Residue-Specific Exchange of Proline by Proline Analogs in Fluorescent Proteins: How “Molecular Surgery” of the Backbone Affects Folding and Stability
Summary
To overcome the limitations of classical site-directed mutagenesis, proline analogs with specific modifications were incorporated into several fluorescent proteins. We show how the replacement of hydrogen by fluorine or of the single by double bonds in proline residues (“molecular surgery”) affects fundamental protein properties, including their folding and interaction with light.
Abstract
Replacement of proline (Pro) residues in proteins by the traditional site-directed mutagenesis by any of the remaining 19 canonical amino acids is often detrimental to protein folding and, in particular, chromophore maturation in green fluorescent proteins and related variants. A reasonable alternative is to manipulate the translation of the protein so that all Pro residues are replaced residue-specifically by analogs, a method known as selective pressure incorporation (SPI). The built-in chemical modifications can be used as a kind of "molecular surgery" to finely dissect measurable changes or even rationally manipulate different protein properties. Here, the study demonstrates the usefulness of the SPI method to study the role of prolines in the organization of the typical β-barrel structure of spectral variants of the green fluorescent protein (GFP) with 10-15 prolines in their sequence: enhanced green fluorescent protein (EGFP), NowGFP, and KillerOrange. Pro residues are present in connecting sections between individual β-strands and constitute the closing lids of the barrel scaffold, thus being responsible for insulation of the chromophore from water, i.e., fluorescence properties. Selective pressure incorporation experiments with (4R)-fluoroproline (R-Flp), (4S)-fluoroproline (S-Flp), 4,4-difluoroproline (Dfp), and 3,4-dehydroproline (Dhp) were performed using a proline-auxotrophic E. coli strain as expression host. We found that fluorescent proteins with S-Flp and Dhp are active (i.e., fluorescent), while the other two analogs (Dfp and R-Flp) produced dysfunctional, misfolded proteins. Inspection of UV-Vis absorption and fluorescence emission profiles showed few characteristic alterations in the proteins containing Pro analogs. Examination of the folding kinetic profiles in EGFP variants showed an accelerated refolding process in the presence of S-Flp, while the process was similar to wild-type in the protein containing Dhp. This study showcases the capacity of the SPI method to produce subtle modifications of protein residues at an atomic level ("molecular surgery"), which can be adopted for the study of other proteins of interest. It illustrates the outcomes of proline replacements with close chemical analogs on the folding and spectroscopic properties in the class of β-barrel fluorescent proteins.
Introduction
Classical site-directed mutagenesis allows permutation of any existing gene-encoded protein sequence by codon manipulation at the DNA level. To study protein folding and stability, it is often desirable to replace similar amino acids with similar counterparts. However, traditional protein mutagenesis is definitely limited to structurally similar replacements among canonical amino acids such as Ser/Ala/Cys, Thr/Val, Glu/Gln, Asp/Asn, Tyr/Phe, which are present in the standard genetic code repertoire. On the other hand, there are no such possibilities for other canonical amino acids such as Trp, Met, His, or Pro, which often play essential structural and functional roles in proteins1. An ideal approach to study these interactions in the context of the highly specific internal architecture of proteins and their folding process is to generate non-disruptive isosteric modifications. Indeed, when isosteric amino acid analogs of these canonical amino acids, also known as non-canonical amino acids (ncAAs), are inserted into proteins, they allow for subtle changes even at the level of single atoms or atom groups such as H/F, CH2/S/Se/Te known as "atomic mutations"2. Such "molecular surgery" produces altered proteins whose properties result solely from the exchange of single atoms or groups of atoms, which in favorable cases can be analyzed, and the detected changes can be rationalized. In this way, the scope of protein synthesis to study protein folding and structure is extended far beyond classical DNA mutagenesis. Note that proteins generated by site-directed mutagenesis are usually referred to as "mutants," whereas proteins with substituted canonical amino acids are referred to as "variants" 3, "alloproteins"4, or "protein congeners"5.
The green fluorescent protein (GFP), first identified in the marine organism Aequorea victoria, exhibits bright green fluorescence when exposed to ultraviolet-to-blue light6,7. Today, GFP is commonly used as a highly sensitive labeling tool for routine visualization of gene expression and protein localization in cells via fluorescent microscopy. GFP has also proven useful in various biophysical8,9,10 and biomedical11,12 studies, as well as in protein engineering13,14,15. Rigorous analysis of the GFP structure enabled the creation of numerous variants characterized by varied stability and fluorescence maxima16,17. Most of the GFP variants used in cell and molecular biology are monomeric proteins both in solution and in the crystal18. Their principal structural organization is typical for all members of the GFP family, independent of their phylogenetic origin, and consists of 11 β-strands forming a so-called β-barrel, while a kinked α-helix is running through the center of the barrel and bears the chromophore (Figure 1A). The autocatalytic maturation of the chromophore (Figure 1B) requires the precise positioning of the side chains surrounding it in the central place of the protein; many of these side chains are highly conserved in other GFP variants19. In most fluorescent proteins from jellyfish such as Aequorea victoria, the green-emitting chromophore consists of two aromatic rings, including a phenol ring of Tyr66 and the five-membered heterocyclic structure of imidazolinone (Figure 1B). The chromophore, when properly embedded in the protein matrix, is responsible for the characteristic fluorescence of the whole protein. It is located in the center of the structure, while the barrel structure insulates it from the aqueous medium20. Exposure of the chromophore to the bulk water would result in fluorescence quenching, i.e., loss of fluorescence21.
