Preparation of Rat Oligodendrocyte Progenitor Cultures and Quantification of Oligodendrogenesis Using Dual-infrared Fluorescence Scanning
Summary
Oligodendrocytes are the myelinating cells of the central nervous system. This protocol describes a method for the isolation and culture of their precursors, oligodendrocyte progenitor cells, from rat cortices, as well as a fast and reliable quantitative method to evaluate oligodendrogenesis in vitro in response to experimental factors.
Abstract
Efficient oligodendrogenesis is the therapeutic goal of a number of areas of research including spinal cord injury, neonatal hypoxia, and demyelinating diseases such as multiple sclerosis and transverse myelitis. Myelination is required to not only facilitate rapid impulse propagation within the central nervous system, but also to provide trophic support to underlying axons. Oligodendrocyte progenitor cells (OPCs) can be studied in vitro to help identify factors that may promote or inhibit oligodendrocyte differentiation. To date, many of the methods available to evaluate this process have either required large numbers of cells, thus limiting the number of conditions that can be investigated at any one time, or labor-intensive methods of quantification. Herein, we describe a protocol for the isolation of large numbers of highly pure OPCs together with a fast and reliable method to determine oligodendrogenesis from multiple conditions simultaneously. OPCs are isolated from P5-P7 neonatal rat cortices and grown in vitro for three days prior to differentiation. Four days after differentiation, oligodendrogenesis is evaluated using a dual-infrared fluorescence-scanning assay to determine expression of the myelin protein.
Introduction
Efficient nerve conduction in the mammalian central nervous system (CNS) requires myelination of axons by oligodendrocytes. During development, oligodendrocytes arise from a pool of oligodendrocyte progenitor cells (OPCs) that are thought to migrate from the ventricular zones of the developing forebrain and neural tube1. After migration OPCs differentiate into mature, myelinating oligodendrocytes that not only facilitate efficient impulse propagation, but also provide axons with trophic support2. The adult CNS maintains an abundant population of OPCs that are dispersed throughout the gray and white matter comprising approximately 5-8% of all cells3. Following a demyelinating insult, OPCs migrate to the site of injury, proliferate, and differentiate to replace lost or damaged myelin sheaths on exposed axons. However, in some disease/injury settings, this process is found to be inefficient or can fail completely4. While chronic demyelination is thought to add to the burden of disease, efficient oligodendrogenesis and remyelination may alleviate symptoms5. It has therefore been of interest to study OPCs in vitro to determine the effect of experimental factors on oligodendrogenesis.
Insight into the different phases of oligodendrocyte differentiation has been made possible by the identification of stage specific cell markers. Self-renewing early progenitor cells are defined by their expression of platelet derived growth factor receptor alpha (PDGFRα), neural/glia antigen 2 (NG2) and A2B56–8. As OPCs initiate their differentiation program and withdraw from the cell cycle, they downregulate expression of these markers and begin to express proteins indicative of premyelinating oligodendrocytes including Cyclic-nucleotide 3'-phosphodiesterase (CNPase) and O4. Finally, their differentiation into more mature oligodendrocytes is characterized by the expression of myelin-associated proteins, including myelin-associated glycoprotein (MAG), proteolipid protein (PLP), and myelin basic protein (MBP). MBP is an intracellular peripheral membrane protein and a major component of the myelin sheath. Mice lacking MBP develop a severe phenotype in which CNS myelination is significantly decreased leading to tremors, convulsions and early death9,10. This important role of MBP in myelination has led to its use as a marker of oligodendrocyte differentiation both in vitro and in vivo11.
Quantification of MBP can be achieved using several different methodologies. RT-PCR and Western blot analysis allow for quantification of MBP levels under different treatment regimens. Immunocytochemistry is a common qualitative approach that can also be quantitative when camera-based microscopy approaches are used. Although these systems are reliable and fundamental to the study of oligodendrocyte differentiation, they each have their own disadvantages that limit their use in drug screening. First, the amount of primary OPCs that are needed for RT-PCR and Western blot analysis reduces the number of variables that can be examined simultaneously. While the cell requirement for immunocytochemistry is much lower, substantial time must be devoted to each experiment if quantification is the goal. Numerous images must be captured and then quantified to facilitate experimental analysis. These caveats become important obstacles to consider for studies that require high throughput assessment. Here we describe a method that utilizes fundamental aspects from both the immunocytochemistry and Western blot methodologies for myelin quantification, while significantly reducing both the number of cells required and time to complete quantitative analysis.
Protocol
Procedures involving animal subjects have been approved by Institutional Animal Care and Use Committee (IACUC) and Johns Hopkins School of Medicine.
1. Preparation of Assay Plates, Stock Solutions, and OPC Base Media
Note: The rat OPC base (Sato) media described here has been derived from previously published studies12,13. Alternative media formulations may also be compatible with this procedure.
- Resuspend poly-L-lysine (PLL) to a concentration of 10 µg/ml in sterile deionized water. Pipette 400 μl of the diluted PLL into each well of a black-walled, clear bottom 24 well tissue culture grade dish.
- Coat tissue culture dishes for 2 hr in a 37 °C tissue culture incubator or overnight in a 4 °C refrigerator.
- Aspirate PLL from coated dishes at least 20 min prior to plating. Allow the remaining PLL to dry from the well by placing 24 well plates with the lid partially ajar in a tissue culture hood. Verify that the wells are completely dry before plating isolated OPCs. Cells will not adhere to plastic that has liquid PLL present.
- Prepare N-Acetyl-L-cysteine (NAC) stock solution (1,000x): Dissolve 100 mg N-Acetyl-L-cysteine (NAC) in 20 ml of Dulbecco's modified Eagle's medium (DMEM). Aliquot and store at -20 °C.
- Prepare Hydrocortisone stock solution (1,000x): Add 1 ml of ethanol to 1 mg Hydrocortisone and swirl to dissolve. Add 19 ml of DMEM, mix the solution, aliquot and store at -20 °C. The addition of hydrocortisone to the base media has been shown to enhance the survival of glial cells14.
- Prepare d-Biotin stock solution (5,000x): Dissolve 2.5 mg of d-biotin in 50 ml of PBS. Add 1-2 x 5 µl drops of 0.1 N NaOH to aid in dissolution. Aliquot and store at -20 °C.
- Prepare Insulin stock solution (100x): Dissolve 25 mg Insulin in 50 ml of tissue culture grade water. Add 250 µl of 1 N HCl and mix until solution is clear. Sterilize with a 0.22 µm filter and store at 4 °C for up to 6 weeks.
- Prepare Sato stock solution (100x):
- Prepare progesterone stock solution (25 µg/µl): Dissolve 2.5 mg progesterone in 100 µl of ethanol.
