A Cryo-pulverization Protocol for Processing Mouse Paws to Evaluate Molecular Pathways of Tissue Inflammation in a Collagen Induced Arthritis Model
Özet
A cryogenic pulverization method to process murine paws using a liquid nitrogen freezer mill was developed to improve the yield and quality of RNA or protein extracted from the tissues and enable the analysis of molecular profiles associated with inflammatory responses.
Abstract
Profiling molecular changes in local tissues is crucial to understand the mechanism(s) of action of therapeutic candidates in vivo. In the field of arthritis research, many studies are focused on inflamed joints that are composed of a complex mixture of bone, cartilage, muscle, stromal cells and immune cells. Here, we established a reliable and robust mechanical method to disrupt inflamed mouse paws into homogeneous pulverized samples in a cryogenically controlled environment. Protein and RNA lysates were processed to enable proteomic and transcriptional endpoints and molecular characterization of relevant disease pathways in local tissue.
Introduction
Rheumatoid arthritis (RA) is a chronic systemic inflammatory disease with persistent symmetric synovitis in joints and extra articular involvement of organs such as the skin, heart, lungs, and eyes1. Although the systemic manifestations of the immune response are evident in human patients, one of the hallmarks of RA pathology is infiltration of immune cells in synovial tissue and proliferation of synovial fibroblast cells2.
Similar to human RA, mouse collagen induced arthritis (CIA) model elicits strong tissue inflammation with active immune responses in synovial tissues and systemic compartments. The susceptibility of different mouse strains to CIA model links to Major Histocompatibility Complex (MHC) haplotype and antigen specific T cell and B cell interactions3,4. In addition, many pathogenic pathways in human RA, including autoantibody production, immune complex deposition, myeloid cell activation, polyarticular manifestations and pannus formation with synovial immune infiltration, are also evident in this model5,6. Investigators have employed this well-established CIA model to investigate effects of anti-inflammatory cytokine treatments7. Many biologics approved for autoimmune or inflammatory diseases, such as anti-TNFα and anti-IL-6, are found to be efficacious in the CIA model8,9.
Profiling the interactions of the immune system in synovial tissue is crucial to elucidate molecular mechanisms associated with the pathogenesis of RA. In the human clinical setting, a common practice is to perform needle synovial biopsies under the guidance of ultrasound imaging. In the preclinical settings, the smaller architecture of the murine joints makes biopsy procedures much more difficult if not impossible. Recently, we demonstrated the utilization of the murine CIA model to evaluate combinations of drugs to impact disparate end-points and resolve disease in a combinatorial approach10. A cryogenic freezer mill-based pulverization method was employed to process inflamed murine paws into homogeneous fine powders and established downstream processes to extract RNA and proteins. This method protects RNA and protein from enzymatic and chemical degrative processes and enables us to apply multiple analytical methods to a single homogenized sample source.
Protocol
All animal experiments were conducted in accordance within the policies of the Institutional Animal Care and Use Committee (IACUC) of Janssen R&D.
1. Cryogenic Freezer Mill-based Pulverization Method
- At termination of the CIA study, sacrifice mice with 1.5% isoflurane followed by cervical dislocation.
- On the day of tissue processing, pre-chill all instruments (spatulas, grinding vials, etc.) on dry ice for a minimum of 10 min. Wear thermal gloves to avoid freeze damage.
- Cut hind paws with a scissor at the fur line as depicted in Figure 1A.
- Transfer each paw into one 1.5 mL microcentrifuge tube, snap freeze in liquid nitrogen immediately, and store frozen paws at -80 °C. Do not keep samples frozen for more than one week.
- Fill freezer mill with liquid nitrogen as shown in Figure 1B and let it equilibrate for at least 10 min.
- Keep unprocessed sample on dry ice and avoid freeze-thaw cycles.
- Transfer one hind paw into pre-chilled large polycarbonate grinding vial with a bottom steel plug, insert the pre-chilled steel impactor and close the polycarbonate grinding vial with the pre-chilled top steel plug (Figure 1C).
