A Scalable, Cell-Based Method for the Functional Assessment of Ube3a Variants
Summary
A simple and scalable method was developed to assess the functional significance of missense variants in Ube3a, a gene whose loss and gain of function are linked to both Angelman syndrome and autism spectrum disorder.
Abstract
The increased use of sequencing in medicine has identified millions of coding variants in the human genome. Many of these variants occur in genes associated with neurodevelopmental disorders, but the functional significance of the vast majority of variants remains unknown. The present protocol describes the study of variants for Ube3a, a gene that encodes an E3 ubiquitin ligase linked to both autism and Angelman syndrome. Duplication or triplication of Ube3a is strongly linked to autism, whereas its deletion causes Angelman syndrome. Thus, understanding the valence of changes in UBE3A protein activity is important for clinical outcomes. Here, a rapid, cell-based method that pairs Ube3a variants with a Wnt pathway reporter is described. This simple assay is scalable and can be used to determine the valence and magnitude of activity changes in any Ube3a variant. Moreover, the facility of this method allows the generation of a wealth of structure-function information, which can be used to gain deep insights into the enzymatic mechanisms of UBE3A.
Introduction
Recent technological advances have made the sequencing of exomes and genomes routine in clinical settings1,2. This has led to the discovery of a large number of genetic variants, including millions of missense variants that typically change one amino acid in a protein. Understanding the functional significance of these variants remains a challenge, and only a small fraction (~2%) of the known missense variants have a clinical interpretation1,3.
A prominent example of this problem is Ube3a, a gene that encodes an E3 ubiquitin ligase that targets substrate proteins for degradation4. Ube3a resides within chromosome 15q11-13 and is expressed exclusively from the maternally inherited allele5,6,7. Observations from disease genetics strongly suggest that insufficient or excessive activity of the UBE3A enzyme causes distinct neurodevelopmental disorders. Deletion of maternal chromosome 15q11-13 causes Angelman syndrome (AS)8, a disorder characterized by severe intellectual disability, motor impairments, seizures, a happy demeanor with frequent smiling, and dysmorphic facial features8,9,10. In contrast, duplication or triplication of maternal chromosome 15q11-13 causes Dup15q syndrome, a heterogeneous condition recognized as one of the most prevalent syndromic forms of autism11,12,13. In addition, there are hundreds of missense variants identified in Ube3a, the majority of which are considered variants of uncertain significance (VUS) as their functional and clinical significance are unknown. Thus, there is considerable interest in developing methods that can empirically assess Ube3a variants to determine whether they contribute to neurodevelopmental disease.
The UBE3A enzyme belongs to the HECT (homologous to E6-AP C-terminus) domain family of E3 ubiquitin ligases that all possess the eponymous HECT domain, which contains the biochemical machinery necessary to accept activated ubiquitin from E2 enzymes and transfer it to substrate proteins14. Historically, the characterization of E3 enzymes has relied on reconstituted in vitro systems that require an ensemble of purified proteins4,15,16. Such methods are slow and labor-intensive and not amenable to assessing the activity of a large number of variants. In previous work, UBE3A was identified to activate the Wnt pathway in HEK293T cells by modulating the function of the proteasome to slow the degradation of β-catenin17. This insight allows the use of Wnt pathway reporters as an efficient and rapid method to identify both loss- and gain-of-function variants of Ube3a18. The protocol below describes in detail a method to generate Ube3a variants as well as a luciferase-based reporter to assess changes in the activity of Ube3a variants.
Protocol
1. Mutagenesis cloning to generate Ube3a variants
- Clone all Ube3a variants into the pCIG2 plasmid (Figure 1A), a bicistronic vector that contains a chicken-β-actin promoter and an internal ribosome entry site (IRES) for EGFP expression19. Full-length Ube3a constructs contain an N-terminal Myc-tag sequence and are cloned into pCIG2 using a 5' SacI site and a 3' XmaI site. In addition, naturally occurring EcoRI, EcoRV, and PstI sites within the Ube3a coding sequence are also used to subclone fragments.
NOTE: De-identified variants for Ube3a can be obtained through the literature and in publicly accessible repositories such as the ClinVar database1. Additional unreported variants can be obtained through personal communication with medical centers. - Use an adapted two-step megaprimer mutagenesis method20 to introduce mutations into the Ube3a reading frame. In the first step, design the mutagenic oligonucleotide that contains the desired mutation (the example shown in this protocol is to generate a UBE3A Q588E variant) flanked by at least 30 base pairs (bp) of homology 5′ and 3′ to the mutation (Figure 1B and Table 1).