The proper folding of the barrel-like structure is essential for protecting the chromophore against fluorescence quenching22. Proline (Pro) residues play a special role in the structural organization of GFP23. Being unable to support a β-strand, they constitute connecting loops responsible for maintaining the protein structure as a whole. Not surprisingly, 10-15 proline residues are found in both Aequorea– and Anthoathecata-derived GFPs; some of them are highly conserved in other types of fluorescent β-barrel proteins. Prolines are expected to critically influence folding properties due to their peculiar geometric features. For example, in Aequorea-derived GFPs, of the ten proline residues (Figure 2A), nine form trans– and only one forms a cis-peptide bond (Pro89). Pro58 is essential, i.e., not interchangeable with the rest of the 19 canonical amino acids. This residue may be responsible for the correct positioning of the Trp57 residue, which has been reported to be crucial for chromophore maturation and the overall GFP folding24. The fragment PVPWP with three proline residues (Pro54, Pro56, Pro58) and Trp57 is the essential part of the "lower lid" in the GFP structure from Figure 1A. The PVPWP structural motif is found in several proteins such as cytochromes and eukaryotic voltage-activated potassium channels25. Proline-to-alanine substitutions at positions 75 and 89 are also detrimental to protein expression and folding and abolish chromophore maturation. Pro75 and Pro89 are part of the "upper lid" burying the chromophore (Figure 1A) and are conserved across 11-stranded β-barrel fluorescent proteins23. These two "lids" keep the chromophore excluded from the aqueous solvent, even when the stable tertiary structure has been partially broken26. Such a specific molecular architecture protects the fluorophore from collisional (dynamic) fluorescence quenching, e.g., by water, oxygen, or other diffusible ligands.
In order to perform molecular engineering of the GFP structure, one should introduce amino acid substitutions in the primary structure of the protein. Numerous mutations have been performed on GFP, providing variants with elevated stability, fast and reliable folding, and variable fluorescence properties17. Nonetheless, in most cases, mutation of proline residues is considered a risky approach due to the fact that none of the remaining 19 canonical amino acids can properly restore the conformational profile of the proline residue27. Thus, an alternative approach has been developed, in which proline residues are replaced with other proline-based structures, dubbed as proline analogs28. Owing to its unique cyclic chemical structure, proline exhibits two characteristic conformational transitions (Figure 1C): 1) the proline ring puckering, a fast process entailing organization of the backbone, which primarily affects the φ torsion angle, and 2) the peptide bond cis/trans isomerization, a slow process impacting backbone folding via the ω torsion angles. Due to its slow nature, the latter transition is commonly responsible for the rate-limiting steps in the folding process of the whole protein. It has been shown previously that peptide bond cis/trans isomerization around some proline residues features slow steps in the folding of GFP variants. For example, the formation of the cis-peptide bond at Pro89 features the slow step in the process of folding because it relies on the bond transition from trans 세스 cis29. A faster refolding can be achieved after replacing Pro89 with an all-trans peptide loop, i.e., by abolishing a cis-to-trans isomerization event30. In addition to the cis/trans isomerization, the pucker transitions may also generate profound changes in protein folding due to the backbone organization and packing within the protein interior27,31.
Chemical modifications result in alteration of the intrinsic conformational transitions of the proline residues, thereby affecting the ability of the protein to fold. Certain proline analogs are particularly attractive candidates for proline substitution in proteins as they allow the manipulation and study of the folding properties. For example, (4R)-fluoroproline (R-Flp), (4S)-fluoroproline (S-Flp), 4,4-difluoroproline (Dfp), and 3,4-dehydroproline (Dhp) are four analogs (Figure 1D) that differ minimally from proline in terms of both molecular volume and polarity32. At the same time, each analog exhibits a distinct ring puckering: S-Flp stabilizes the C4–endo pucker, R-Flp stabilizes the C4–exo pucker, Dfp exhibits no apparent pucker preference, while Dhp abolishes the puckering (Figure 1D)33. By using these analogs in the protein structure, one can manipulate with the conformational transition of the proline residues, and with this, affect the properties of the resulting GFP variants.