- Transfer large polycarbonate grinding vials with the samples into the freezer mill pre-filled with liquid and close the lid with the rubber clasp found at the front of the instrument.
- Let samples cool in liquid nitrogen for 1 minute.
- Set the freezer mill to the 1 minute program with 10 cycles per second and press the start button. Wait for the freezer mill to complete its cycle.
NOTE: The machine makes a drumming sound as the steel impactor moves back and forth. Use ear plugs to avoid hearing loss. - Open the lid of the freezer mill, take out the large polycarbonate grinding vial and place it in an extractor as depicted in Figure 1D. Remove the top steel plug by placing downward pressure on the black handle until the steel plug slides out of the polycarbonate tube.
- Transfer the opened large polycarbonate grinding vial onto dry ice. Remove the steel impactor with pre-chilled forceps.
- Transfer the frozen powder into pre-chilled 50 mL conical tube.
- Weight 30-50 mg of frozen powder into a tared frozen 1.5 mL microcentrifuge tube for RNA extraction and 100-200 mg of frozen powder for protein extraction.
NOTE: The frozen powder should look like white or light pink fine sand as depicted in Figure 1E. - Store pulverized samples at -80 °C and proceed to RNA and protein isolations within 24 h.
2. RNA Extraction
- Add 10 µL of β-mercaptoethanol (β-ME) per 1 mL of RLT buffer.
- Calculate the volume of RLT Buffer corresponding to a ratio of 23.3 mL/mg of tissue and add the appropriate volume of RLT Buffer with β-ME to the tube with frozen powder. This ratio was empirically determined to be ideal for mouse paws.
- Vortex the sample at 3,000 RPM for 10 s.
- Mix well by pipetting up-and-down 10 times vigorously with a 1 mL pipettor.
- Vortex the sample again at 3,000 RPM for 20 s.
- Centrifuge the tube at 13,000 x g for 2 min.
- Transfer 700 µL of the supernatants to a fresh tube and add 700 µL of 70% ethanol.
NOTE: If <700 µL is in the tube, it is acceptable to proceed, still adding equivalent volume of 70% ethanol. - Load 700 µL of the sample onto a RNA purification column.
- Centrifuge the tube at ≥10,000 x g for 30 s.
- Discard the flow-through and load the remaining part of the sample to the RNeasy column.
- Centrifuge the tube at ≥10,000 x g for 30 s.
- Discard the flow through and wash the column by adding 700 µL of RW1 buffer with centrifugation of ≥10,000 x g for 30 s. If performing option DNASE digestion, 350 µL of RW1 is added prior to DNASE treatment. And an additional 350 µL of RW1 is added following DNASE treatment.
- Optional) Perform a DNase digestion step follow manufacturer's instructions.
- Wash the tube twice by adding 500 µL of RPE buffer followed by centrifugation at ≥10,000 x g for 30 s. Discard flow through each time.
- Dry the column by centrifugation at ≥10,000 x g for 2 min.
- Elute RNA by adding 50 µL water with centrifugation at ≥10,000 x g for 2 min. Collect flow through and transfer into a new 1.5 mL tube.
- Determine the quantity of RNA using the researcher's method of choice and store at -80 °C before further analysis.
3. Protein Extraction
- Dilute the 10x cell lysis stock solution in 1x cell lysis stock solution using cell culture grade water.
- Reconstitute the Protease Inhibitor Cocktail Set I with 1 mL of water to make a 100x protease inhibitor stock.
- Add 100 µL of protease inhibitor to 9.9 mL of 1x Cell lysis to make a 1x final stock solution.
- Add 4 µL of ice cold 1x Cell Lysis Buffer per mg tissue powder (e.g., 800 µL of lysis buffer to 200 mg of tissue powder).
- Add one 5 mm stainless steel bead to the tube.
