Sand Fly (Phlebotomus papatasi) Embryo Microinjection for CRISPR/Cas9 Mutagenesis
Özet
This protocol details the steps of CRISPR/Cas9 targeted mutagenesis in sand flies: embryo collection, injection, insect rearing, and identification as well as selection of mutations of interest.
Abstract
Sand flies are the natural vectors for Leishmania species, protozoan parasites producing a broad spectrum of symptoms ranging from cutaneous lesions to visceral pathology. Deciphering the nature of the vector/parasite interactions is of primary importance for better understanding of Leishmania transmission to their hosts. Among the parameters controlling the sand fly vector competence (i.e. their ability to carry and transmit pathogens), parameters intrinsic to these insects were shown to play a key role. Insect immune response, for example, impacts sand fly vector competence to Leishmania. The study of such parameters has been limited by the lack of methods of gene expression modification adapted for use in these non-model organisms. Gene downregulation by small interfering RNA (siRNA) is possible, but in addition to being technically challenging, the silencing leads to only a partial loss of function, which cannot be transmitted from generation to generation. Targeted mutagenesis by CRISPR/Cas9 technology was recently adapted to the Phlebotomus papatasi sand fly. This technique leads to the generation of transmissible mutations in a specifically chosen locus, allowing to study the genes of interest. The CRISPR/Cas9 system relies on the induction of targeted double-strand DNA breaks, later repaired by either Non-Homologous End Joining (NHEJ) or by Homology Driven Repair (HDR). NHEJ consists of a simple closure of the break and frequently leads to small insertion/deletion events. In contrast, HDR uses the presence of a donor DNA molecule sharing homology with the target DNA as a template for repair. Here, we present a sand fly embryo microinjection method for targeted mutagenesis by CRISPR/Cas9 using NHEJ, which is the only genome modification technique adapted to sand fly vectors to date.
Introduction
Vector-borne diseases are a major public health threat in constant evolution. Hundreds of vector species spread across very distinct phylogenic families (e.g., mosquitoes, ticks, fleas) are responsible for the transmission of a huge number of microbial pathogens, resulting in more than 700,000 human deaths a year, according to the World Health Organization. Among vector insects, phlebotomine sand flies (Diptera, Psychodidae) constitute a vast group, with 80 proven vector species exhibiting distinct phenotypic traits and vectorial capacities found in different geographical regions. They are vectors for the protozoan parasites of the genus Leishmania, causing around 1.3 million new cases of Leishmaniases and between 20,000 and 30,000 deaths a year. Leishmaniases clinical outcomes are diverse, with symptoms ranging from self-limiting cutaneous lesions to visceral dissemination which is fatal in the absence of treatment.
Sand flies are strictly terrestrial insects. Their life cycle, relatively long compared to other Diptera, lasts up to three months, depending on different parameters such as temperature, humidity, and nutrition. It consists of one embryonic stage (6 to 11 days), four larval stages (lasting a total of 23 to 25 days) and one pupal stage (9 to 10 days) followed by metamorphosis and then adulthood. Sand flies require a humid and warm environment for rearing. Both males and females feed on sugars, obtained in the wild from flower nectars. Only females are blood-feeders, as they require proteins obtained from the blood meal for egg production1.
An important focus of research is to identify the nature of the vector/parasite interactions that lead to the development of transmissible infections. As with other vector insects, parameters intrinsic to sand flies have been shown to impact their vector competence, which is defined as their ability to carry and transmit pathogens to their hosts. For example, the expression of galectins by the Phlebotomus papatasi sand fly midgut cells, acting as receptors recognizing parasite surface components, can directly influence their vector competence for Leishmania major2,3. The insect immune response pathway, Immune Deficiency (IMD), is also crucial for the Phlebotomus papatasi sand fly vector competence for Leishmania major4. A critical role for vector insect immune response pathways in controlling their transmission of infectious pathogens has been similarly reported in Aedes aegypti mosquitos5,6,7, in the tsetse fly Glossina morsitans8, and in Anopheles gambiae mosquitoes9,10.
Studies of sand fly/Leishmania interactions have been limited by the lack of gene expression modification methods adapted for use in these insects. Only gene downregulation by small interfering RNA (siRNA) had been performed11,12,13,14 until recently. The technique, limited by the mortality associated with the microinjection of adult females, leads only to a partial loss of function, which cannot be transmitted from generation to generation.