In this work, we set out to incorporate the designated set of proline analogs (Figure 1D) into the structure of GFP variants using the selective pressure incorporation method (SPI, Figure 3)34. Replacement of amino acid residues with their closest isostructural analogs is an applied biotechnological concept in protein design35,36. Thus, the effects of proline analogs in a model protein illustrate their potential to serve as tools in protein engineering37. The production of proteins containing desired analogs was performed in modified E. coli strains that are not able to produce proline (proline-auxotrophy). Thus, they could be forced to accept replacement of substrates in the process of protein biosynthesis38. This global substitution of proline is enabled by the natural substrate flexibility of endogenous aminoacyl-tRNA synthetases39, the key enzymes catalyzing the esterification of tRNAs with appropriate amino acids40. In general, as outlined in Figure 3, cellular growth is performed in a defined medium until the mid-logarithmic growth phase is reached. In the next step, the amino acid to be replaced is intracellularly depleted from the expression system during fermentation and subsequently exchanged by the desired analog or ncAA. Target protein expression is then induced for residue-specific non-canonical amino acid incorporation. The substitution of the cognate amino acid with its analog occurs in a proteome-wide manner. Although this side effect may have a negative impact on the growth of the host strain, the quality of target protein production is mostly not affected, since, in recombinant expression, the cellular resources are mainly directed to the production of the target protein41,42. Therefore, a tightly regulated, inducible expression system and strong promoters are crucial for high incorporation efficiency43. Our approach is based on multiple residue-specific incorporation of ncAAs in response to sense codons (sense codon reassignment), whereby within the target gene, the number of positions for Pro analog insertion can be manipulated via site-directed mutagenesis44. A similar approach was applied in our previous report on the preparation of recombinant peptides with antimicrobial properties45. In this work, we have applied the SPI method, which allows all proline residues to be replaced by related analogs, to generate proteins expected to possess distinct physicochemical properties not present in proteins synthesized with the canonical amino acid repertoire. By characterizing the folding and fluorescence profile of resulting variants, we aim to showcase the effects of atomic substitutions in variants of GFP.
Protocol
1. Introduction of expression plasmids into competent Pro-auxotrophic E. coli cells
- Mix 1 ng of each sample plasmid, pQE-80L H6-EGFP, pQE-80L H6-NowGFP, and pQE-80L H6-KillerOrange, with 50 µL of chemically competent or electrocompetent cells of the Pro-auxotrophic E. coli K12- derived strain JM83 (Addgene #50348, or ATCC #35607) for transformation by the heat shock method or electroporation according to the protocols available46,47.
NOTE: The expression vector pQE-80L EGFP-H6 encodes a C-terminally 6xHis-tagged enhanced green fluorescent protein (EGFP), the expression vectors pQE-80L H6-NowGFP and pQE-80L H6-KillerOrange encode an N-terminally 6xHis-tagged NowGFP48 or an N-terminally 6xHis-tagged KillerOrange49, respectively, each in a pQE-80L plasmid backbone. All target genes are under the control of a bacterial T5 promoter regulated by the lac operator. Ampicillin resistance was used as a selection marker and colE1 as the origin of replication. See the Table of Materials for further information on the pQE-80L plasmid. Alternative Pro-auxotrophic E. coli originating from strains K-12 and B can be used for expression as well and should yield similar results. The calculated molecular mass of the H6-tagged singly protonated ([M+H]+) wild-type proteins (after chromophore maturation) are 27,745.33 Da (EGFP- H6), 27,931.50 Da (H6-NowGFP), and 27,606.09 Da (H6-KillerOrange). The primary structures of the target proteins are given in Table 1 (His-tag underlined). The glutamine (Q) within the His-tag sequence of NowGFP was identified by DNA sequencing after subcloning of the NowGFP cDNA received from Pletnev et al.51 into the pQE-80L plasmid. It does not impede protein purification or the properties of the fluorescent protein. - Recover the cells in 950 µL of SOC medium at 37 °C for 1 h.
- Spread 50 µL of the recovered cells onto Luria Agar (LA) medium plates (see Supplementary Material) containing glucose (10 g/L) and ampicillin (100 µg/mL).
- Incubate the LA medium plates at 37 °C overnight or 30 °C for 24 h.
2. Production of recombinant wild-type fluorescent proteins (harboring canonical proline) and procedure for selective pressure incorporation (SPI) to produce fluorescent proteins with proline analogs (S-Flp, R-Flp, Dfp, Dhp)
- Overnight culture of Pro-auxotrophic E. coli K12-derived strain JM83 harboring pQE-80L EGFP- H6, pQE-80L H6-NowGFP, and pQE-80L H6-KillerOrange
- Use a sterile pipette tip or inoculation loop to select a single colony from an LA medium plate and resuspend the cells in 5 mL of Lysogeny Broth (LB) medium (see Supplementary Material; containing 10 g/L glucose, 100 µg/mL ampicillin) in a sterile 14 mL polystyrene culture tube.