- Prepare sodium selenite stock solution (400 ng/µl): Dissolve 4 mg sodium selenite in 100 µl of 0.1 N NaOH. Add 10 ml of DMEM and mix.
- To 100 ml of DMEM, add 1 g of human apo-Transferrin, 1 g of bovine serum albumin, and 160 mg of putrescine and mix to fully dissolve. Add 25 µl of progesterone stock solution, 1,000 µl of sodium selenite stock solution, and mix. Aliquot and store at -20 °C.
- Prepare OPC Base Media: per 100 ml of DMEM (with 4.5 g/L D-glucose, L-glutamine, and 110 mg/L sodium pyruvate), add 100 µl NAC, 100 µl hydrocortisone, 20 µl d-Biotin, 1 ml insulin, 1 ml Sato stock, 100 µl trace elements B, 2 mL B-27 supplement (50x), and 1 ml of penicillin/streptomycin (100x). Sterilize with a 0.22 µm filter and store at 4 °C.
- Prepare PDGF-AA stock solution (1,000x): Dissolve 250 μg in 12.5 ml of PBS with 0.1% bovine serum albumin (BSA) to make a 20 μg/ml solution. Aliquot and store at -80 °C.
- Prepare OPC proliferation media: OPC base media supplemented with 20 ng/ml PDGF-AA.
- Prepare OPC differentiation media: OPC base media supplemented with 45 nM triiodothyronine.
- Prepare Column Buffer: To 500 ml of PBS, add 2.5 g of BSA and 2 ml of 0.5 M ethylenediaminetetraacetic acid (EDTA) and adjust the pH to 7.2. Sterilize with a 0.22 µm filter and store at 4 °C.
2. Rat Brain Dissection
Note: P5-P7 rat pups are used in this protocol. Each rat pup cortex yields approximately 1.5-2.0 x 106 A2B5 positive cells.
- Sterilize all dissection equipment using a 250 °C heated bead sterilizer.
- Add 15-20 ml of PBS (without Ca2+ and Mg2+) to 50 ml conical tubes. One 50 ml conical tube is sufficient for 3-4 rat pup cortices. Place 50 ml conical tubes on ice. Use 50 ml conical tubes to hold diced cortex tissue during dissection.
- Add cold PBS to a 10 cm Petri dish. This dish will be used for dissection under the light microscope.
- Sacrifice rat pups by decapitation with large surgical scissors. Spray head with 70% ethanol.
- Extract the Whole Brain
- Using small scissors cut the skin down the midline and behind the ears and peel skin flaps back. Cut the cranium down the midline starting from the brainstem and ending at the eyes. Be careful not to cut too deeply in order to preserve the structure of the underlying cortex.
- Make two lateral cuts inferior to the cerebellum by inserting small surgical scissors into the foramen magnum. Make an additional cut between the eyes, anterior to the olfactory bulbs.
- Carefully peel each half of the cranium back. Remove the whole brain and place in the 10 cm Petri dish filled with cold PBS.
- Under the dissection light microscope, remove the olfactory bulbs and cerebellum with fine surgical forceps.
- Flip the brain so that the ventral surface is visible under the light microscope.
- Using fine straight forceps perform a blunt dissection by placing closed forceps tips between the cortex and hypothalamus to a depth about 2/3 of the brain. Once forceps are in place allow the ends to open. Repeat for the other hemisphere.
- Tease the cortex from the hypothalamus and midbrain regions.
- Remove the hypothalamus, thalamus, and midbrain by holding the midbrain at its posterior surface and peeling it towards the anterior end of the brain. Sever the anterior connections.
- Remove the hippocampus by pealing it outward and then severing its connection to the cortex using fine bent dissection forceps.
- Remove remaining striatum by scrapping it from the underlying cortex in an outward diagonal motion using fine bent dissection forceps.
- Clear any blood vessels and meninges from the ventral surface of the cortex.
- Flip the brain so that the dorsal surface is visible. Meninges and blood vessels should be apparent. Peel meninges from the underlying cortex using fine dissection forceps. Start peeling from the olfactory bulb attachment location.
- Complete steps 2.4-2.14 for a total of 3-4 rat pups.
- Place 3-4 dissected rat pup cortices into a dry petri dish and chop with a sterilized razor blade until 1 mm x 1 mm chunks are achieved. Collect tissue by rinsing dish with PBS from one 50 ml conical tube (step 2) then place back on ice.
3. Enzymatic and Mechanical Tissue Dissociation
Note: For this step, a papain-based neural dissociation kit is recommended. Using Neural Dissociation Kit (P), special steps that are optimal for OPC isolation are described.
- Prepare the first dissociation enzyme by diluting enzyme P with the appropriate buffer. The contents of each 50 ml conical (1 sample) will be resuspended with a total of 1,950 µl of enzyme mix. Place in the enzyme mix in a 37 °C bath for 10 min.
1 Sample: 1,900 µl of Buffer + 50 µl of papain enzyme - Spin all 50 ml conical tubes containing 3-4 diced rat pup cortices at 300 x g for 3 min.
- Aspirate the supernatant and add 1,950 µl of enzyme solution to each sample tube and break apart the pellet by inverting the tube and shaking gently.
- Incubate in a 37 °C tissue culture incubator for 15 min with continuous tube rotation.
- During the last 5 min of the incubation, prepare the second enzyme mix A. Dilute the enzyme in its appropriate buffer. A total of 30 µl will be added to each 50 ml conical sample tube.
1 Sample: 20 µl of Buffer + 10 µl of enzyme - Once incubation is completed add 30 µl of the second enzyme mix to each tube and invert to mix.
- Using a glass Pasteur pipette attached to a pipette controller, mechanically dissociate the tissue by pipetting 10-15 times or until the tissue pieces are reduced in size and the solution has become cloudy. Return the sample to the 37 °C tissue culture incubator for 15 min with continuous rotation.
- Pipette each tissue homogenate sample 10-15 times using a 1 ml pipette.
- Pipette each tissue homogenate sample 15-20 times using a 200 µl pipette
- Return all samples to the 37 °C tissue culture incubator for 10 min with continuous rotation.
- Add 10 ml of PBS (with Ca2+ and Mg2+) to each sample and filter the homogenate through a 40 µm pore filter placed over a 50 ml tube. Rinse the filter with an additional 1-2 ml of PBS.
- Pool all of the homogenate samples into one 50 ml conical tube and acquire an overall cell count.
- After performing the cell count, split the homogenate evenly into 15 ml conical tubes (1 tube for each sample). Centrifuge for 10-12 min at 300 x g.
4. Anti-A2B5 Bead Application, Column Purification, and Plating
- Perform calculations for column buffer and anti-A2B5 microbeads. Add 7 μl Column Buffer for every 1 x 106 cells. Add 2 μl Anti-A2B5 microbeads for every 1 x 106 cells.