- Vortex the tube at 3,000 RPM for 60 s. Transfer the tube to wet ice and continue to the next sample.
- Place all sample tubes in a box and shake the centrifuge box on a rocker at 1,000 RPM, 4 °C for 1 h.
NOTE: Researchers may find that placing the box vertical can facilitate better mixing with the bead. - Centrifuge the tubes at 10,000 x g and 4 °C for 15 min.
- Transfer the supernatants into a fresh tube and avoid the fat layer on the top.
- Determine the total protein concentration. 10 to 30-fold dilutions of protein lysate are ideal for a BCA test.
- Aliquot the sample into 1.5 mL microcentrifuge tubes and store at -80 °C before further analysis.
Representative Results
Here, we show a representative gel image visualization of RNA extracted from front paws of CIA mice in Figure 2A. The 28S rRNA and the 18S rRNA band indicate all samples have sufficient amount of intact RNA. Next, we show a representative scatter plot of total protein concentrations based on protein BCA analysis in Figure 2B. Total protein concentrations from naïve mice, CIA mice or CIA mice under various treatments are comparable across groups. To determine the concentrations of inflammatory cytokines and chemokines in the protein extract, Luminex analyses were conducted. We show a representative scatter plot of the concentrations of normalized cytokines and chemokines (pg/mg total protein) in Figure 2C. Compared to naïve mice, several cytokines are elevated in CIA mice and treatment with anti-IL17A antibody significantly inhibits production of several cytokines. To evaluate transcriptome changes in CIA and treatment-related effects, microarray analysis was performed. We show a representative heatmap plot of genes significantly increased in CIA mice compared to naïve mice in Figure 2D.
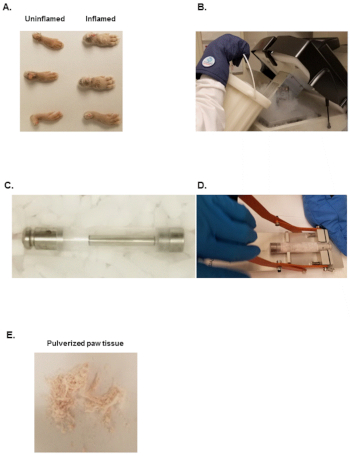
Figure 1: Equipment needed to pulverize murine paws. (A) Inflamed or uninflamed paws from CIA or naïve mice. (B) Liquid nitrogen Dewar used to fill the stainless-steel bath of the freezer mill. (C) Assembled freezer mill tube set. (D) Freezer mill tube opener. (E) Pulverized tissue from murine paws. Please click here to view a larger version of this figure.
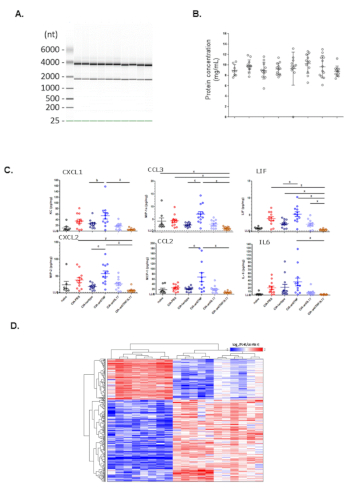
Figure 2: Representative data on analysis of protein and RNA from pulverized murine paws. (A) Gel image visualization of RNA integrity. (B) Total protein concentrations from different groups. (C) Chemokine and cytokine expression (Luminex) from different groups. (D) Hierarchical clustering heatmap of microarray analysis from different treatment groups. Please click here to view a larger version of this figure.
Discussion
Although there is strong scientific rationale to evaluate molecular pathways in synovial tissues, many reports on immune profiling of murine CIA model were focused on peripheral blood, while protein and RNA analysis data of inflamed paws are rather limited. There are several possible reasons for this bias: murine ankle joints are no larger than ~2 cm; the affected areas consist of skin, bone and connective tissues which are often difficult to homogenize using traditional methods like tissue grinders, pestle and mortar, silica bead disruption, trituration or enzymatic digestion. These methods typically result in incomplete homogenization, low protein or RNA yield, or inconsistent quality due to proteolytic and nucleic acid degradation. The robust method described herein provides a detailed procedure to generate a homogeneous pulverized frozen powder to enable downstream proteomic and transcriptional profiling efforts to establish molecular signatures of disease from a single uniform sample source.