NOTE: Freshly transformed colonies are recommended for inoculation. Cells on LA medium plates (from step 2.2.) stored at 4 °C should be used within a few days. - Grow the cell culture overnight at 37 °C in an orbital shaker at 200-250 rpm.
- Use a sterile pipette tip or inoculation loop to select a single colony from an LA medium plate and resuspend the cells in 5 mL of Lysogeny Broth (LB) medium (see Supplementary Material; containing 10 g/L glucose, 100 µg/mL ampicillin) in a sterile 14 mL polystyrene culture tube.
- Production of recombinant EGFP- H6, H6-NowGFP and H6-KillerOrange with native proline and the corresponding protein variants with proline analogs
- Inoculate 200 mL of fresh NMM medium (7.5 mM (NH4)2SO4, 50 mM K2HPO4 and 22 mM KH2PO4, 8.5 mM NaCl, 1 mM MgSO4, 20 mM D-glucose, 1 µg/mL FeCl2, 1 µg/mL CaCl2, 10 µg/mL thiamine, 10 µg/mL biotin, 0.01 µg/mL trace elements (CuSO4, ZnCl2, MnCl2, (NH4)2MoO4), pH ~7.2) with all canonical l-amino acids (50 mg/L); see Supplementary Material) supplemented with 100 µg/mL ampicillin with 2 mL of the overnight culture in a 2-L Erlenmeyer flask.
NOTE: Depending on the type of protein, alternative cultivation media like MOPS medium52, glucose-mineral salts medium53, Davis minimal medium54, M9 minimal medium55, or GMML56 can be tested to optimize protein yield. - Incubate the cells at 37 °C in an orbital shaker at 220 rpm for ~3 h 30 min.
- Measure the optical density at 600 nm (OD600) in a spectrophotometer every 30 min until an OD600 value of ~ 0.7 is reached.
NOTE: For OD600 determination in a spectrophotometer, the cuvette should have a path length of 1 cm. The corresponding cultivation medium is used for the reference ("zero") measurement. The incubation time until an OD600 value of ~ 0.7 is reached may depend on culture volume. 3 h 30 min is an approximate value. In a variation of the protocol substeps 2.2.1-2.2.3., the culture from substep 2.1.2. is used to inoculate fresh NMMΔPro medium (see Supplementary Material) supplemented with a limited concentration of proline (e.g., 5 mg/L instead of 50 mg/L), and the cells are grown overnight at 37 °C in an orbital shaker at 220 rpm. The next day, the OD600 determination is performed every 30 min until the differences between the measurements are less than 0.05. The maximum OD600 value should be around 1 (± 0.3). The initial, limited proline concentration can be adjusted depending on the expression strain and cultivation medium (see Discussion). - Spin down the cell suspension for 10 min at 3,000 x g and 4 °C.
- Gently decant the supernatant into waste.
- Washing step: Resuspend the cells in 50 mL of ice-cold NMMΔAA (without any amino acid, see Supplementary Material) or NMMΔPro (without proline, see Supplementary Material) medium by careful pipetting.
NOTE: For this washing step, either of the two stated buffers can be used, since it is only important to get rid of residual proline in the incubation medium. - Separate the cells from the medium by sedimentation at 4 °C in a centrifuge at 3,000 x g for 10 min.
- Gently decant the supernatant into waste.
- Gently pipette up and down to resuspend the cell pellet in 200 mL of NMMΔPro medium supplemented with 100 µg/mL ampicillin into a 2-L Erlenmeyer flask.
NOTE: The resulting cell suspension can be separated into several samples of equal volume to obtain identical start cultures for comparing protein expression in the presence of Pro or proline analogs (e.g., separation into 4 x 50 mL in 100 mL Erlenmeyer flasks). - Incubate for 30 min at 37 °C in an orbital shaker at 220 rpm for the complete depletion of Pro.
NOTE: This step is critical to completely deplete Pro from cells. - Add an appropriate volume of either L-proline or R-Flp, S-Flp, Dfp, and Dhp from 50 mM stock solution to adjust 1 mM final concentration in the cell suspension.
NOTE: As a general rule, fresh 50 mM stock solutions of Pro or proline derivatives should always be prepared prior to use. Only if hydrolysis of the particular canonical or non-canonical amino acid in an aqueous solution is not a concern, frozen stocks may be used as well. - Add 0.5 mM IPTG from a 1 M stock solution to induce target protein expression.
- Express the target protein overnight (12 h) at 37 °C in an orbital shaker at 220 rpm.