- Use the mutagenic oligonucleotide along with a complementary oligonucleotide for the first round PCR to generate a 200-400 bp megaprimer containing the mutation (Figure 1C; Table 2 and Table 3). Use the entire 50 μL of the PCR reaction volume for agarose gel electrophoresis using a 1% gel and subsequent gel purification (Table of Materials). Elute the PCR product in 30 μL of deionized H2O (diH2O) for use in the second round PCR step below.
- Set up the second round PCR as indicated (Figure 1C; Table 2 and Table 3). This step will generate larger DNA fragments (typically ~1-1.5 kb in length) containing the mutation and terminal restriction sites suitable for sub-cloning (Figure 1C).
- After completion of the PCR reaction, purify the resulting product using a PCR purification kit (Table of Materials). Elute in 30 μL and digest all of the purified product and the pCIG2 Ube3a WT construct with the appropriate restriction enzymes (the example shows digestion with EcoRI and XmaI).
- After 2 h of digestion at 37 °C, resolve the digestion products through agarose gel electrophoresis (1% gel), and gel-purify the appropriate products. Set up ligations according to the manufacturer's protocol (Table of Materials), transform the ligation products into a DH10B bacterial strain (Table of Materials), and then plate with ampicillin (100 μg/mL) selection.
- The next day, pick two to four colonies to inoculate 3 mL cultures containing Luria Broth (LB) and 100 μg/mL ampicillin. Let the cultures grow overnight at 37 °C in an orbital incubator with shaking (225 rpm).
- The following day, use 1-1.5 mL of the bacterial culture to miniprep the plasmid DNA (Table of Materials). Save the remaining culture by placing them at 4 °C. Screen the miniprepped plasmids by Sanger sequencing using an oligonucleotide that binds ~200 bp from the desired mutation.
- Use the bacterial cultures saved at 4 °C to re-inoculate 50 mL cultures to midiprep the correct plasmid (Table of Materials). Fully sequence the Ube3a coding sequence to validate the correct sequence of the DNA.
- Create working stocks of Ube3a variant plasmids, as well as the β-catenin activated reporter (BAR)21, TK-Renilla luciferase, ligase-dead Ube3a (UBE3A C820A mutation), and pCIG2 empty vector at a concentration of 100 ng/μL using sterile H2O for the subsequent transfection steps.
2. Preparation and transfection of human embryonic kidney 293T (HEK293T) cells
- Perform the assays in HEK293T cells grown in Dulbecco's modified Eagle medium (DMEM) pre-supplemented with glutamine, 10% fetal bovine serum (FBS), and antibiotic-antimycotic agent as described previously17. Grow the cells in a sterile, humidified incubator at 37 °C with 5% CO2.
- Using a biosafety cabinet, first plate the suspended HEK293T cells onto tissue culture-treated 96-well flat-bottomed plates at a density of 2.2 × 104 cells per well in 100 μL of culture media and allow them to grow overnight.
- On the second day, transfect the cells in a biosafety cabinet with Firefly and Renilla luciferase reporter plasmids (Table 4) and Ube3a plasmids in triplicate. Perform the transfections in 10 μL of total volume per well using a 10:1:8 ratio of plasmids (50 ng of BAR, 5 ng of TK-Renilla, and 40 ng of Ube3a plasmid per well, respectively), as previously described18, and 0.4 μL of transfection reagent (Table 4).
NOTE: For each experiment, an empty vector (GFP only) and a plasmid encoding ligase-dead Ube3a are also transfected to serve as negative controls. - Create a master mix for the transfections for each variant in a 1.5 mL microcentrifuge tube (Table 4) and incubate at room temperature (RT) for 15 min. After the incubation, gently agitate the transfection mixture by tapping the tube, and then add 10 μL of the transfection mixture directly to the existing growth media in the wells.
- Afterward, return the cells to the incubator and allow the plasmids to express for 48 h. Replacement of the growth media is not necessary.
- Monitor the transfection efficiency by GFP fluorescence using a fluorescence microscope. HEK293T cells are efficiently transfected by this method, and >80% of cells should express GFP after 48 h.
