Methods. 12 (1), 51-54 (2015).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 9 (7), 676-682 (2012).
- Cardona, A., et al. Trakem2 software for neural circuit reconstruction. PLoS One. 7 (6), e38011 (2012).
- Redmann, M., et al. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 11, 73-81 (2017).
- Lu, F., et al. Identification of npc1 as the target of u18666a, an inhibitor of lysosomal cholesterol export and ebola infection. eLife. 4, e12177 (2015).
- Jung, M., Choi, H., Mun, J. Y. The autophagy research in electron microscopy. Appl Microsc. 49 (1), 11 (2019).
- Hao, T., et al. Hypoxia-reprogramed megamitochondrion contacts and engulfs lysosome to mediate mitochondrial self-digestion. Nat Commun. 14 (1), 4105 (2023).
- Hoglinger, D., et al. Npc1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat Commun. 10 (1), 4276 (2019).
- Jung, M., Mun, J. Y. Mitochondria and endoplasmic reticulum imaging by correlative light and volume electron microscopy. J Vis Exp. (149), e59750 (2019).
- Ohta, K., Hirashima, S., Miyazono, Y., Togo, A., Nakamura, K. I. Correlation of organelle dynamics between light microscopic live imaging and electron microscopic 3d architecture using fib-sem. Microscopy (Oxf). 70 (2), 161-170 (2021).
- Wang, H., et al. Lysosome-targeted biosensor for the super-resolution imaging of lysosome-mitochondrion interaction. Front Pharmacol. 13, 865173 (2022).
- Chen, Q., et al. Quantitative analysis of interactive behavior of mitochondria and lysosomes using structured illumination microscopy. bioRxiv. , (2018).
- Wong, Y. C., Ysselstein, D., Krainc, D. Mitochondria-lysosome contacts regulate mitochondrial fission via rab7 gtp hydrolysis. Nature. 554 (7692), 382-386 (2018).
- Perkins, G., et al. Electron tomography of neuronal mitochondria: Three-dimensional structure and organization of cristae and membrane contacts. J Struc Biol. 119 (3), 260-272 (1997).
- Sharma, N., Jung, M., Mishra, P. K., Mun, J. Y., Rhee, H. -. W. Flex: Genetically encodable enzymatic fluorescence signal amplification using engineered peroxidase. Cell Chem Biol. 31 (3), 502-513.e506 (2024).
- Jung, M., Choi, H., Kim, J., Mun, J. Y. Correlative light and transmission electron microscopy showed details of mitophagy by mitochondria quality control in propionic acid treated sh-sy5y cell. Materials (Basel). 13 (19), (2020).
- Mourik, M. J., Müller-Reichert, T., Verkade, P., et al. . Methods Cell Biol. 124, 71-92 (2014).
- Hegermann, J., et al. Volume-clem: A method for correlative light and electron microscopy in three dimensions. Am J Physiol Lung Cell Mol Physiol. 317 (6), L778-L784 (2019).

.