- Measure OD600 on the next day.
NOTE: OD600 determination is done to quantify the amount of cells after protein expression. A lower OD600 value compared to the value measured before the washing step 2.2.3 indicates cytotoxicity of the supplied amino acid. In this case, the procedure should be repeated with the concentration of the supplied amino acid minimized (down to 0.1 mM). - Centrifuge and collect the bacterial cells at 5,000 x g and 4 °C for 10 min and decant the supernatant into waste.
- Washing step: Resuspend the cells by careful pipetting in 50 mL of binding buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT) containing 10% glycerol and transfer the cell suspension into a 50 mL conical polystyrene tube.
- Centrifuge and collect the bacterial cells at 5,000 x g and 4 °C for 10 min and decant the supernatant into waste.
- Store the cell pellet in a 50 mL conical polystyrene tube at -20 °C or -80 °C until further use (protein purification, see below).
- Inoculate 200 mL of fresh NMM medium (7.5 mM (NH4)2SO4, 50 mM K2HPO4 and 22 mM KH2PO4, 8.5 mM NaCl, 1 mM MgSO4, 20 mM D-glucose, 1 µg/mL FeCl2, 1 µg/mL CaCl2, 10 µg/mL thiamine, 10 µg/mL biotin, 0.01 µg/mL trace elements (CuSO4, ZnCl2, MnCl2, (NH4)2MoO4), pH ~7.2) with all canonical l-amino acids (50 mg/L); see Supplementary Material) supplemented with 100 µg/mL ampicillin with 2 mL of the overnight culture in a 2-L Erlenmeyer flask.
3. Purification procedure of protein samples by immobilized metal ion affinity chromatography (IMAC)
- Bacterial cell lysis
NOTE: Perform all steps of cell lysis on ice or at 4 °C to prevent degradation of the target protein.- Thaw the bacterial cell pellet on ice or at 4 °C in a 50 mL conical polystyrene tube for 10-20 min.
- Add 10 mL of ice-cold binding buffer (see Supplementary Material) and gently pipette up and down to resuspend the cell pellet.
- Add 100 µL of 50 mg/mL lysozyme, 100 µL of 1 mg/mL DNase I, 100 µL of 1 mg/mL RNase A, 30 µL of 1 M MgCl2. Carefully invert the cell suspension five times, and keep the closed tube on ice or at 4 °C for 60 min.
NOTE: Lysozyme induces chemical cell lysis by disrupting the bacterial cell wall. - Sonicate the sample for cell disruption using an ultrasound homogenizer (e.g., 3 times for 3 min in a 50 mL polystyrene tube on an ice-water mixture, pulse 2 s/pause 4 s, 45% amplitude).
NOTE: Alternatively, other cell disruption methods can be used, e.g., high-pressure homogenization in 20 cycles at 14,000 psi. If necessary, dilute using a binding buffer (see Supplementary Material) to reach the minimal instrument volume. Moreover, protein extraction reagents can be used for cell disruption. See the Table of Materials for examples. - Centrifuge for 60 min at 18,000 x g, 4 °C.
- Note down the liquid volume for substep 3.1.9. and pour the supernatant into a fresh 50 mL polystyrene tube.
- Clear the supernatant using a membrane filter with 0.45 µm pore diameter.
- Take a sample of "lysate" for SDS-PAGE (see section 4. below); this corresponds to "soluble protein fraction", the supernatant from substep 3.1.6.
- Add an equal volume of ddH2O as determined in substep 3.1.6. to resuspend the cell debris in order to maintain the same dilution of the samples for subsequent SDS-PAGE analysis.
- Take a sample of "pellet" for SDS-PAGE (see section 4. below); this corresponds to "insoluble protein fraction", the cell debris suspension from substep 3.1.9.
- Immobilized metal affinity chromatography (IMAC) purification
NOTE: Purification of the target fluorescent protein can be performed at 4 °C or at room temperature (RT). For the latter option, wait for the lysate, column, and all buffers to equilibrate at RT to prevent air bubble formation due to vaporization of air trapped in cold solution upon placement into a warm column.- Purify the sample using a 1 mL prepacked or self-packed IMAC FPLC (fast protein [or performance] liquid chromatography) column according to the manufacturer's instructions; set maximum column pressure to 0.3 MPa, and flow rate to 1 mL/min; use binding buffer (see Supplementary Material) for column equilibration, wash buffer (see Supplementary Material) for the wash step, and elution buffer (see Supplementary Material) for target protein elution.
NOTE: Alternatively, an automated FPLC system can be applied to elute the target protein with elution buffer running a linear imidazole concentration gradient (20-200 mM). - Collect and pool the eluate fractions with fluorescent proteins (choose by visible green or orange color).