3. Measurement of luciferase activity
NOTE: Luciferase activity is assessed using a commercially available system that assays both Firefly and Renilla luciferase (Table of Materials) according to the manufacturer's protocol.
- Carefully aspirate the culture media from transfected HEK293T cells, and then wash the cells with cold phosphate-buffered saline (PBS). Aspirate the PBS and lyse the cells by adding 25 μL of non-denaturing lysis buffer and incubate for 15 min with gentle rocking on ice.
- Afterward, add 20 μL of the resulting lysate (no centrifugation is necessary) into a new 96-well assay plate containing 100 μL of a Firefly luciferase substrate reagent. Mix the plate gently by tapping.
- Load the plate onto a plate reader and measure the luminescence using a top detector with a read height of 1 mm and an integration time of 1 s.
NOTE: The gain of the detector may need to be adjusted based on the strength of the signal. - Immediately after reading the Firefly luciferase luminescence, add 100 µL of Renilla luciferase substrate to the wells. This step simultaneously quenches the Firefly luciferase activity while assessing Renilla luciferase activity. Mix the samples by gentle agitation of the plate and measure the luminescence again to assess the Renilla luciferase activity.
NOTE: While other plates can be used for this assay, using plates with black frames and white wells will provide the most sensitive results as this amplifies the luciferase signal while minimizing noise. - Quantify the reporter responses as the Firefly luciferase to Renilla luciferase ratio (Firefly/Renilla), and average the values of the triplicate measurements. Afterward, normalize the Firefly/Renilla value for each variant against the values for wild-type (WT) Ube3a expressing cells to compare the relative activity of variant proteins.
Representative Results
Large-scale functional screening of Ube3a missense variants identifies a broad landscape of loss- and gain-of-function mutations
Previous work with Ube3a mutants suggested that the Wnt response can serve as a reporter of cellular UBE3A protein activity. These observations were expanded, and additional validation experiments were performed to investigate whether the BAR assay is suitable to report a range of UBE3A activities in the cell. First, HEK293T cells were transfected with varying amounts of plasmid DNA encoding human WT Ube3a. This experiment demonstrated that the BAR response changes linearly with the quantity of Ube3a transfected into cells (Figure 2A). Secondly, the BAR assay was tested using plasmid DNA encoding disease-linked variants previously characterized in the literature22. Among these were benign variants (R39H and A178T), one autism-linked gain-of-function variant (T458A), and a collection of confirmed Angelman syndrome-linked variants that cause a loss of UBE3A function through various mechanisms15,22. The BAR assay correctly identified each variant (Figure 2B), including all loss-of-function variants, demonstrating the accuracy and generality of this method18.
The BAR assay was used to screen 152 missense variants, most of which were identified from the ClinVar database. Similar to genetic evidence regarding copy number variations in Ube3a-dependent disorders, the functional impact of missense variants was broad and included benign variants as well as both loss-of-function and gain-of-function variants (Figure 3)18. Of particular interest, the gain-of-function variants identified using this method were all novel, and subsequent validation strongly suggested that they define a sub-class of Ube3a-dependent disorders with phenotypes that differ from classic Angelman syndrome18.
The effect of missense variants is often unpredictable, underscoring the need for functional assessment
An intriguing observation from the functional screening of Ube3a variants is that changes at some amino acid positions produce drastically different effects, thereby underscoring the importance of empirical variant assessment18. For example, three individuals were identified with variants at the glutamine 588 (Q588) position in UBE3A that included a gain-of-function glutamate substitution (Q588E), a loss-of-function arginine substitution (Q588R), and another loss-of-function proline substitution (Q588P; Figure 4A). A comprehensive mutation approach was used to mutate the Q588 position to every possible amino acid. When the activity of these variants was measured using the BAR assay, it showed that any negatively charged amino acid at the 588 position produces a hyperactive enzyme (Figure 4B). This insight provided important functional clues that allowed the discovery of a novel site within the HECT domain of UBE3A that facilitates ubiquitin chain elongation18.