Several critical steps should be considered when performing this method. Firstly, murine paws should be cut at the fur line during tissue harvest and large amounts of hair should be avoided. Excessive hair can clog columns during RNA extraction and interfere with protein extraction. Secondly, tubes, forceps and spatulas should be kept on dry ice throughout the procedure because elevated temperature in tissue powders will quickly turn the material into amorphous "mud" and the integrity of protein and RNA will be compromised. Thirdly, the amount of tissue powder used for protein or RNA extraction should be carefully optimized. More tissue powder may not always provide higher yield but can cause additional problems in downstream processing including clogging RNA isolation columns, altering the DNase/DNA ratio (if this step is employed) to prevent complete DNA degradation. Finally, normalization of protein and RNA input should be an integral part of the workflow and data analysis. Variability in tissue load can significantly mask changes in relative protein and RNA levels. In addition to total protein or total RNA, expression of housekeeping genes can also be considered as anchors for normalization.
Through extensive trial and error process, we have identified some potential frequent issues for first-time users of this method. There can be low yield of RNA. This is often a result of overloading the column with too much tissue extract. The amount of the tissue per column and ratio of RLT/powder are crucial for efficient RNA isolations. We have found that some commercial 96 well extraction kits may not work for this process as the samples will not spin through the column at an equivalent rate, thus compromising these samples in downstream washing and elution steps. There can be high variability in protein concentration in BCA analysis and cytokine concentrations in Luminex analysis. If there are no issues with the BCA or the Luminex process, it is typically caused by fat or other contaminants in the protein extract. It is important to avoid the fat layer at the top or insoluble matter at the bottom of the microcentrifuge tubes when harvesting for protein extraction. If needed, repeat the spin and collect supernatants for downstream analysis.
As this method involves a specialized cryogenic freezer mill and large quantities of dry ice and liquid nitrogen, additional safety measures should be considered. Proper personal protection equipment including safety glasses, lab coats and cryogenic gloves should always be worn to prevent potential freeze burns and injury due to explosion. Generation of aerosols should also be minimized and using of vented balance enclosures is advised. Finally, similar to any other protocols handling animal tissues, care should be taken to properly dispose of all unused tissue samples, while reusable tubes need to be decontaminated prior to cleaning.
While the method has many advantages over conventional methods, it still harbors some limitations. The transcriptome profile from a murine whole paw significantly overlaps with transcriptome profile from human synovial biopsy (data not shown). Additional mRNA from muscle, skin and bone marrow could dilute the signals from synovial tissue. An alternative solution is to collect murine synovial tissue through laser microdissection, which can be performed on OCT embedded mouse joints. However, low throughput and the relatively small quantity of tissue limits the broad application for laser microdissection. Additionally, even with significant automation, this method is still quite labor intensive. It takes at least 8 h to process 30-60 samples with the help of 3-4 investigators. Finally, this method requires large amounts of liquid nitrogen (~10-15 L for processing 30-60 samples).
In the preceding method description, the evaluation of proteomic and transcriptional endpoint analyses was demonstrated. However, additional endpoint, such as lipidomic, metabolomics and small RNA profiling could be of interest to the wider arthritis research community.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The authors wish to thank Edith Janssen for the critical review of the manuscript and Navin Rao and Jennifer Towne for their support of the publication of this manuscript.