- Transfer the pooled fractions into a dialysis membrane (molecular weight cutoff (MWCO) of 5,000-10,000) according to the manufacturer's instructions and dialyze at least three times against dialysis buffer or MS buffer (see Supplementary Material). For instance, perform dialysis of a 1-mL sample three times against 100 mL of buffer for at least 2 h each round. For a detailed protocol, refer to Budisa et al.34.
- Prepare a 1:100-fold dilution of the dialyzed elution fraction in PBS buffer (see Supplementary Material).
- Record the absorbance spectrum of the diluted samples in a UV-Vis spectrophotometer.
- Calculate the protein concentration based on the Lambert-Beer law as follows, using literature values of the molar extinction coefficients ε at specific wavelength (for EGFP at 488 nm ε488 = 55,000 cm-1·M-1, NowGFP at 493 nm ε493 = 53,600 cm-1·M-1, KillerOrange at 514 nm ε514 = 22,600 cm-1·M-1):
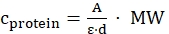 (Lambert-Beer law)
(Lambert-Beer law)
cprotein = protein concentration [mg/mL]
A = absorbance at specific wavelength
ε = molar extinction coefficient at specific wavelength [M-1·cm-1]
d = cuvette path length, here 1 cm
MW = molecular weight of protein [g/mol]
Use dialysis buffer (see Supplementary Material) for the reference ("zero") measurement. - Take a sample of "eluate" for SDS-PAGE (see section 4 below), load 1-10 µg of protein (calculated according to the previous step) per sample well for Coomassie Brilliant Blue-stained gels.
NOTE: Adjust SDS sample amounts depending on the applied staining method and sensitivity of the dye if compounds different from Coomassie Brilliant Blue are used for protein band staining. - Freeze and store the protein sample in dialysis buffer (see Supplementary Material) at -80 °C.
NOTE: Under this storage condition, protein samples should be stable for at least 6 months. Alternative laboratory UV-Vis and fluorescence spectrophotometers can be used for the recording of absorption and fluorescence emission spectra of target proteins. The following excitation wavelengths can be applied for fluorescence emission measurements: 488 nm (EGFP), 493 nm (NowGFP), and 510 nm (KillerOrange).
- Purify the sample using a 1 mL prepacked or self-packed IMAC FPLC (fast protein [or performance] liquid chromatography) column according to the manufacturer's instructions; set maximum column pressure to 0.3 MPa, and flow rate to 1 mL/min; use binding buffer (see Supplementary Material) for column equilibration, wash buffer (see Supplementary Material) for the wash step, and elution buffer (see Supplementary Material) for target protein elution.
4. SDS-PAGE sample preparation
- Determine absorbance A at 280 nm (A280nm) for samples "eluate", "lysate" and "pellet" from section 3. Adjust the probe volume to achieve A280nm = 2 by adding an appropriate amount of elution buffer (final probe volume should be at least 80 µL).
- Mix the sample at a ratio of 4:1 (v/v) with 5x SDS loading dye buffer (see Supplementary Material) by pipetting, e.g., 80 µL of the sample with 20 µL of 5x SDS loading buffer.
- Boil the SDS samples at 95 °C for 5 min in a water bath or heat block to denature the proteins.
- Allow the samples to cool down to RT and spin down the probes at 13,000 x g for 1 min in a microcentrifuge prior to loading onto the gel.
- Use 5-10 µL for Coomassie Brilliant Blue-stained SDS-PAGE. For details of the SDS-PAGE procedure, consult57.
NOTE: Adjust the SDS sample amounts depending on the applied staining method and sensitivity of the dye. The result of the SDS-PAGE should be checked carefully to ensure that the samples contain more than 95% of the total protein in a band corresponding to the expected molecular weight of the desired protein. For this, take a photograph of the Coomassie-stained gel and compare the intensity of the protein band of desired molecular weight with the intensity of all other bands in the lane (if any) by densitometry. For densitometric evaluation of band intensities, the software ImageJ can be used58. To experimentally prove the incorporation of the desired ncAAs into the protein of interest, intact protein mass analysis by high-performance liquid chromatography (HPLC) coupled to electrospray ionization time-of-flight mass spectrometry (LC-ESI-TOF-MS) should be carried out as described59 (see protocol section 5 and Table of Materials therein for exemplary equipment).
5. Fluorescence emission of protein variants
- Prior to the procedure, check the result from the SDS-PAGE experiment to make sure that sample purity is >95% (see the NOTE after step 4.5.)
- Adjust the samples of each purified protein variant to a concentration of 0.3 µM, taking the calculated absorbance value at the appropriate wavelength as a reference (substep 3.2.5.). Ensure that the approximate final sample volume is 200 µL.
- Let the diluted samples equilibrate for 1 h at RT.