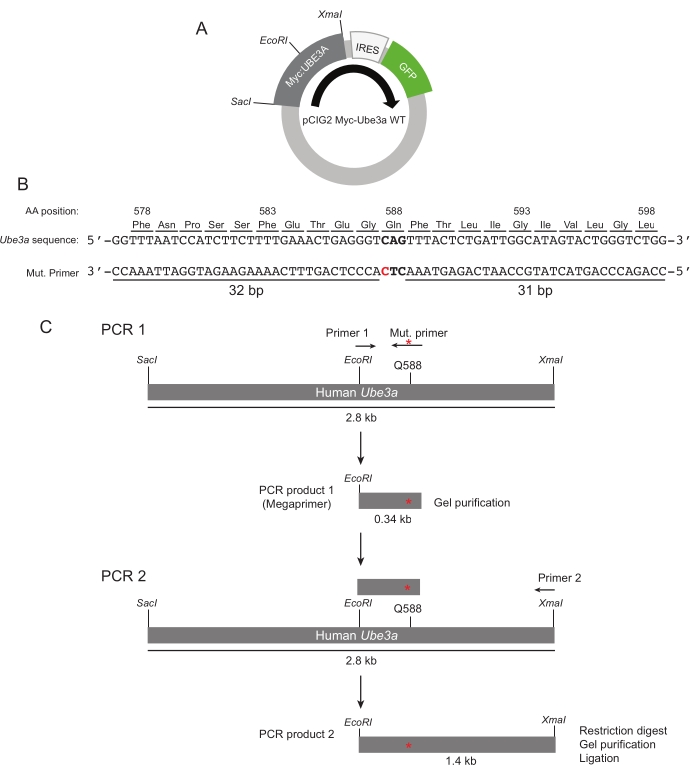
Figure 1: PCR-dependent mutagenesis of Ube3a. (A) Representation of the Ube3a expression vector indicating relevant restriction sites. (B) The DNA sequence encoding UBE3A is shown at the top. The Q588 codon is indicated by bold lettering. The Q588E mutagenesis primer (Mut. primer) is shown aligned to the UBE3A sequence. The red lettering indicates the mutagenized site. (C) A schematic of the PCR to generate the megaprimer and the DNA fragment used to subclone the Q588E fragment is shown. The position of the Q588E mutation is indicated by the red asterisk. Please click here to view a larger version of this figure.
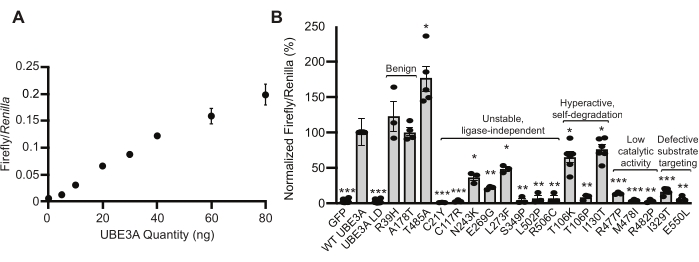
Figure 2: Validation of the BAR assay as a reporter of UBE3A ubiquitin ligase activity. (A) HEK293T cells transfected with increasing amounts of plasmid encoding WT UBE3A show a linear increase in the BAR response. Mean values are shown for Firefly/Renilla luciferase ratios ± standard error (SE). N= 3 independent experiments. (B) Previously characterized Ube3a variants were tested in the BAR assay. Responses were normalized to WT UBE3A, and the values are shown as the mean ± SE. The number of experiments (n) and p-values calculated using a one-sample t-test (two-tailed) with Benjamini-Hochberg multiple comparisons correction (FDR = 0.05) are as follows: GFP, n = 19, ***p = 1.733 × 10−31; WT UBE3A, n = 19; UBE3A LD, n = 19, ***p = 1.519 × 10−31; R39H, n = 3, p = 0.567; A178T, n = 4, p = 0.828; T485A, n = 5, *p = 0.019; C21Y, n = 3, ***p = 8.725 × 10−6; C117R, n = 3, ***p = 3.419 × 10−4; N243K, n = 3, **p = 0.0072; E269G, n = 3, **p = 7.787 × 10−4; L273F, n = 3, **p = 0.0052; S349P, n = 3, **p = 0.0016; L502P, n = 3, **p = 0.0033; R506C, n = 3, **p = 0.0033; T106K, n = 6, **p = 0.0.0078; T106P, n = 3, **p = 0.0013; I130T, n = 6, *p = 0.016; R477P, n = 3, ***p = 4.24 × 10−4; M478I, n = 3, ***p = 3.39 × 10−4; R482P, n = 3, **p = 5.82 × 10−4; I329T, n = 5, ***p = 1.22 × 10−5; E550L, n = 3, *p = 0.0013. Data reprinted with permission from Weston et al.18. Please click here to view a larger version of this figure.