CRISPR/Cas9 technology has revolutionized functional genomic research in non-model organisms such as sand flies. Modified from the adaptive immune system in prokaryotes for defense against bacteriophages15,16, the CRISPR Cas9 system has been rapidly adapted as a genome editing tool for superior eukaryotic organisms, including insects. The principle of CRISPR/Cas9 targeted genome-editing is based on the complementarity of a single guide RNA (sgRNA) to a specific genomic locus. The Cas9 nuclease binds to the sgRNA and creates a double-strand DNA (dsDNA) break in the genomic DNA where the sgRNA associates with its complementary sequence. The Cas9-sgRNA complex is guided to the target sequence by 17 to 20 complementary bases in the sgRNA to the chosen locus, the dsDNA break can then be repaired by two independent pathways: nonhomologous end joining (NHEJ) or homology-directed repair (HDR)17. NHEJ repair involves a simple closure of the break but frequently leads to small insertion/deletion events. DNA repair through HDR uses a donor DNA molecule sharing homology with the target DNA as a template for repair. Insects possess both machineries.
CRISPR/Cas9 technology can generate mutations in a chosen locus, through the NHEJ repair pathway; or for more complex genome editing strategies, such as knock-ins or expression reporters, through the HDR pathway with an appropriate donor template. In sand flies, null mutant alleles of the immune response factor Relish were generated through NHEJ-mediated CRISPR in Phlebotomus papatasi4. Sand fly embryos were also injected in another study with a CRISPR/Cas9 mix targeting the gene encoding Yellow. Still, no adults carrying the mutation were produced18. We describe here a detailed method of sand fly targeted mutagenesis by NHEJ-mediated CRISPR/Cas9, with a particular focus on the embryo microinjection, a critical step of the protocol.
Protocol
The use of mice as a source of blood for sand fly feeding was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). The protocol was approved by the Animal Care and Use Committee of the NIAID, NIH (protocol number LPD 68E). Invertebrates are not covered under NIH guidelines.
1. Needle preparation (Figure 1)
- Pull needles and bevel as described in Meuti and Harrell19. Briefly, pull needles on a dual-stage glass micropipette puller, using borosilicate glass capillaries.
- Sharpen the needles by wet beveling on the micropipette beveller, using an extra fine stone.
2. Embryo collection and micromanipulation (Figure 2)
- Five days before the day of injection, blood feed sand fly females as performed routinely for colony maintenance. For details on sand fly colonies rearing procedures and required material, please refer to Lawyer et al.1 Maintain the insects in incubators at 70% humidity and 26 °C, during their entire development.
- One day post blood-feeding, capture blood-fed females by groups of 100-150 in plaster pots with a side port. Maintain the females by feeding them with sugar solution (30% sucrose – enough volume to soak a small cotton ball) which is usual for colony maintenance.
- At day 2-3 post blood-feeding, do NOT moisten the plaster pots. Keeping the females on a dry substrate will prevent premature egg laying, since females prefer to lay eggs on a moist environment.
- On day 5 post blood-feeding, perform the embryo collection, micromanipulation, and microinjection. Introduce a moist filter paper into the egg-laying chamber. Freshly laid embryos do not have a fully developed chorion and appear white. Use black filter paper during the procedure to facilitate visualizing the white embryos.
- Transfer a small group of females from the plaster pot to the egg-laying chamber through the side ports by using a mouth aspirator. The presence of a moist substrate (the filter paper) induces females to oviposit.
- After 30-60 min, carefully retrieve the filter paper with newly laid embryos from the chamber. Keep the filter paper and embryos in a Petri dish for up to 3 hours. During this time, regularly assess the moisture level of the filter paper to ensure that it remains moist. During this time, repeat steps 2.4 to 2.6 with new groups of females and collect as many embryos as possible.
- Prepare microscope slides for the injection: manually collect the eggs one by one with a very fine paintbrush and carefully transfer them onto another piece of moist black filter paper placed on a microscope slide topped with a glass coverslip.
- Add water to the filter paper, enough to keep the embryos moist but not causing them to float away from the coverslip or being sucked under the coverslip. Line up the embryos against the coverslip. The coverslip acts as a backstop preventing the eggs from rolling away during injection.
3. Embryo Injections (Figure 3)
- Start injections 2.5 to 3 hours post start of egg collection. Perform injections at room temperature and ambient lab humidity.
- Make sure the aligned embryos have enough water so they are kept moist.