- Transfer the samples into a 1-cm quartz cuvette and measure a fluorescence emission spectrum of the samples using a fluorescence spectrometer (see Table of Materials) applying the following excitation wavelengths: 488 nm (for EGFP), 493 nm (for NowGFP), 510 nm (for KillerOrange).
6. Denaturation and refolding of EGFP variants
- Prior to the procedure, check the result from the SDS-PAGE experiment to make sure that sample purity is >95% (see the NOTE after step 4.5.)
- Prepare for each purified protein variant two samples of 2 µL final volume at a concentration of 300 µM (see protein concentration determination in substeps 3.2.4-3.2.6).
- Add 18 µL of 1.11x PBS buffer (see Supplementary Material) containing 8.89 M urea and 5.56 mM DTT to 2 µL of each purified protein variant (to obtain 1x PBS containing 8 M urea and 5 mM DTT) to induce denaturation.
NOTE: For steps 6.4-6.6, process each sample separately. - Incubate the samples for 5 min at 95 °C.
- Dilute the 20 µL samples 100-fold by adding 1980 µL of 1x PBS (see Supplementary Material) containing 5 mM DTT to induce renaturation, yielding 0.3 µM final protein concentration and immediately transfer 200 µL of the samples into a 1-cm quartz cuvette.
NOTE: It is very important to work fast here since renaturation starts immediately. - Insert the quartz cuvette into an appropriate fluorescence spectrometer (see Table of Materials) and monitor protein refolding in the samples by acquiring a fluorescence spectrum every 3 s over 30 min. For each protein variant, use 295 nm fluorescence excitation for the first sample and 488 nm fluorescence excitation for the second one.
- Transfer the refolding samples into 1.5 mL microcentrifuge tubes, close the lid and store the samples at RT in the dark for 24 h to allow complete refolding of EGFP variants.
- Measure fluorescence emission of refolded protein samples according to step 6.6 using the same excitation wavelength as before to capture the temporal endpoint of fluorescence recovery.
Representative Results
At the beginning of the study, we selected three different fluorescent protein variants sharing the parent GFP architecture. The first protein selected was EGFP, which is an engineered variant derived from the original GFP from the jellyfish Aequorea victoria containing Phe64Leu/Ser65Thr mutations. The second selected protein was NowGFP51,60. It is also a variant of A. victoria GFP derived by mutagenesis in several steps via preceding fluorescent proteins. NowGFP contains 18 mutations compared to its immediate predecessor fluorescent protein "Cerulean"61. In turn, the "Cerulean" protein is a derivative of the enhanced cyan fluorescent protein (ECFP)62,63, a protein previously selected by directed laboratory evolution and containing a tryptophan-based chromophore. Both, EGFP and NowGFP are widely used in cell biology and biophysical studies, and they contain ten conserved proline residues in their structures. In addition, NowGFP has an eleventh proline residue at position 230, which appeared due to the extensive mutation history of this protein variant. The third protein selected was the KillerOrange fluorescent protein64,65. It is a derivative of the chromoprotein anm2CP from the hydrozoan genus Anthoathecata. The protein sequence contains 15 proline residues, and the chromophore is based on a tryptophan rather than a tyrosine residue. High-resolution X-ray structures have been reported for all three selected proteins (Figure 2)51,65,66.
In the first step, proline analogs (Figure 1D) were incorporated into all proline positions of three model proteins (EGFP, NowGFP, and KillerOrange) by selective pressure incorporation (SPI, a scheme of the procedure is given in Figure 3). Instrumentally, the proline-auxotrophic E. coli K12 strain JM8367 was used for expression of the proteins in the presence of proline and analogs (Figure 1D), yielding wild-type and modified proteins, respectively. Pellets from cells expressing the native protein and variants bearing S-Flp and Dhp had the typical bright color due to the intact chromophore, whereas variants containing R-Flp and Dfp remained colorless, indicating misfolding and deposition of unfolded protein in inclusion bodies (Figure 4A). SDS-PAGE analysis of the expressed samples verified the presence of insoluble R-Flp-containing proteins (Figure 4B–D), which precluded further investigations. Although this is beyond the scope of the present study, it should be noted that protein solubility and misfolding issues can be alleviated to some extent by in vitro refolding procedures68. In contrast, native proteins as well as S-Flp- and Dhp-bearing variants were found mainly in the soluble fractions (Figure 4B–D). The wild-type, as well as S-Flp- and Dhp-containing variants, could be further isolated and characterized in fluorescence studies. Soluble proteins were purified by immobilized metal ion affinity chromatography (IMAC), yielding 20-30 mg/L of culture volume for EGFP, 60-80 mg for NowGFP and KillerOrange, whose yields for wild-type and modified proteins were very similar. Liquid chromatography-mass spectrometry (LC-MS)-coupled analysis confirmed the expected identity and purity of the isolates obtained in this fashion (Figure 5). In the mass spectra, each proline replacement with S-Flp produced a +18 Da shift per each proline residue in the sequence, while for the proline-to-Dhp replacement, the shift was −2 Da per residue.