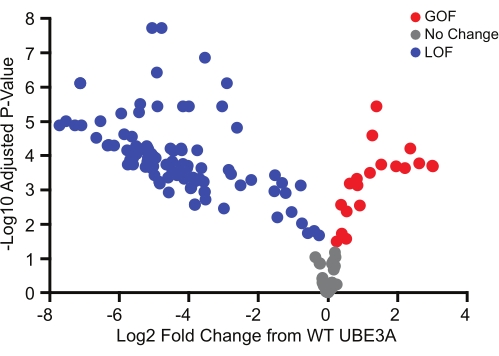
Figure 3: UBE3A variants encompassing a wide range of functional effects. BAR assay screen of 152 Ube3a variants showing benign (gray), loss-of-function (blue), and gain-of-function (red) mutations relative to WT UBE3A. Significance was determined using a one-sample t-test (two-tailed) with Benjamini-Hochberg multiple comparisons correction (FDR = 0.05). Red, gain-of-function; Gray, no change from WT UBE3A; Blue, loss-of-function. Data reprinted with permission from Weston et al.18. Please click here to view a larger version of this figure.
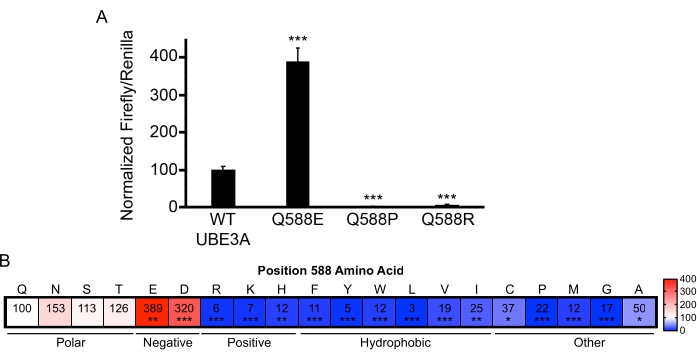
Figure 4: UBE3A variants encompassing a wide range of functional effects. (A) BAR assay results of variants at position 588 of UBE3A showing both loss- and gain-of-function substitutions. Q588E, n = 6; Q588P, n = 3; Q588R, n = 4; ***p < 0.0005, One-sample t-test (two-tailed) with Benjamini-Hochberg multiple comparison correction (FDR = 0.05). (B) Heat plot showing normalized BAR results for comprehensive mutational analysis at position 588 in UBE3A. The white shading represents WT UBE3A activity levels, the blue shading indicates loss-of-function, and the red shading indicates gain-of-function. The scale bar shows the percent change relative to WT UBE3A. N = 3 independent experiments for Q588S, Q588T, Q588N, Q588K, Q588P, Q588M, Q588G, Q588A, Q588F, Q588Y, Q588W, Q588L, Q588V, Q588I, Q588C; n = 8 for Q588E, n = 6 for Q588D, n = 5 for Q588R, n = 4 for Q588H, *p < 0.05, **p < 0.005, ***p < 0.0005. One-sample t-test (two-tailed) with Benjamini-Hochberg multiple comparisons correction (FDR = 0.05). Data reprinted with permission from Weston et al.18. Please click here to view a larger version of this figure.
| Name | Sequence | Used for | |
| Primer 1 | GATTATATTATGACAATAGAA TTCGCATGTACAGTGAAC |
Ube3a EcoRI sense | |
| Mut. Primer | CCAGACCCAGTACTATGCCAAT CAGAGTAAACTCACCCTCAGTT TCAAAAGAAGATGGATTAAACC |
UBE3A Q588E mutagenesis antisense | |
| Primer 2 | ATTATATTCCCGGGTTACAGCAT GCCAAATCCTTTGG |
Ube3a XmaI antisense | |
Table 1: Primers used for UBE3A Q588E mutagenesis.