- Combine all of the samples by resuspending the cells in a total volume of 10 ml of column buffer and centrifuge at 300 x g for 10-12 min.
- Resuspend the cell pellet in the appropriate amount of column buffer as calculated in step 4.1. If the pellet is large and loose, only add approximately half of the volume of column buffer to keep the antibody at the appropriate concentration in the cell resuspension.
- Incubate the cell suspension for 10 min at 4 °C.
- Add anti-A2B5 microbeads (calculated in step 4.1), invert several times and incubate at 4 °C for 30-60 min. Gently invert the tube several times every 10 min. Longer incubation times may increase cell yields but may increase non-specific binding.
- Add 5 volumes of column buffer. If the cells are resuspended in a volume of 1 ml, add 5 ml of column buffer, whereas if the cells are resuspended in 2 ml, add 10 ml of column buffer.
- Centrifuge for 10 min at 300 x g.
- Determine how many LS columns will be needed for the purification. One LS column is sufficient for 15-20 x 106 total cells.
- Resuspend the cell pellet in 1,000 µl of column buffer per LS column being used.
- Prime each LS column by adding 5 ml of column buffer. Collect the flow through in a 50 ml conical tube. Once the column buffer has completely passed through the upper chamber, apply 1,000 µl of the cell suspension to each column and allow the cells to run through by gravity.
Note: If the density of cells is too high, the column may run slow. - Immediately after the cell suspension has passed through the column, slowly add 3 ml of column buffer to each column to wash the unbound cells. If the wash is readily running through the column (1 drop for every 30-45 sec), wash two additional times with 3 ml of column buffer.
- Remove each column and place it on a 15 ml conical tube. Add 5 ml of column buffer and plunge the buffer through the column at a fast rate using the plastic plunger that is packaged with the column. Plunging will dislodge the bound cells releasing them from the column.
- Increase purity by running the sample through a second round of LS columns. Use one third of the number of columns used during the first pass. Prime the columns with 5 ml of column buffer. Allow the column buffer to pass through the upper chamber.
- Since the volume of cell suspension will be much greater, evenly apply 1,000 µl of cell suspension to each column and allow the suspension to pass through the upper chamber of the column before replenishing with an additional 1,000 µl.
- Once the cell suspension has been completely applied to the second round of columns, wash 2-3 times with 3 ml of column buffer.
- Remove each column and place it over a sterile 15 ml conical tube. Add 5 ml of column buffer and plunge the buffer using the plastic plunger that is packaged with the column. Plunging will dislodge the bound cells releasing them from the column.
- Centrifuge the cell suspension at 300 x g for 12-15 min. The pellet should appear compact.
- Resuspend the cell pellet in 5-10 ml of pre-warmed OPC proliferation media (OPC base media supplemented with PDGF-AA 20 ng/ml).
- Count the cells using a hemocytometer.
- Plate the cells. Dilute the cells to 60,000 cells/ml with OPC proliferation media. Into each black, clear bottom 24 well, pipet 500 µl of cells along the side of the well to obtain 30,000 cells per well. Mix the cells by shaking the plate horizontally using a figure eight motion. If performing the assay in 48, 96, or 384-well plates, add 15,000, 7,500, or 1,500 cells per well, respectively. These numbers may be adjusted depending on individual plate specifications.
- Prepare a separate plate for the Day 0 baseline control cultures. This control plate should be fixed with 4% paraformaldehyde as described in section 5.7, when differentiation is initiated on Day 0.
- Culture the cells in OPC proliferation media at 37 °C for 3 days with no media change.
5. Induction of OPC Differentiation and Fixation with 4% Paraformaldehyde
Note: When testing the effect of small molecules on oligodendrogenesis, we routinely dissolve drugs in dimethyl sulfoxide (DMSO) to prepare 1,000x stocks that can then be stored at -80 °C and diluted directly into SATO media to obtain a final concentration of 0.1% DMSO. We have observed no changes in either OPC proliferation or differentiation using this concentration of DMSO (data not shown).
- Pre-warm OPC base media to 37 °C and thaw drug treatment stocks to room temperature. When performing treatments in duplicate, prepare approximately 1.25 ml of media per treatment group in a 1.5 ml tube. For triplicate samples, use 2 ml capacity centrifuge tubes to account for slack.
- Prepare the treatment master mixes for replicate wells by diluting treatment stocks directly into OPC base media. Briefly vortex or gently mix each mastermix to ensure homogeneous distribution of the treatment.
- Prepare 45 nM triiodothyronine (T3) as the positive control and the appropriate vehicle as the negative control. Dilute 10 mM T3 stock 1:1,000 in 1 ml of OPC base media and then further dilute in the treatment master mix to reduce the final concentration of DMSO if necessary.
- Aspirate the media from the culture wells one treatment group at a time so as to avoid drying the cells. Use a 200 µl pipet tip on the end of the glass Pasteur pipet to avoid scraping off black material from the sides of the well and into the culture.
- Slowly add 500 µl of treatment mastermix along the side of the well using a 1 ml pipet.
- Incubate the cultures for the desired timeframe, performing a full media exchange with fresh treatment media every 2-3 days. We routinely observe a 30-40% MBP+ culture after 4 days in differentiation media.
- At the end of the desired treatment period, prepare fresh 4% paraformaldehyde (PFA) or thaw a frozen stock to room temperature. CAUTION: PFA is extremely toxic and must be prepared in a fume hood.
- Aspirate the media and slowly add 400 µl of PFA to the side of the well to fix the cells. Incubate the plate for 20 min at room temperature.
- Aspirate the PFA and slowly add 500 µl of sterile D-PBS (with Ca2+ and Mg2+) to the side of the well to wash the cells. Repeat the wash a total of two more times. Do not aspirate the final wash.
- Wrap the plate in parafilm and store at 4 °C for later processing or proceed directly to the next step. Plates can be stored for several weeks.
6. Immunocytochemistry and Quantification of Myelin Basic Protein
Note: When performing liquid handling procedures in the 24 well plates, it is recommended to work fast to avoid drying the cells. Here, the use of a multi-channel vacuum aspirator and multi-channel pipet are recommended.
- Bring the plate to room temperature, aspirate the wells, and add 250 μl of D-PBS (with Ca2+ and Mg2+) containing 0.1% Triton X-100 and 5% normal goat serum (NGS). Incubate the plate at room temperature for 1 hr with gentle orbital shaking.
- Prepare the primary incubation solution by diluting mouse monoclonal anti-MBP antibody 1:1,000 and rabbit polyclonal anti-Actin antibody 1:200 in D-PBS (with Ca2+ and Mg2+) containing 5% NGS. Alternatively, rabbit polyclonal anti-Olig2 at 1:1,000 can be used for oligodendrocyte specific normalization in mixed glial cultures. Prepare enough primary solution to accommodate 250 µl per well.