NOTE: It is critical that the embryos are kept moist during injections. If they are not moist during the injection process, injections become difficult because the needle has trouble piercing the embryo. The correct amount of water is the point at which the embryos have a meniscus of water around them, but the coverslip is not floating on top of the water causing the embryos to push under the coverslip. It is critical to monitor the amount of water around the embryos during injections. Add water as necessary to keep the embryos moist. - Add water carefully by wetting a brush with water and transferring the water to the back end of the filter paper. Adding water in this fashion allows water to be added in a slow and controlled fashion so that just enough is added to keep the embryos moist. Too much water can cause the embryos to float out of alignment, making it difficult to inject the embryos.
- Back load injection mix into the microinjection needle. Add approximately 0.5 to 1 µL of injection mix into the needle using a hand-drawn borosilicate needle filler, gel loading pipet tips can also be used. The injection mix is composed of one or several guide RNA (80 ng/µL each) designed to target a specific locus mixed with commercial recombinant Cas9 protein (300 ng/µL).
- Starting with the injection pressure at 30 psi, press the injection trigger to expel air out of the tip of the needle. This will force the injection mix to the tip of the needle allowing the injection mix to flow. At this point, slowly increase the hold/constant pressure until injection material starts to flow, then back off the hold/constant pressure so that it is just below the point of the injection mix flowing from the needle. The gated setting should deliver just enough injection material so that a small amount of material can be seen to enter the embryo.
NOTE: Injection of sand fly embryos under halocarbon oil similar to other protocols used for Drosophila and mosquitoes was initially tested, however the survival percentage for these injections was very low. Survival was improved by switching to injections on damp filter paper as described above. - Insert the needle into the side of the embryo. Use the coverslip behind the embryo as a backstop which will aid the needle to pierce the embryo. Deliver a small amount of injection mix into the embryo before gently removing the needle.
- Immediately after removal of the needle, press the injector to remove any backfilled material from the needle tip. This will help to prevent the needle from clogging.
- Proceed to the next embryo. Between each injection, ensure that the filter paper is moist and the needle is not clogged.
- Once all embryos have been injected, count the number of injected embryos to keep a running tally.
- Remove the coverslip by adding water to the filter paper so the coverslip floats. At this point, use a probe (wood applicator stick with insect pin glued to the tip) to hold the filter paper in place while a finger is used to pull the coverslip away from the injected embryos.
- Blot away excess water from the filter paper once the coverslip has been removed.
4. Post-injection rearing (G0) (Figure 4)
- Transfer the filter paper with the embryos on top of several layers of moist filter paper placed in a Petri dish to maintain humidity. Keep the embryos here during the time needed to inject other embryos. Embryos can be held for a maximum of 3 hours like this. After injection they will slowly turn brown.
- Once all embryos have been injected, manually transfer them onto previously moistened small size plaster pots. Do not put too many embryos in each pot and maintain distance between them, to avoid possible fungal contamination between individuals. Cover the pots with a screen and store them as would typically be done for sand fly colony eggs, maintained in incubators at 70% humidity and 26 °C, during their entire development.
- On day 1 post-injection, remove all the damaged and dead embryos with a paintbrush. Add 100 µL of 0.5% propionic acid, drop by drop, onto the plaster pots containing the injected embryos to limit fungal contamination. The release of cytoplasm from embryos damaged during the injection process is particularly favorable to fungal growth.
- Check the plaster pots every day and remove any bad or dead embryos. If fungus is present, remove all infected embryos and add a few drops of 0.5% propionic acid.
- Larvae hatch between day 8 and 12 post-injection. Twice a day, transfer the newly emerged larvae onto new plaster pots, in groups of 5-10 maximum. Add a small amount of food and check the food 2-3 times a week.
- Carefully monitor the quantity of larval food, as too much greatly increases the risk of fungal contamination. Sand fly larvae survive better in groups however they can be cannibalistic if there is not enough food.
- When the larvae pupate, separate them by sex to maintain the females as virgins. If there are a lot of survivors, discard the males and keep only the females.
NOTE: Both males and females can carry and transmit mutations. However, as females have to be kept virgins before they are to be crossed, it is easier to use only the G0 injected females to avoid having to separate wt insects by sex. The G0 males can be kept as a backup if the number of adult survivors is low and will in this case be mated with sorted wt virgin females.
5. Selection and screening of mutant alleles (Figure 5)
- Mass-cross the G0 females with wt males in a cage. Blood-feed them altogether, as would be done for normal colony maintenance.
- One day post blood-feeding, transfer each G0 female with 2-3 wt males into individual plaster tubes using a mouth aspirator. Feed the flies with sugar on a small cotton ball changed every day and use a syringe of water to ensure the plaster moisture is suitable for egg-laying.