| 1st Round PCR | |
| Reagent | Reaction Quantity |
| 5x high-fidelity buffer | 10 µL |
| dNTP (10 mM stock) | 1 µL |
| Primer 1 (10 mM stock) | 1 µL |
| Primer 2 (10 mM stock) | 1 µL |
| WT-Ube3a DNA (150 ng/µL stock) | 1 µL |
| H2O | 35 µL |
| High-fidelity DNA polymerase | 1 µL |
| Total Volume | 50 µL |
| 2nd Round PCR | |
| Reagent | Reaction Quantity |
| 5x high-fidelity buffer | 10 µL |
| dNTP (10 mM stock) | 1 µL |
| Primer 3 (10 mM stock) | 1 µL |
| Purified PCR product (Megaprimer) | 30 µL |
| WT-Ube3a DNA (150 ng/µL stock) | 1 µL |
| H2O | 6 µL |
| High-fidelity DNA polymerase | 1 µL |
| Total Volume | 50 µL |
Table 2: PCR parameters for mutagenesis.
| 1st Round PCR | ||
| Step | Temperature | Time |
| Initial denaturation | 95 °C | 2 min |
| (1 cycle) | ||
| Denaturation | 95 °C | 30 s |
| Annealing | 50 °C | 20 s |
| Extension | 72 °C | 15 s |
| (30 cycles) | ||
| Final extension | 72 °C | 1 min |
| Hold | 4 °C | Infinite |
| 2nd Round PCR | ||
| Step | Temperature | Time |
| Initial denaturation | 95 °C | 2 min |
| (1 cycle) | ||
| Denaturation | 95 °C | 30 s |
| Annealing | 50 °C | 20 s |
| Extension | 72 °C | 35 s |
| (30 cycles) | ||
| Final extension | 72 °C | 1 min |
| Hold | 4 °C | Infinite |
Table 3: PCR Program Parameters for Mutagenesis.
| Reagent | Working concentration | 1x transfection | 3.5x master mix |
| pGL3 BAR plasmid | 100 ng/µL | 0.5 µL | 1.75 µL |
| pTK Renilla plasmid | 100 ng/µL | 0.05 µL | 0.175 µL |
| Ube3a plasmid | 100 ng/µL | 0.4 µL | 1.4 µL |
| DMEM supplemented with glutamine | 8.65 µL | 30.275 µL | |
| Transfection reagent | 0.4 µL | 1.4 µL |
Table 4: Transfection mixtures.
Discussion
The protocol described here provides an efficient and scalable method to assess the enzymatic activity of Ube3a variants. There are several technical details that warrant careful consideration when using this assay. One consideration is the choice of Wnt reporter plasmids used in this assay. The protocol described here specifically uses the β-catenin activated reporter (BAR)21, a reporter that contains a concatemer of 12 T-cell factor (TCF) response elements separated by specifically designed linker sequences to minimize recombination and loss of TCF-binding sites. There are additional luciferase-based Wnt reporter plasmids described in the literature23, and although previous studies indicate that some can be used to assess UBE3A activity, the BAR plasmid has been most thoroughly characterized for this purpose17,18. Secondly, the choice of Renilla luciferase plasmid is critical. A plasmid encoding Renilla luciferase is included during the transfection step to normalize the transfection efficiency. The plasmid used in this protocol constitutively expresses Renilla luciferase under the control of a thymidine kinase (TK) promoter (TK-Renilla). The use of plasmids containing other promoters, such as the widely used cytomegalovirus (CMV) promoter, may lead to variable outcomes in Renilla luciferase expression.
A second consideration is the behavior of cells depending on the type and lot of FBS used for cell culture. For instance, FBS can variably affect the growth rate of HEK293T cells. The current protocol was optimized with cells exhibiting a typical doubling time of 18-24 h, making the cell density at the time of transfection approximately ~4 × 104 cells per well. As cell density may affect transfection efficiencies, the cell growth rates should be carefully assessed by the user and the cell numbers adjusted accordingly at the time of plating. Additionally, the UBE3A-dependent BAR response requires some amount of Wnt ligand to be present during the experiment. In most standard conditions, there is enough Wnt in the FBS to drive this response; however, this response can also vary. The titration of plasmid ratios used for transfection is recommended to ensure the BAR response is measured within the appropriate linear range. An alternative approach is to stimulate Wnt signaling using a recombinant Wnt ligand or Wnt-supplemented growth media17.
An advantage of the BAR assay is that it can report the ubiquitin ligase activity of all Ube3a isoforms. Humans express three isoforms of Ube3a that differ at their extreme N-termini due to the use of alternative transcriptional start sites24. The BAR assay provides the same readout agnostic to isoforms, and thus, this assay can be used to assess all Ube3a variants. In addition, the large-scale characterization of Ube3a variants provides deep structure-function data that can uncover novel domains and mechanisms that are critical for enzyme function. A previous study utilized variant data to discover a ubiquitin-binding domain within UBE3A that facilitates ubiquitin chain elongation18. Additional structure-function studies will likely provide new insights into the biochemical mechanisms of UBE3A that may be leveraged for therapeutic development.