- Incubate primary antibodies at 4 °C overnight for 16-18 hr with gentle orbital shaking.
- Aspirate the primary incubation solution and wash the cells 3 x 10 min with 500 µl of D-PBS (with Ca2+ and Mg2+) at room temperature with gentle orbital shaking.
- While washing the plate, prepare the secondary incubation solution by diluting anti-mouse 680 and anti-rabbit 800 antibodies 1:500 in D-PBS (with Ca2+ and Mg2+) containing 5% NGS. Minimize ambient light exposure when preparing this solution.
- Aspirate the final wash and add 250 µl of secondary incubation solution. Protect the plate from light and incubate it at room temperature for 1 hr with gentle orbital shaking.
- Aspirate the secondary incubation solution and wash the wells 3 x 10 min with 500 µl of D-PBS (with Ca2+ and Mg2+) at room temperature with gentle orbital shaking.
- Scan the plate using an imaging system capable of detecting 700 nm and 800 nm fluorescence emissions. As a starting point, set the focal offset to 3 and sensitivities to 1.5 for MBP, 5.0 for Actin, and 3.5 for Olig2. Adjust the sensitivity values as needed to avoid signal overexposure.
- Quantify the extent of oligodendrogenesis using semi-automated software-based methods.
- Using the software, set the plate analysis mode to "24 Well" and align circles around the outer perimeter of the scanned wells. Set the normalization channel to "800" and export the total and relative fluorescence intensities for the 700 nm (MBP) and 800 nm (Olig2/Actin) channels.
- Divide the intensity at 700 nm by the intensity at 800 nm to give the normalized MBP expression in relative fluorescence units. Calculate the average of replicate measurements and scale the expression to the Day 0 +PDGF controls as needed for the analysis.
Representative Results
A2B5-positive OPCs are isolated through positive cell selection using a magnetic column separation system. Prior to the isolation procedure, the whole brain is removed from rat pups that are between P5 and P7. Once the whole brain is successfully detached from the skull, the olfactory bulbs and cerebellum are removed using fine surgical forceps (Figure 1A). In order to isolate cerebral cortical tissue the hypothalamus, thalamus and midbrain are excised by careful dissection (Figure 1B). Next, the hippocampus and striatum are removed using bent surgical forceps (Figure 1C–D). The isolation process efficiently eliminates meningeal and fibroblast cells, but enzymatic digestion is improved when meninges and vasculature are removed (Figure 1E). Finally, tissue blocks of 1 mm x 1 mm are prepared for the subsequent tissue dissociation steps (Figure 1F).
A good OPC isolation should result in a compact cell pellet after the final spin step (Figure 2A). Once plated these cultures will be relatively free of cellular debris when viewed under a microscope (Figure 2B). A suboptimal isolation may result in a large and fluffy pellet due to the presence of large amounts of cellular debris. This debris will adhere strongly to the PLL-coated vessel and may interfere with the culture (Figure 2C). If a large and fluffy pellet materializes, it may help to resuspend the cells in 10 ml of PBS and repeat the final spin for 10 min at <300 x g. Once the isolation is complete, the purity of the cultures can be assessed by flow cytometry to identify the percentage of A2B5-positive cells. A successful isolation should result in a pool of cells that are >95% positive for A2B5 (Figure 2D–F). Compared to cells stained with a fluorescent dye-conjugated anti-A2B5 antibody, unstained cells should appear as a negative population and can be used as a baseline reference for gating. Moreover, we have previously shown that selection of OPCs using the A2B5 antigen yields cultures that stain positive for PDGFRα by immunocytochemistry11.
As demonstrated by immunocytochemistry, OPCs at the start of treatment (Day 0) are Olig2+/MBP– whereas cells are Olig2+/MBP+ by Day 4 (Figure 3). The optimal confluency of OPCs and oligodendrocytes at Day 0 and Day 4 are shown in representative bright field images (Figure 4A). The absence of MBP expression at Day 0 serves as a baseline for quantifying oligodendrogenesis. Spontaneous differentiation as a result of PDGF-AA withdrawal serves as a negative control, whereas 45 nM thyroid hormone (triiodothyronine; T3) can be used as a positive control11,15. Quetiapine, also known as Seroquel, and clemastine, also known as Tavist, are FDA-approved drugs that have been shown to accelerate in vitro oligodendrogenesis and can be used as additional positive controls16,17. Figure 4B shows a representative two-channel infrared fluorescent scan demonstrating the expected results for each of these conditions in triplicate. Actin and MBP signals were pseudocolored green and red respectively so that highly differentiated cultures appear yellow in merged images. Results show that MBP expression at Day 4 in the PDGF withdrawal condition was 4.5 ± 1.4-fold higher compared to Day 0, whereas the fold changes in MBP due to treatment with thyroid hormone, quetiapine, and clemastine were 33.9 ± 4.1, 33.4 ± 1.0, and 32.8 ± 0.5 respectively (Figure 4C). The robustness and accuracy of the assay are demonstrated by the small standard deviations among the triplicate samples and the large treatment activation window that is about 20 standard deviations above the PDGF withdrawal control.
While Actin serves as a reliable reference gene for normalization in oligodendrocyte cultures, alternative reference genes can also be used. Olig2 is a nuclear transcription factor that is constitutively expressed in cells of the oligodendrocyte lineage. Its specific and constitutive expression pattern thus may provide a more preferable normalization strategy in heterogeneous culture situations. Figure 5 shows that in OPC cultures differentiated with 10 nM thyroid hormone, the calculated fold change in MBP expression relative to PDGF withdrawal is the same when using either Olig2 or Actin for normalization. Presumably, since the OPC cultures in this experiment were very pure, both proteins can be used reliably for normalization. However, we often observe a small population of Olig2-negative cells by Day 4, but the ratio of this population to the Olig2-positive population does not seem to significantly differ between the PDGF withdrawal and T3 treated group. Thus, the fold-changes are not affected. Given a scenario where a treatment might cause proliferation of Olig2-negative cell types, the choice of normalization antibody should be carefully considered.