- After the G0 females have laid eggs (corresponding to G1), collect their bodies in individual, annotated Eppendorf tubes for later genotyping. Maintain the embryos as usually done for wt colonies.
- Extract the DNA from the G0 females by the method of choice.
- Screen the DNA for the presence of mutations, for example by designing a PCR assay using primers surrounding the region of expected CRISPR cuts.
- Keep only the G1 embryos laid by G0 females showing evidence of transformation. When these G1 embryos develop into pupae, separate them by sex and keep only the females. Cross the G1 females from the same G0 female to wt males, blood-feed them, then transfer in small groups of 2-3 females and 4-5 males into small size containers.
NOTE: All G1 individuals have a chance to carry a unique mutation, so if the number of surviving individuals is not too high then it is better to cross G1 females individually with wt males. The case of several females carrying a different mutation present in a same tube is rare but can occur. If such case is observed after genotyping, the G2 progeny females should then be mated individually to wt males. This mating scheme avoids the possibility of multiple mutations in the progeny. - After egg-laying, collect the G1 female bodies and screen them for mutations. Keep only the tubes where at least one female showed evidence of mutation.
- For later generations make single pair in-crosses. After egg-laying, screen parents for presence of mutation. Repeat at each generation until both parents are homozygous for the same mutation. Once identified, they will be the founders of a homozygous mutant stock.
Representative Results
The CRISPR/Cas9 microinjection protocol described here to generate sand fly mutants was established in a previous publication4. This approach produced highly efficient mutagenesis, as 11 out of 540 individuals survived the procedure, of which 9 were mutant. When designing guides for CRISPR/Cas9 mutation, a critical first step is to sequence the region around the area to be targeted. The template for sequencing should be from the strain that is going to be used as a source of embryos for injection. It is risky to rely only on published genome sequences to design guides. It is not unusual to have differences between published genome and pre-guide design sequences. In some cases, guides designed from published genome sequence are not present when the region around the target gene is sequenced (Harrell, personal observation). In addition, the sequence may include Single Nucleotide Polymorphisms (SNPs) that could be mistaken as real CRISPR edits during genotyping. Therefore, confirming the sequence for the target region is critical for the rest of the protocol to succeed.
Successful genetic modification of insects by microinjection depends mainly on two critical aspects: delivery of materials (protein, plasmid, or mRNA) at the appropriate time in development with as little damage to the embryos as possible; and rearing of robust, healthy insects that will survive the procedure and produce offspring. The initial phase of this procedure, described in Figure 2, starts with blood-feeding of the females from the wt colony and proceeds to embryo collection and microinjection five days later. The second phase consists of rearing the injected embryos until adulthood, making the appropriate crosses and identifying and isolating mutations of interest.
Embryos for injection are collected for 30 – 60 min so that the relative age in hours post oviposition can be determined. Embryos are then allowed to develop for 3 h before the start of the injections. This time window of insect development allows the embryos to survive the injections. After this aging period, the embryos are collected onto a prepared injection slide and the embryos are rolled into place against a coverslip edge using a fine brush. The final configuration is shown in Figure 3A. It is important to wet the filter paper enough so that a meniscus forms at the coverslip edge where the embryos sit. Too much water and the embryos will be pushed away from the coverslip edge. Too little will cause the embryos to be drawn under the coverslip. The embryos need to be kept damp, otherwise the embryo membranes become difficult to inject. The coverslip edge acts as a back stop, allowing the needle to penetrate the embryos when pressed against the edge. It is the combination of a sharp needle and a backstop that allows for the needle to penetrate smoothly.
As with most successful microinjection protocols, a good microinjection needle suited to the embryos being injected is important. Good, sharp needles are defined as needles that easily penetrate the embryo without allowing the material to escape post-injection. Good penetration is evident when the needle slips into the embryo, causing little to no indentation of the embryo membrane during penetration and no material leaks from the embryo after the needle is withdrawn. Good sharp needles are produced using needle puller settings that produce a needle that comes to a fine point (Figure 1A). The pulled needle should not have a taper that is too long. Otherwise, the lumen of the needle becomes very narrow for a major portion of the taper (Figure 1B), and it becomes difficult to get the injection pressure high enough to force the material through the needle. In this protocol, beveled needles were used. The process for pulling and beveling needles is described in Meuti and Harrell19. In the case of sand fly embryos that develop over a long period of time, it is particularly critical to have sharp needles that minimize damage to the embryo, thus preventing material from leaking at the injection site. When embryoplasm leaks from an embryo post-injection, this material is a rich medium for mold and fungal growth. In embryos that develop over a shorter period of time the embryo can develop and hatch before the mold becomes an issue. Any embryos that are visibly leaking embryoplasm after injection should be removed.