There are some limitations to the BAR assay that warrant careful consideration when determining the pathogenicity of a variant. Due to the epigenetic imprinting of Ube3a in neurons, this assay cannot discriminate maternal and paternal alleles, and additional genetic evidence must be weighed alongside functional data to determine the pathogenicity of a variant. Moreover, additional variables beyond the ubiquitin ligase activity of UBE3A must be considered in the context of disease. For example, previous work showed that nuclear-localized UBE3A contributes to Angelman syndrome pathology25 and that some variants perturb this localization pattern of the enzyme26. Thus, additional downstream characterizations must be performed to gain a complete understanding of the contribution of Ube3a variants to neurodevelopmental pathology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a Simons Foundation Bridge to Independence Award (SFARI Award #387972; J.J.Y.), a NARSAD Young Investigator Award from the Brain and Behavior Research Foundation (J.J.Y.), a Research Fellowship from the Alfred P. Sloan Foundation (J.J.Y.), and research grants from the Angelman Syndrome Foundation (J.J.Y.), the Whitehall Foundation (J.J.Y.), and the NIMH (R01MH122786; J.J.Y.).
Materials
| 0.05% Trypsin-EDTA (1x), phenol red | Gibco | 25300-054 | |
| 1 Kb DNA ladder | Lambda Biotech | M108-S | |
| 100 bp DNA Ladder | Lambda Biotech | M107 | |
| 10x Buffer for T4 DNA Ligase with 10 mM ATP | New England BioLabs | B0202A | |
| 5x Phusion HF Reaction Buffer | New England BioLabs | B0518S | |
| Antibiotic-Antimycotic Solution | Corning | 30004CI | |
| Black/White Isoplate-96 Black Frame White Well plate | PerkinElmer | 6005030 | |
| Carbenicillin Disodium Salt | Midwest Scientific | KCC46000-5 | |
| Countess cell counting chamber slides | Invitrogen by Thermo Fisher Scientific | C10283 | |
| Countess II Automated Cell Counter | life technologies | Cell counting machine | |
| Custom DNA oligos | Integrated DNA Technologies (IDT) | ||
| Deoxynucleotide (dNTP) Solution Mix | New England BioLabs | N0447S | |
| DMEM, high glucose, GlutaMAX Supplement, pyruvate | Gibco | 10569044 | Basal medium for supporting the growth of HEK293T cell line |
| DPBS (1x) | Gibco | 14190-136 | |
| Dual-Luciferase Reporter Assay System | Promega | E1910 | |
| EcoRI-HF | New England BioLabs | R3101S | Restriction enzyme |
| Fetal Bovine Serum, qualified, heat inactivated | Gibco | 16140071 | Fetal bovine serum |
| Fisherbrand Surface Treated Tissue Culture Dishes | Fisherbrand | FB012924 | |
| FuGENE 6 Transfection Reagent | Promega | E2691 | |
| Gel Loading Dye Purple (6x) | New England BioLabs | B7024A | |
| HEK293T cells | ATCC | CRL-3216 | |
| High Efficiency ig 10B Chemically Competent Cells | Intact Genomics | 1011-12 | E. coli DH10B cells |
| HiSpeed Plasmid Midi Kit | Qiagen | 12643 | Midi prep |
| pCIG2 plasmid | |||
| pGL3 BAR plasmid | |||
| Phusion HF DNA Polymerase | New England BioLabs | M0530L | DNA polymerase |
| ProFlex 3 x 32 well PCR System | Applied biosystems by life technologies | Thermocycler | |
| pTK Renilla plasmid | |||
| QIAprep Spin Miniprep Kit (250) | Qiagen | 27106 | Mini prep |
| QIAquick Gel Extraction Kit (250) | Qiagen | 28706 | Gel purification |
| QIAquick PCR Purification Kit (250) | Qiagen | 28106 | PCR purification |
| rCutSmart Buffer | New England BioLabs | B6004S | |
| SacI-HF | New England BioLabs | R3156S | Restriction enzyme |
| Synergy HTX Multi-Mode Reader | BioTek | Plate reader runs Gen5 software v3.08 (BioTek) | |
| T4 DNA Ligase | New England BioLabs | M0202L | Ligase |
| TAE Buffer, Tris-Acetate-EDTA, 50x Solution, Electrophoresis | Fisher Scientific | BP13324 | |
| Tissue Culture Plate 96 wells, Flat Bottom | Fisherbrand | FB012931 | |
| UltraPure Ethidium Bromide Solution | Invitrogen by Thermo Fisher Scientific | 15585011 | |
| XmaI | New England BioLabs | R0180S | Restriction enzyme |
References
- Landrum, M. J., et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research. 42, 980-985 (2014).