Materials
| 5 mm stainless steel bead | Qiagen | 69989 | |
| beta-mercaptoethanol | Sigma | M6250 | Sample reducing agent that inhibits RNASE enzymes |
| Bioanalyzer Kit | Agilent | 5067-1511 | RNA qualification kit |
| b-mercaptoethanol | Sigma | M6250 | |
| Cell Culture Grade Water | Corning | 25-055-CI | Water |
| Cell lysis stock solution | Cell Signaling | 9803 | |
| Eppendorf Tube | Eppendorf | 22363204 | Microfuge tubes |
| Eppendorf tube centrifuge box | Nalgene | 5055 | Box for holding eppendorf tubes in horizontal tube arrangement |
| Everlast 247 Variable Speed Rocker | Benchmark Scientific | BR5000 | |
| Freezer Mill | Spex Sample Prep | 6875 | Freezer/Mill for processing paws into pulverized powder |
| Grinding Vial | Spex Sample Prep | 6801 | Polycarbonate vial for processing paws into pulverized powder |
| Pierce BCA kit | Pierce | 23225 | Kit for Total Protein Quantification |
| Protease Inhibitor Cocktail set 1 | Calbiochem | 539131 | Protease Inhibitors |
| Protein BCA Kit | Pierce | 23225 | |
| Quantigene Kit | Thermofisher | QP1013 | bDNA analysis Kit |
| Refrigerated microcentrifuge | Eppendorf | 5417R | Centrifugation |
| RLT Buffer | Qiagen | 79216 | RNA extraction buffer |
| RNeasy mini kit | Qiagen | 74104 | including RNeasy column, RLT Buffer and RW1 Buffer |
| Shaker | Benchmark Scientific | BR5000 | Rocker/Shaker |
| Spatula | VWR | 10806-412 | Spatula for powder transfer |
| Stainless Steel Bead | Qiagen | 69989 | Bead for mixing during protein extraction |
| Tube Extractor | Spex Sample Prep | 6884 | Extractor for removing the top of grinding vial |
| Vortexer | VWR | 10153-838 | Sample mixing |
Referanslar
- Firestein, G. S. Immunologic mechanisms in the pathogenesis of rheumatoid arthritis. Journal of Clinical Rheumatology. 11, S39-S44 (2005).
- McInnes, I. B., Schett, G. The pathogenesis of rheumatoid arthritis. New England Journal of Medicine. 365, 2205-2219 (2011).
- Ahlqvist, E., Hultqvist, M., Holmdahl, R. The value of animal models in predicting genetic susceptibility to complex diseases such as rheumatoid arthritis. Arthritis Research and Therapy. 11, 226 (2009).
- David, C. S., Taneja, V. Role of major histocompatibility complex genes in murine collagen-induced arthritis: a model for human rheumatoid arthritis. American Journal of the Medical Sciences. 327, 180-187 (2004).
- Tarkowski, A., Holmdahl, R., Klareskog, L. Rheumatoid factors in mice. Monographs in Allergy. 26, 214-229 (1989).
- Schurgers, E., Billiau, A., Matthys, P. Collagen-induced arthritis as an animal model for rheumatoid arthritis: focus on interferon-gamma. Journal of Interferon & Cytokine Research. 31, 917-926 (2011).
- Joosten, L. A., Helsen, M. M., van de Loo, F. A., van den Berg, W. B. Anticytokine treatment of established type II collagen-induced arthritis in DBA/1 mice. A comparative study using anti-TNF alpha anti-IL-1 alpha/beta, and IL-1Ra. Arthritis & Rheumatology. 39, 797-809 (1996).
- Asquith, D. L., Miller, A. M., McInnes, I. B., Liew, F. Y. Animal models of rheumatoid arthritis. European Journal of Immunology. 39, 2040-2044 (2009).
- Williams, R. O. Collagen-induced arthritis as a model for rheumatoid arthritis. Methods in Molecular Medicine. 98, 207-216 (2004).
- Shen, F., et al. Combined Blockade of TNF-alpha and IL-17A Alleviates Progression of Collagen-Induced Arthritis without Causing Serious Infections in Mice. Journal of Immunology. 202, 2017-2026 (2019).