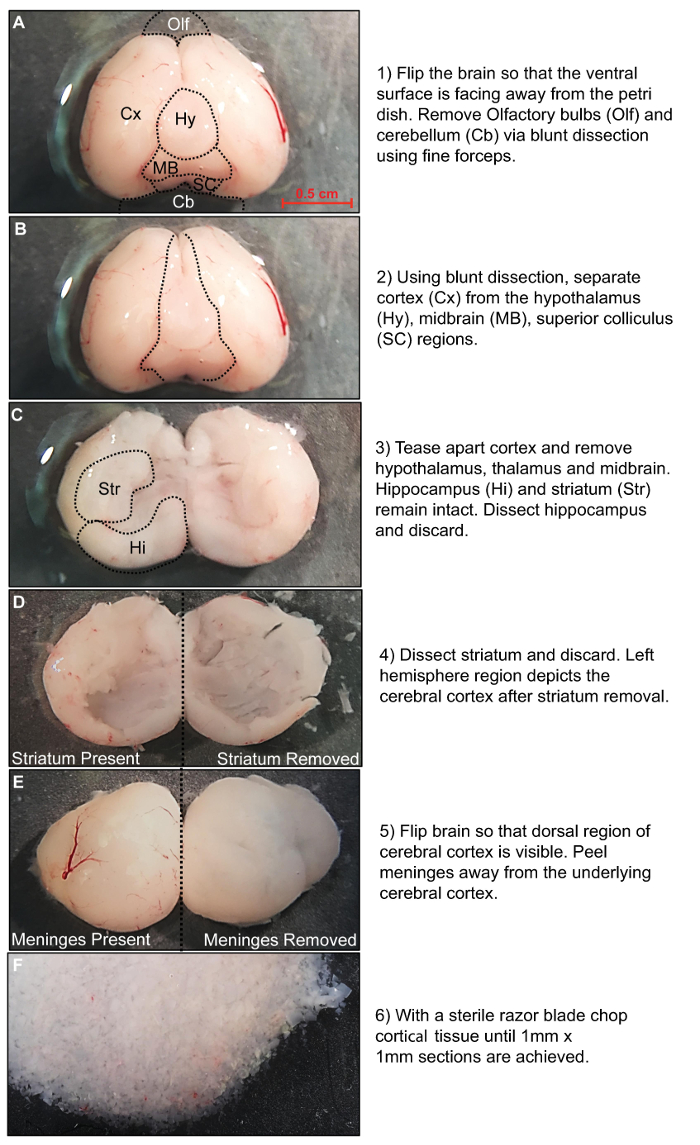
Figure 1: Rat brain dissection procedure. Stepwise description of the dissection procedure. Please click here to view a larger version of this figure.
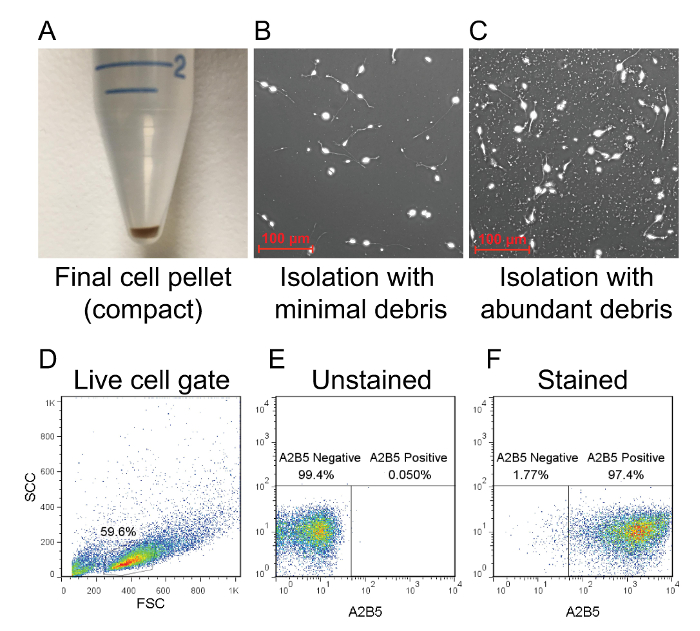
Figure 2: Flow cytometry analysis of isolated OPCs. (A) After magnetic column purification, the bound cell population is plunged out of the column and centrifuged to form a compact pellet. The amount of debris is indicated by the size of the pellet. (B) If the pellet is compact, then debris will be minimal in the culture. (C) A large and fluffy pellet typically results in a culture with heavy debris. The purity of the culture is determined by flow cytometry using a rat anti-mouse A2B5 antibody that is conjugated to APC. (D) Dead cells and debris are excluded from the analysis as shown in forward scatter and side scatter plots. (E) The A2B5 positive population can be determined by using an unstained sampled as a baseline reference for gating. (F) Successfully isolated OPCs should be >95% positive for A2B5. Please click here to view a larger version of this figure.
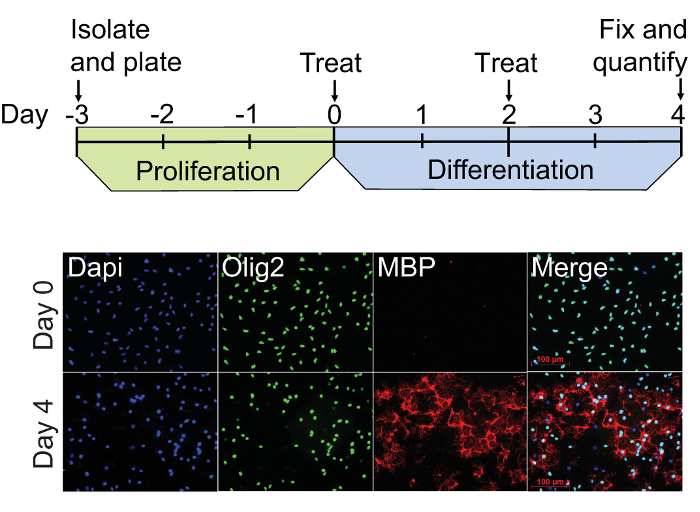
Figure 3: OPC culture and differentiation procedure. The dual-infrared fluorescence-scanning (DIFS) assay begins with freshly isolatedA2B5-positive rat OPCs plated on PLL-coated culture vessels (Day 3). These cultures are proliferated for 3 days in the presence of 20 ng/ml PDGF-AA, and then treated with experimental factors in fresh PDGF-free media on Day 0. Treatment media is fully replenished on Day 2, and the cells are fixed with 4% paraformaldehyde on Day 4. Immunocytochemistry for Olig2 (green) and MBP (red) was performed on representative cultures at Day 0 and Day 4 using Dapi (blue) as a nuclear counter stain. These cultures exhibit constitutive staining for the pan oligodendrocyte lineage cell marker Olig2 at Days 0 and 4, whereas staining for the mature oligodendrocyte marker MBP is only evident by Day 4. Please click here to view a larger version of this figure.