- Lek, M., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536 (7616), 285-291 (2016).
- Starita, L. M., et al. Variant interpretation: Functional assays to the rescue. American Journal of Human Genetics. 101 (3), 315-325 (2017).
- Scheffner, M., Huibregtse, J. M., Vierstra, R. D., Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 75 (3), 495-505 (1993).
- Albrecht, U., et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature Genetics. 17 (1), 75-78 (1997).
- Rougeulle, C., Glatt, H., Lalande, M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nature Genetics. 17 (1), 14-15 (1997).
- Vu, T. H., Hoffman, A. R. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nature Genetics. 17 (1), 12-13 (1997).
- Kishino, T., Lalande, M., Wagstaff, J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature Genetics. 15 (1), 70-73 (1997).
- Jiang, Y. H., et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 21 (4), 799-811 (1998).
- Mabb, A. M., Judson, M. C., Zylka, M. J., Philpot, B. D. Angelman syndrome: Insights into genomic imprinting and neurodevelopmental phenotypes. Trends in Neuroscience. 34 (6), 293-303 (2011).
- Hogart, A., Wu, D., LaSalle, J. M., Schanen, N. C. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiology of Disease. 38 (2), 181-191 (2010).
- Urraca, N., et al. The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Research. 6 (4), 268-279 (2013).
- de la Torre-Ubieta, L., Won, H., Stein, J. L., Geschwind, D. H. Advancing the understanding of autism disease mechanisms through genetics. Nature Medicine. 22 (4), 345-361 (2016).
- Scheffner, M., Staub, O. HECT E3s and human disease. BMC Biochemistry. 8, (2007).
- Cooper, E. M., Hudson, A. W., Amos, J., Wagstaff, J., Howley, P. M. Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. Journal of Biological Chemistry. 279 (39), 41208-41217 (2004).
- Yi, J. J., Barnes, A. P., Hand, R., Polleux, F., Ehlers, M. D. TGF-beta signaling specifies axons during brain development. Cell. 142 (1), 144-157 (2010).
- Yi, J. J., et al. The autism-linked UBE3A T485A mutant E3 ubiquitin ligase activates the Wnt/beta-catenin pathway by inhibiting the proteasome. Journal of Biological Chemistry. 292 (30), 12503-12515 (2017).
- Weston, K. P., et al. Identification of disease-linked hyperactivating mutations in UBE3A through large-scale functional variant analysis. Nature Communications. 12 (1), 6809 (2021).
- Hand, R., Polleux, F. Neurogenin2 regulates the initial axon guidance of cortical pyramidal neurons projecting medially to the corpus callosum. Neural Development. 6, 30 (2011).
- Karginov, A. V., Ding, F., Kota, P., Dokholyan, N. V., Hahn, K. M. Engineered allosteric activation of kinases in living cells. Nature Biotechnology. 28 (7), 743-747 (2010).
- Biechele, T. L., Moon, R. T. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods in Molecular Biology. , 99-110 (2008).
- Yi, J. J., et al. An Autism-linked mutation disables phosphorylation control of UBE3A. Cell. 162 (4), 795-807 (2015).
- Kuhnle, S., et al. Angelman syndrome-associated point mutations in the Zn(2+)-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. Journal of Biological Chemistry. 293 (47), 18387-18399 (2018).
- Yamamoto, Y., Huibregtse, J. M., Howley, P. M. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 41 (2), 263-266 (1997).
- Avagliano Trezza, R., et al. Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome. Nature Neuroscience. 22 (8), 1235-1247 (2019).
- Bossuyt, S. N. V., et al. Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations. Human Molecular Genetics. 30 (6), 430-442 (2021).