During injections, it is important to add water as needed by wetting a fine paintbrush and touching it to the filter paper, repeating as necessary until the meniscus of water is just at the base of the embryos. Attention to relative humidity on the injection day needs to be noted. On low humidity days, more water will need to be added. Once injections are completed, a little extra water is added to the filter paper so that the coverslip slightly floats, making it easier to remove. Once the coverslip has been removed, the filter paper with injected embryos can be blotted, so that the filter paper is just barely damp. Embryos can then be transferred to a moist plaster pot for hatching using a fine paintbrush. At this stage, embryos are very fragile so the process has to be undertaken very carefully. It is also important to prevent the embryos from touching each other to be able to remove moldy embryos and limit the spread of fungal contamination.
Post-injection, the G0 injected embryos are kept on plaster pots as per normal rearing procedures. Until they hatch, injected embryo pots should be checked once a day to remove unhealthy embryos. Figure 4A presents the expected timeline of G0 individual rearing from the day of egg-laying and injection to adulthood. Figure 4B shows examples of G0 embryos that are healthy and should be retained; or damaged, contaminated by fungus, desiccated, or deformed and should be discarded. The G0 injected individuals are supposedly mosaic for mutant alleles. A method for identifying potential mutant alleles should be designed, such as a PCR-assay of the region surrounding the expected cutting site(s). Figure 4C presents an example of a PCR-assay showing a mosaic G0 individual, exhibiting a PCR product additional to the expected wt product, representing a mutant allele.
Once the adults emerge, the G0 females developing from G0 injected embryos are crossed with wt males, allowed to lay eggs, and are genotyped later. Only the tubes containing a G0 fly showing evidence of mutation from the genotyping method of choice are retained. The flies from the next generations (G1 females) are crossed either with wt males or individually between siblings (from G2). These crosses are allowed to lay eggs and then genotyped by the method of choice. The last step is repeated until homozygous mutant males and females are obtained, establishing a homozygous mutant line. A schematic representation of the appropriate succession of crosses to establish a mutant line is given in Figure 5A. Figure 5B shows an example of a genotyping PCR allowing the identification of a homozygous mutant sibling cross.

Figure 1: Microinjection needles. A. Good needle. B. Needle with extreme taper not good for injections. Please click here to view a larger version of this figure.
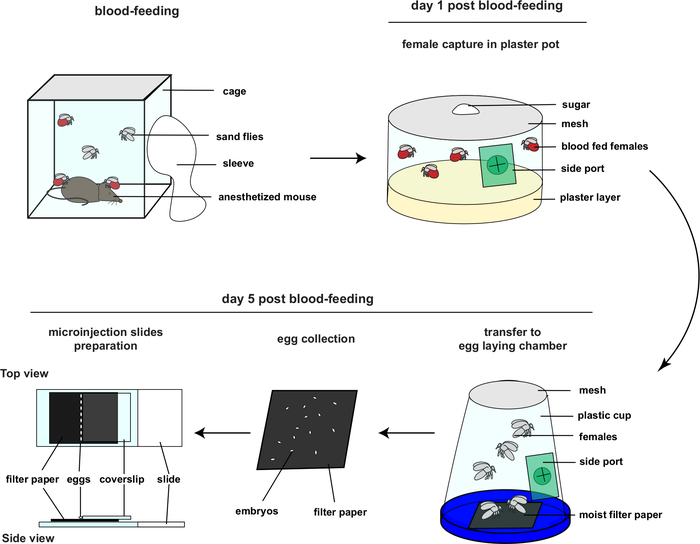
Figure 2: Overview of embryo collection and micromanipulation (This figure had been adapted from4). Please click here to view a larger version of this figure.
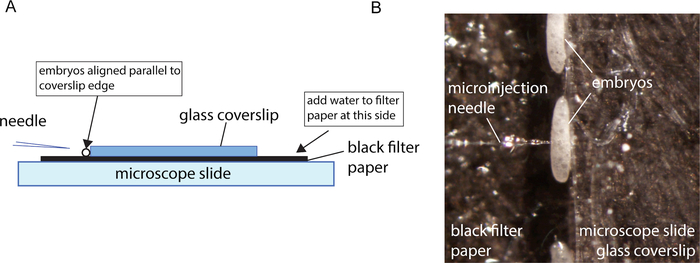
Figure 3: Microinjection setup. A. Schematic representation of the microinjection set-up. B. Close-up of aligned embryo for microinjection. Please click here to view a larger version of this figure.