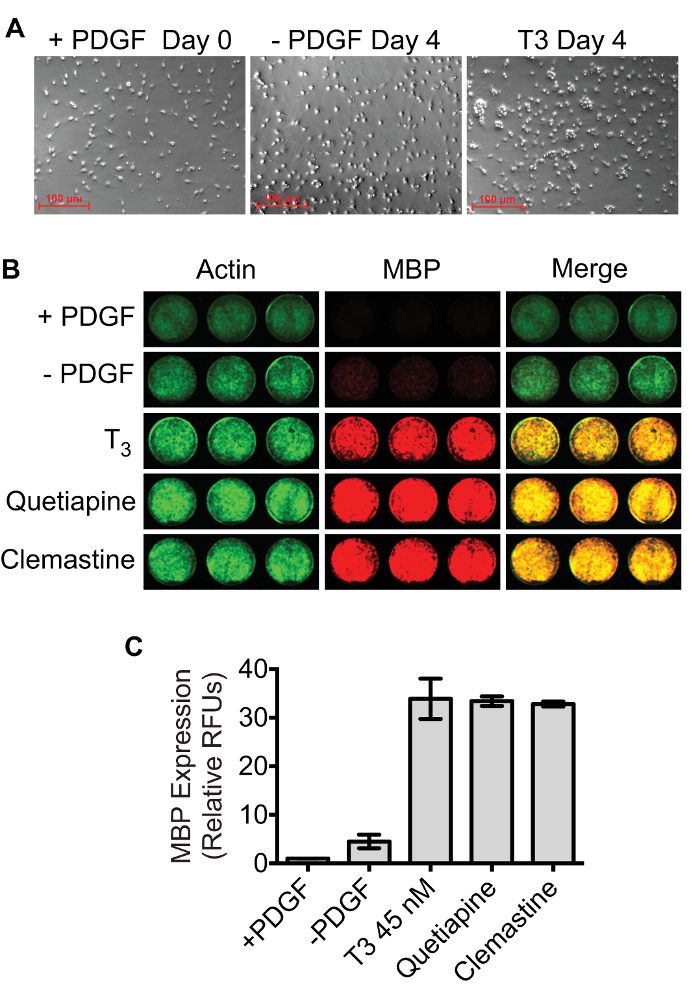
Figure 4: MBP quantification using the dual-infrared fluorescence-scanning (DIFS) assay. (A) The density of OPC cultures at Day 0 and Day 4 required for optimal assay performance. (B) Representative dual-infrared fluorescence scans. OPCs were differentiated in 24 well plates for 4 days in the presence of 45 nM T3, 1 µM quetiapine, 1 µM clemastine, or 0.1% DMSO in triplicate. A separate plate containing OPCs that were fixed on day 0 (+PDGF) was processed in parallel. Actin (pseudocolored green) and MBP (pseudocolored red) were simultaneously quantified using a Licor Odyssey scanner set to detect 700 nm and 800 nm emissions. (C) MBP signals were normalized to Actin signals, and results were scaled to the Day 0 +PDGF control. The mean relative fluorescence units ± SD were plotted using GraphPad Prism 6. Please click here to view a larger version of this figure.
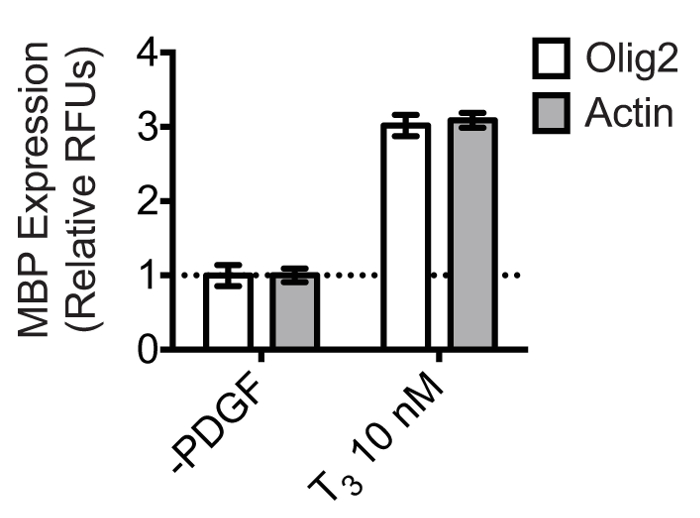
Figure 5: Olig2 is a reliable reference protein for normalizing MBP expression in oligodendrocytes. OPCs were differentiated in triplicate in the presence of 10 nM T3 or 0.1% DMSO vehicle control in 24 well plates for 4 days. The dual-infrared fluorescence-scanning assay was performed using anti-Olig2 or anti-Actin primary antibodies for normalization. Olig2/Actin and MBP were simultaneously quantified using a Licor Odyssey scanner set to detect 700 nm and 800 nm emissions. MBP expression was normalized to either Olig2 or Actin expression, and results were scaled to the 0.1% DMSO control. The mean relative fluorescence units ± SD were plotted using GraphPad Prism 6. Results show that the calculated fold-change is the same when normalizing to either of the two reference proteins.

Figure 6: Effect of Triton X-100 on MBP staining. OPCs were differentiated in 24 well plates for 4 days in the presence of 45 nM T3 or 0.1% DMSO. Cultures were then fixed with 4% PFA and stained for Olig2 and MBP using two different immunocytochemistry protocols. For the high Triton X-100 sample, the cells were permeabilized for 1 hr with 0.4% Triton X -100 and all antibody incubation steps were performed in the presence of 0.1% Triton X-100. For the low Triton X-100 sample, the cells were permeabilized for 1 hr with 0.1% Triton X-100, while antibody incubations were performed without Triton X-100. Olig2 and MBP were visualized with fluorescent dye-conjugated secondaries and images were acquired using a camera-based fluorescence microscope. Please click here to view a larger version of this figure.
Discussion
The protocol presented here describes a fast and reliable method for isolating rat OPCs and quantifying in vitro oligodendrogenesis in response to experimental factors. Using positive selection, high yields of viable and pure OPCs are used directly for experimental purposes. This method streamlines the isolation, culturing, and differentiation steps into a 1 week time frame, and fixed cells can be conveniently stored at 4 °C for several weeks without any loss in assay sensitivity.
A major advantage with scanner-based fluorescence assays is that data acquisition and analysis is greatly expedited versus traditional camera-based microscopy techniques. Although microscopy has been used successfully in high-throughput screens to identify inducers of OPC differentiation, data acquisition is inherently slower because it requires programming of image capture sequences, several hours of data collection, analysis of hundreds to thousands of image files, and user-defined algorithms to distinguish individual cell bodies for normalization or analysis18,19. Furthermore, with camera-based microscopy systems it is not feasible to acquire data from the entire surface of the culture vessel. Instead, representative images must be captured from different areas within a single well, which may lead to positional bias and false-positive hits. By comparison, this assay detects MBP expression from all cells within a given well, and multiple plates can be scanned simultaneously. While this method can be adapted for use in 96-well or 384-well formats, the 24-well plate may be preferred when fewer treatments are being tested. We have observed that cell loss in the 24-well plate due to liquid handling is minimized, which is a complication that should be considered when designing higher-throughput experiments.