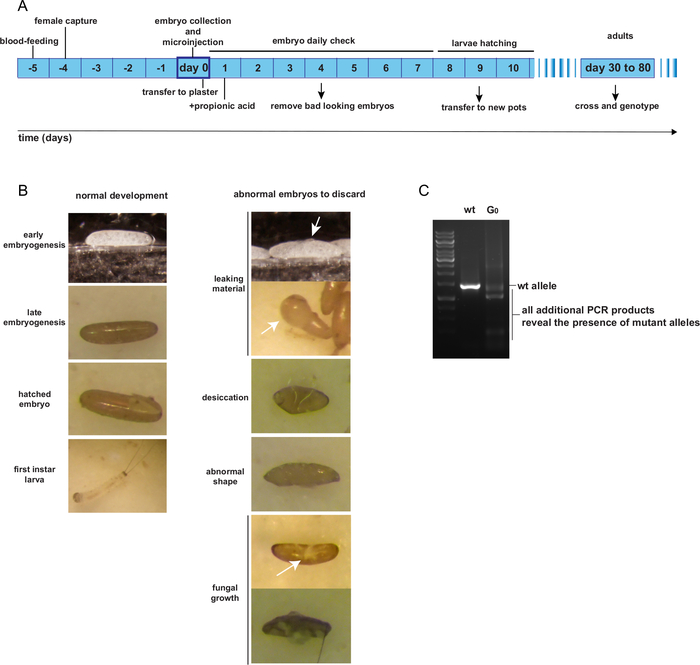
Figure 4: Rearing and identification of mosaic G0 adult individuals. A. Expected time schedule from embryo microinjection to G0 adulthood. B. Examples of good and bad looking embryos post-injection. C. PCR genotyping of one transformed G0 mosaic individual. Please click here to view a larger version of this figure.
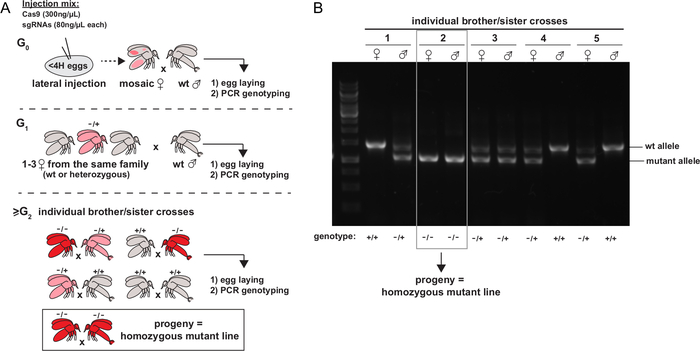
Figure 5: Identification and isolation of mutations of interest. A. Schematic representation of the experimental strategy to isolate mutant alleles and establish homozygous mutant stocks (figure from 4). The color represents the presence of cells carrying mutant alleles, at the heterozygous (pink) or homozygous (red) state. B. Example of a PCR screening strategy for genotyping individual sibbling crosses of sand flies. Please click here to view a larger version of this figure.
Discussion
We present here a recently developed embryo microinjection method for targeted mutagenesis by CRISPR/Cas9 in Phlebotomus papatasi sand flies. Embryo microinjection for insect genetic modification was developed in Drosophila in the mid-1980s21 and is now routinely used in a wide variety of insects. Other methods for delivery of genetic modification materials have been developed for use in insects, such as ReMOT20,21,22,23 and electroporation23. However, embryo microinjection is presently the most versatile and efficient method for delivery of these materials. ReMOT is extremely promising as a delivery method and should be explored for use in sand flies, owing to the relative ease of adult injections as compared to embryo microinjections. However, thus far ReMot has not been successfully used for CRISPR/Cas9 HDR or transposon transgenesis. Although the technique for embryo microinjections in insects is basically the same as initially developed, for each new species refinements of the techniques may be needed. These species differences include the pattern and duration of embryo development, embryo structure, and the environment in which the embryo normally develops. Sand fly embryos are particularly small (0.3-0.5 mm long and 0.1- 0.15 mm wide, representing roughly 1/3 of the size of Drosophila egg1). This reduced size increases the difficulty of handling the eggs without damaging them. The moisture level at each step of the protocol has to be monitored very carefully, as sand fly embryos are particularly sensitive to desiccation. However, they are not aquatic and will die if completely submerged in water for an extended period. Drosophila embryo microinjection requires injection at the posterior end of the embryo. Because of the near symmetrical shape of sand fly embryos, determining embryo polarity is difficult. Finally, while sand fly embryo development proceeds similar to Drosophila, it does so at a much slower rate. Embryogenesis alone lasts between 6 to 11 days, depending on the sand fly species, whereas Drosophila eggs hatch only one day post egg laying. Drosophila egg cellularization occurs about 2 hours post oviposition, whereas sand fly embryos reach this stage at proximately 9 hours post oviposition. Given this difference in developmental timing, injections are targeted to the center of the developing sand fly embryo on the assumption that developing nuclei in the syncytial embryo are located near the center of the developing embryo at this early stage of development, thus obviating the need to distinguish embryo polarity.