A critical parameter of the assay is the density of the cells when differentiation is initiated at Day 0. Cultures with too few cells appear to differentiate more slowly and die off, while high-density cultures exhibit spontaneous differentiation. Both situations may reduce the dynamic range and reliability of the assay. To achieve an optimal balance between cell health and cell density, we culture the OPCs for 3 days without a media exchange or supplementation of fresh PDGF-AA. We hypothesized that since the concentration of PDGF-AA in the media declines by Day 3 in vitro (Day 0) as a result of cellular catabolism, the proliferative rate of the cells will slow such that proliferation is reduced through the first day of PDGF-AA withdrawal. This technique works best when the cells are evenly distributed during plating. Pipetting the cells along the side of the well and mixing them using a figure eight motion should promote an even distribution. Pipetting into the center of the well tends to produce clusters of cells around the edges of the well with fewer cells adhering to the center. This phenomenon may be due to disruption of the freshly dried PLL layer by liquid pipetting forces. Accumulation of cells around the perimeter may reduce assay sensitivity because of density-dependent differentiation. It is important to note that significant differences in the intensity of the Actin/Olig2 signal can occur between drug-treated and control, PDGF withdrawal cultures. Significantly decreased Actin/Olig2 signal may indicate drug toxicity, whereas significantly increased Actin/Olig2 signal may indicate drug-induced proliferation. We normally observe a slightly higher Actin/Olig2 signal from cells treated with pro-differentiation drugs compared to the PDGF withdrawal controls. We attribute this difference to a higher rate of spontaneous apoptosis in the PDGF withdrawal group over the 4 day differentiation procedure. When assessing oligodendrogenesis, normalization of MBP to Actin/Olig2 should correct for cell number differences. To assess toxicity and proliferation using this assay, the anti-MBP antibody can be exchanged for primary antibodies directed against the appropriate markers.
When experiments require both quantitative and qualitative assessment, it is important to limit the exposure of the fixed rat oligodendrocyte cultures to Triton X-100 when using 4% PFA to fix the cells. Other immunocytochemistry protocols for oligodendrocyte cultures, which typically include Triton X-100 during both of the antibody incubation steps to improve antibody epitope accessibility, may be used successfully depending on experimental requirements. On the other hand, although MBP is an intracellular protein it may be possible to exclude Triton X-100 entirely if alternative fixation methods are employed that adequately permeabilize the cells. Using the conditions described here, we have observed that Triton X-100 affects the observed morphology of the oligodendrocytes as well as the localization of MBP staining when used at high concentrations. We do not see a significant decline in antibody staining when Triton X-100 is omitted from the antibody incubation steps. As shown in Figure 6, including 0.4% Triton X-100 in the permeabilization and blocking step, and 0.1% Triton X-100 during antibody incubation, reduces the detection of myelin processes and results in discontinuous MBP staining. Therefore, for quantitative and qualitative assessment it may be sufficient to use a lower concentration of Triton X-100 as described here.
While the use of A2B5-selected OPCs from rats can be advantageous, we have observed comparable results in the dual-infrared fluorescence-scanning (DIFS) assay using A2B5-selected OPCs from C57BL/6 mice, although cell yield per mouse is typically much lower (data not shown). Other protocols for OPC purification may also be compatible with this assay. For example, oligodendrocyte marker O4 may be used as an antigen to purify intermediate stage oligodendrocyte precursors using magnetic beads20. As an alternative to magnetic-based methods, OPCs can be enriched from 9 day old mixed glial cultures containing astrocytes and microglia by hi-speed orbital shaking and differential adhesion methods21. Furthermore, immuno-panning procedures which incorporate both negative and positive cell selection may be used if culture purity is preferred over high cell yields22. In addition to examining how therapeutics may directly enhance the differentiation of OPCs in pure cultures, it is sometimes necessary to consider the indirect effects of drugs through other cell types in a co-culture system. In MS, the infiltration of autoreactive T effector cells into the CNS is thought to play a role in disease progression23. Effector T cells may secrete pro-inflammatory factors that may inhibit the differentiation of OPCs at sites of demyelination. It is therefore of interest to identify therapeutics that may protect the OPCs and block these pro-inflammatory factors from exacerbating injury. The DIFS assay may be ideal for modeling this activity as T cells grow in suspension and may be washed away from the oligodendrocyte culture prior to MBP quantification. For co-culture studies involving adherent cells such as neurons, astrocytes, and microglia, Olig2 normalization may be employed to collect data from the oligodendrocytes specifically. Thus, the flexibility of the DIFS assay may facilitate a variety of experimental designs.
In conclusion, this protocol provides a fast and reliable method to quantify the oligodendrogenesis of A2B5-positive rat OPCs in vitro. This isolation method consistently delivers high yields of viable OPCs that are responsive to established differentiation cues. Sensitive analysis can be performed in multi-well format vessels using a variety of experimental paradigms, which extends the utility of this system to many different applications. Furthermore, the DIFS assay can be adapted for high-throughput drug screening, or may be used to verify results of low-throughput assays such as quantitative PCR and Western blot. Importantly, this system may offer a simple yet effective means for the identification of OPC targeted therapies in demyelinating diseases such as multiple sclerosis.
Declarações
The authors have nothing to disclose.
Acknowledgements
Grant sponsor: National Institutes of Health; Grant number: R37 NS041435.