CRISPR/Cas9 creates dsDNA breaks at a chosen genomic locus. These double-strand breaks are repaired in the cell by either NHEJ or by HDR. The method we present here was previously used only for NHEJ-based mutagenesis, which generated small insertion/deletion events, indels, leading to a frameshift in gene sequences, resulting in premature stop codons and a protein lacking all functional domains4. Insects also possess the cellular machinery for dsDNA break repair through HDR, which could then be rerouted to design more complex genome editing strategies. HDR-based CRISPR/Cas9 genome editing still needs to be set up for use in sand flies, and should allow the development of reporters of expression and conditional expression mutants, among other constructions.
The development of this microinjection protocol now opens the door to use other genome modification methods, such as CRISPR/Cas9 HDR, transposon transgenesis, binary expression systems such as UAS/Gal4, and site-specific recombination by phiC31 or Cre/lox. The development of these other methods will further expand the toolbox for sand fly gene expression modification by allowing for more complex genomic manipulation. However, before these other methods can be employed, work identifying and isolating regulator elements to drive expression of inserted genes in sand flies will be needed, as well as markers of insertion and other components of these methods, such as promoters to drive expression of transposases and recombinases.
Finally, genome editing in non-model insects has now become a possibility, thanks in particular to the revolution of CRISPR/Cas9 discovery and adaptation15,16. In insects, most genome editing techniques, to the notable exception of the newly developed ReMOT technique, require embryo microinjection, a step both crucial and technically demanding. We hope that this publication will help in expanding the range of gene expression modification techniques adapted to sand flies, and lead to new discoveries on their biology as well as vector competence to Leishmania parasites.
Açıklamalar
The authors have nothing to disclose.
Acknowledgements
The authors thank Vanessa Meldener-Harrell for critical reading of the manuscript.
Materials
| Black Filter Paper 4.25CM PK100 | VWR | 28342-012 | Cut into rectangles that are approximately 46 X 22mm. These are placed between the slide and the coverslip and act as a moist base layer for the embryos during injection. |
| Coverslips | Fisher Scientific | 12-543A | |
| Dissecting Microscope | Any brand | For aligning embryos | |
| Glass slides | Fisher Scientific | 12-550-A3 | Base layer of the microinjection set up Figure 2A |
| Insect cage | custom made or several commercial options | polycarbonate cage for adults holding and mating Lawyer, Phillip, Mireille Killick-Kendrick, Tobin Rowland, Edgar Rowton, and Petr Volf. “Laboratory Colonization and Mass Rearing of Phlebotomine Sand Flies (Diptera, Psychodidae).” Parasite 24. Accessed August 6, 2020. https://doi.org/10.1051/parasite/2017041. | |
| Larval food | custom made | a mix of rabbit chow and rabbit feces Lawyer, Phillip, Mireille Killick-Kendrick, Tobin Rowland, Edgar Rowton, and Petr Volf. “Laboratory Colonization and Mass Rearing of Phlebotomine Sand Flies (Diptera, Psychodidae).” Parasite 24. https://doi.org/10.1051/parasite/2017041. | |
| Microcaps 100 ml | Drummond | 1-000-1000 | Used to back fill microinjection needles |
| Mouth aspirator | John W. Hock Company | Model 612 | mouth aspirator with HEPA filter |
| Olympus SZX12 | Olympus Life Sciences | Microinjection microscope | |
| Ovipots | Nalge company | ovipots are made from 125-ml or 500-ml straigh-sided plolypropylene jars modified by drilling 2.5cm holes in the bottom and filled with 1cm of plaster of Paris. Lawyer, Phillip, Mireille Killick-Kendrick, Tobin Rowland, Edgar Rowton, and Petr Volf. “Laboratory Colonization and Mass Rearing of Phlebotomine Sand Flies (Diptera, Psychodidae).” Parasite 24. Accessed August 6, 2020. https://doi.org/10.1051/parasite/2017041. | |
| Paint Brush 6-0 | Any Art Supply Company | n/a | Used for aligning embryos |
| Propionic acid | Sigma-Aldrich | 402907 | antifungal agent |
| Standard Glass Capillaries | World Precision Instruments | 1B100-3 | Used for making microinjection needles |
| Trio-MPC100 Controller and MP845 Manipulator | Sutter Instruments | Microinjection Controller and Micromanipulator |
Referanslar
- Lawyer, P., Killick-Kendrick, M., Rowland, T., Rowton, E., Volf, P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, Psychodidae). Parasite. 24, 42 (2017).
- Pelletier, I., et al. Specific recognition of Leishmania major poly-beta-galactosyl epitopes by galectin-9: possible implication of galectin-9 in interaction between L. major and host cells. Journal Biological Chemistry. 278 (25), 22223-22230 (2003).
- Kamhawi, S., et al. A role for insect galectins in parasite survival. Cell. 119 (3), 329-341 (2004).
- Louradour, I., Ghosh, K., Inbar, E., Sacks, D. L. CRISPR/Cas9 Mutagenesis in Phlebotomus papatasi: the Immune Deficiency Pathway Impacts Vector Competence for Leishmania major. MBio. 10 (4), (2019).
- Xi, Z., Ramirez, J. L., Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathogens. 4 (7), 1000098 (2008).
- Ramirez, J. L., Dimopoulos, G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Development and Comparative Immunology. 34 (6), 625-629 (2010).
- Ramirez, J. L., et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Neglected Tropical Diseases. 6 (3), 1561 (2012).
- Hu, C., Aksoy, S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Molecular Microbiology. 60 (5), 1194-1204 (2006).
- Meister, S., et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathogens. 5 (8), 1000542 (2009).
- Meister, S., et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences U S A. 102 (32), 11420-11425 (2005).
- Telleria, E. L., et al. Caspar-like gene depletion reduces Leishmania infection in sand fly host Lutzomyia longipalpis. Journal of Biological Chemistry. 287 (16), 12985-12993 (2012).
- Sant’Anna, M. R., Alexander, B., Bates, P. A., Dillon, R. J. Gene silencing in phlebotomine sand flies: Xanthine dehydrogenase knock down by dsRNA microinjections. Insect Biochemistry and Molecular Biology. 38 (6), 652-660 (2008).
- Sant’anna, M. R., Diaz-Albiter, H., Mubaraki, M., Dillon, R. J., Bates, P. A. Inhibition of trypsin expression in Lutzomyia longipalpis using RNAi enhances the survival of Leishmania. Parasites and Vectors. 2 (1), 62 (2009).
- Diaz-Albiter, H., Mitford, R., Genta, F. A., Sant’Anna, M. R., Dillon, R. J. Reactive oxygen species scavenging by catalase is important for female Lutzomyia longipalpis fecundity and mortality. PLoS One. 6 (3), 17486 (2011).
- Barrangou, R., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315 (5819), 1709-1712 (2007).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Pawelczak, K. S., Gavande, N. S., VanderVere-Carozza, P. S., Turchi, J. J. Modulating DNA Repair Pathways to Improve Precision Genome Engineering. ACS Chemical Biology. 13 (2), 389-396 (2018).
- Martin-Martin, I., Aryan, A., Meneses, C., Adelman, Z. N., Calvo, E. Optimization of sand fly embryo microinjection for gene editing by CRISPR/Cas9. PLoS Neglected Tropical Diseases. 12 (9), 0006769 (2018).
- Meuti, M., Harrell, R. Preparing and Injecting Embryos of Culex Mosquitoes to Generate Null Mutations Using CRISPR/Cas9. JoVE. (163), e61651 (2020).
- Chaverra-Rodriguez, D., et al. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nature Communications. 9 (1), 3008 (2018).
- Chaverra-Rodriguez, D., et al. Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Molecular Biology. , (2020).
- Heu, C. C., McCullough, F. M., Luan, J., Rasgon, J. L. CRISPR-Cas9-Based Genome Editing in the Silverleaf Whitefly (Bemisia tabaci). CRISPR Journal. 3 (2), 89-96 (2020).
- Macias, V. M., et al. Cas9-Mediated Gene-Editing in the Malaria Mosquito Anopheles stephensi by ReMOT Control. Genes, Genomes and Genetics (Bethesda). 10 (4), 1353-1360 (2020).