Materials
| Dissection Materials | |||
| PBS without Ca2+ and Mg2+ | Mediatech | 21-040-CV | 1 x |
| 10 cm Polystyrene Petri Dishes | Fisherbrand | 0857512 | |
| Light Dissection Microscope | Motic | SM2-168 | |
| Dissociation Materials | |||
| Tube Rotator | Miltenyi Biotec | 130-090-753 | Dissociation Tube Rotator |
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | 130-092-628 | Enzymatic Dissociation Kit |
| PBS with Ca2+ and Mg2+ | Corning | 20-030-CV | 1 x |
| 40 μm Cell Strainer | Corning | 352340 | |
| A2B5 Column Purification | |||
| Anti-A2B5 Microbeads | Miltenyi Biotec | 130-097-864 | Bead Bound A2B5 Antibody |
| LS Columns | Miltenyi Biotec | 130-042-401 | Magnetic Selection Columns |
| EDTA | Corning | 46-034-Cl | 2mM |
| PBS with Ca2+ and Mg2+ | Corning | 20-030-CV | 1 x |
| Bovine Serum Albumin | Sigma-Aldrich | A9647 | 0.50% |
| Culture Materials | |||
| PDGF-AA | PeproTech Inc | 100-13A | 20 ng/mL |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650 | Hybri-Max 100 mL |
| Triiodothyronine | Sigma-Aldrich | T6397 | 45 nM |
| Clemastine fumarate salt | Sigma-Aldrich | SML0445-100MG | 1 μM |
| Quetiapine hemifumarate salt | Sigma-Aldrich | Q3638-10MG | 1 μM |
| N-acetyl cysteine | Sigma-Aldrich | A9165 | 5 μg/mL |
| Progesterone | Sigma-Aldrich | P8783 | 60 ng/mL |
| Poly-L-lysine | Sigma-Aldrich | P1524 | 10 μg/mL |
| Putrecine | Sigma-Aldrich | P7505 | 16 μg/mL |
| Sodium Selenite | Sigma-Aldrich | S5261 | 40 ng/mL |
| Hydrocortisone | Sigma-Aldrich | H0135 | 50 ng/mL |
| d-Biotin | Sigma-Aldrich | B4639 | 10 ng/mL |
| Insulin | Sigma-Aldrich | I6634 | 5 μg/mL |
| Apo-Transferrin | Sigma-Aldrich | T1147 | 100 μg/mL |
| Bovine Serum Albumin | Sigma-Aldrich | A4161 | 100 μg/mL |
| Trace Elements B | Corning | 25-022-Cl | 1 x |
| B-27 Supplement | Life Technologies | 17504044 | 1 x |
| Penicillin/Streptomycin | Life Technologies | 15140122 | 1 x |
| DMEM | Life Technologies | 11995-065 | 1 x |
| 24-well Black Visiplate with lid | Perkin Elmer | 1450-605 | Tissue culture grade |
| Immunocytochemistry and Quantification | |||
| PFA | Sigma-Aldrich | 100-13A | 4% |
| Triton X-100 | Sigma-Aldrich | 78787 | 0.10% |
| Normal Goat Serum | Jackson Immuno Research | 005-000-121 | 5% |
| mouse monoclonal anti-MBP | Biolegend | SMI-99P | 1:1000 Dilution |
| rabbit polyclonal anti-Actin | Santa Cruz Biotecnology | SC-7210 | 1:200 Dilution |
| rabbit polyclonal anti-Olig2 | Millipore | AB9610 | 1:1000 Dilution |
| goat anti-mouse 680RD | Licor | 926-68070 | 1:500 Dilution |
| goat anti-rabbit 800CW | Licor | 926-32211 | 1:500 Dilution |
| Alexa Fluor 594 goat anti-mouse | Life Technologies | A11032 | 1:000 dilution |
| Alexa Fluor 488 goat anti-rabbit | Life Technologies | A11008 | 1:000 dilution |
| Prolong Gold antifade with DAPI | Life Technologies | P36931 | |
| DPBS with Ca2+ and Mg2+ | Corning | 21-030-CV | |
| Licor Odyssey Clx Infared Imaging System | Licor | ||
Referências
- Rowitch, D. H., Kriegstein, A. R. Developmental genetics of vertebrate glial-cell specification. Nature. 468 (7321), 214-222 (2010).
- Nave, K. A. Myelination and the trophic support of long axons. Nat Rev Neurosci. 11 (4), 275-283 (2010).
- Dawson, M. R. L., Polito, A., Levine, J. M., Reynolds, R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 24 (2), 476-488 (2003).
- Goldschmidt, T., Antel, J., König, F. B., Brück, W., Kuhlmann, T. Remyelination capacity of the MS brain decreases with disease chronicity. Neurology. 72 (22), 1914-1921 (2009).
- Duncan, I. D., Brower, A., Kondo, Y., Curlee, J. F., Schultz, R. D. Extensive remyelination of the CNS leads to functional recovery. Proc Natl Acad Sci USA. 106 (16), 6832-6836 (2009).
- Nishiyama, A., Chang, A., Trapp, B. D. NG2+ glial cells: a novel glial cell population in the adult brain. J Neuropath Exp Neur. 58 (11), 1113-1124 (1999).
- Baracskay, K. L., Kidd, G. J., Miller, R. H., Trapp, B. D. NG2-positive cells generate A2B5-positive oligodendrocyte precursor cells. Glia. 55 (10), 1001-1010 (2007).
- Ffrench-Constant, C., Raff, M. C. Proliferating bipotential glial progenitor cells in adult rat optic nerve. Nature. 319 (6053), 499-502 (1986).
- Popko, B., et al. Myelin deficient mice: expression of myelin basic protein and generation of mice with varying levels of myelin. Cell. 48 (4), 713-721 (1987).
- Bourre, J. M., et al. Density profile and basic protein measurements in the myelin range of particulate material from normal developing mouse brain and from neurological mutants (Jimpy; quaking; Trembler; shiverer and its mld allele) obtained by zonal centrifugation. J Neurochem. 35 (2), 458-464 (1980).
- Baxi, E. G., et al. A selective thyroid hormone β receptor agonist enhances human and rodent oligodendrocyte differentiation. Glia. 62 (9), 1513-1529 (2014).
- Watkins, T. A., Emery, B., Mulinyawe, S., Barres, B. A. Distinct stages of myelination regulated by gamma-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 60 (4), 555-569 (2008).
- Chen, Y., et al. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2 (5), 1044-1051 (2007).
- Warringa, R. A., et al. Hydrocortisone stimulates the development of oligodendrocytes in primary glial cultures and affects glucose metabolism and lipid synthesis in these cultures. Brain Res. 431 (1), 79-86 (1987).
- Tosic, M., Torch, S., Comte, V., Dolivo, M., Honegger, P., Matthieu, J. M. Triiodothyronine has diverse and multiple stimulating effects on expression of the major myelin protein genes. J Neurochem. 59 (5), 1770-1777 (1992).
- Xiao, L., et al. Quetiapine facilitates oligodendrocyte development and prevents mice from myelin breakdown and behavioral changes. Mol Psychiatry. 13 (7), 697-708 (2008).
- Mei, F., et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med. 20 (8), 954-960 (2014).
- Deshmukh, V. A., et al. A regenerative approach to the treatment of multiple sclerosis. Nature. 502 (7471), 327-332 (2013).
- Najm, F. J., et al. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature. 522 (7555), 216-220 (2015).
- Zhang, S. C. Defining glial cells during CNS development. Nat Rev Neurosci. 2 (11), 840-843 (2001).
- O’Meara, R. W., Ryan, S. D., Colognato, H., Kothary, R. Derivation of enriched oligodendrocyte cultures and oligodendrocyte/neuron myelinating co-cultures from post-natal murine tissues. J Vis Exp. (54), (2011).
- Dugas, J. C., Emery, B. Purification of oligodendrocyte precursor cells from rat cortices by immunopanning. Cold Spring Harb Protoc. 2013 (8), 745-758 (2013).
- Fletcher, J. M., Lalor, S. J., Sweeney, C. M., Tubridy, N., Mills, K. H. G. T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin Exp Immunol. 162 (1), 1-11 (2010).