In the next step, light absorption and emission spectra were recorded to analyze the potential effects of non-canonical proline analogs incorporation on the spectroscopic properties of the parent fluorescent proteins (Figure 6). UV-Vis absorption spectra showed a typical band around 280 nm characteristic for aromatic residues, tyrosine, and tryptophan, while the chromophore absorbance was found at 488 nm for EGFP, and 493 nm for NowGFP (Figure 6A,B). In KillerOrange, the chromophore absorbance region comprised two bands (Figure 6C), which correspond to two possible configurational and charge states of the complex chromophore. The band around 510 nm is known as the state from which fluorescence occurs with high quantum yield49,65. In the proline replacement variants, the following was observed: Incorporation of Dhp did not change the absorbance spectra of EGFP and NowGFP, while S-Flp produced an enhanced UV absorption. The latter can be explained by induced differences in the tryptophan residue microenvironments, particularly Trp57 sandwiched between three S-Flp in the PVPWP motif (Figure 6A,B)69. A more trivial explanation for a higher UV absorption, however, may stem from an increased fraction of improperly folded protein. Since the concentration of the protein was assessed by quantification of absorbance features, the presence of a protein with an improperly mature chromophore can increase the absorbance, while this fraction is not counted in the overall concentration (Figure 6A,B). Supporting this hypothesis, we observed that the S-Flp-containing EGFP exhibited a markedly reduced ratio of chromophore versus combined tryptophan and tyrosine absorbance (ε(CRO)/ε(Tyr+Trp) = 0.96) as compared to a higher value (1.57) in the parent protein (Table 2)70. The presence of a non-fluorescent fraction in the S-Flp-containing EGFP will be an important contributing factor in further analysis of the protein properties. In the KillerOrange variant containing S-Flp, an enhanced absorbance alongside a red-shift in the chromophore band was observed. This fact indicated that the chromophore formation favored a configuration with a large fluorescence quantum yield (Figure 6C).
Subsequently, we analyzed the fluorescence spectra of the proteins recorded upon excitation at the corresponding maximum absorbance wavelengths. The results show that the spectra remained essentially identical for the examined fluorescent protein variants bearing proline and replacements, S-Flp and Dhp. This outcome implies that the analogs did not alter the chemical environment of the chromophore in any case (Figure 6G–I). Despite this fact, marked differences were seen in the fluorescence spectra of KillerOrange recorded upon excitation at 295 nm, hence upon tryptophan excitation. This experiment tracks fluorescence resonance energy transfer (FRET) or direct excitonic coupling that occurs between the tryptophan side chains and the mature chromophore as both are located at a short distance of not more than 25 Å. For EGFP and NowGFP variants, when the emission spectra were measured using 295 nm excitation, a strong chromophore emission was observed alongside hardly any tryptophan emission (Figure 6D,E). However, the variants containing S-Flp exhibited a slightly larger tryptophan-specific emission. This observation can be linked to an uncounted contribution of the unfolded apoprotein that contains tryptophans but not the mature chromophore. Substantially increased tryptophan-specific emission was seen in KillerOrange, indicating a lack of fluorescence quenching via the expected mechanism of excitation energy transfer or excitonic coupling. The protein variants containing proline and S-Flp exhibited comparable tryptophan emission alongside the favored red-shifted fluorescence feature of a high quantum yield. In contrast, the variant that contained Dhp showed a drastic decrease in chromophore fluorescence intensity, presumably due to minor structural effects (Figure 6F).
Next, we compared the folding properties of the proteins by performing an unfolding/renaturation experiment. Fluorescence emission spectra were recorded in the folded state (protocol section 5), after chemical denaturation and, subsequently, in the process of refolding monitored over a period of 24 h (protocol section 6). The spectra were recorded upon excitation at both relevant wavelengths, 295 nm, and at the maxima of the chromophores' absorbance spectra, while the resulting fluorescence is presented as normalized to the maximum value for each protein (Figure 7). At the end of the protocol, we observed that EGFP variants could refold, while the NowGFP and KillerOrange variants – once denatured – remained unfolded (data not shown). Thus, refolding capacities of the original fluorescence proteins varied substantially. Of note, KillerOrange has been developed as a photosensitizer starting from the hydrozoan chromoprotein variant KillerRed65,71, and its refolding typically lags behind in spite of the robust β-barrel structure. In our experiments, we found that the wild-type EGFP chromophore fluorescence recovered only partially, although the tryptophan-specific fluorescence was larger after renaturation (Figure 7A,D). Essentially similar behavior was observed in the variant containing Dhp (Figure 7C,F). In S-Flp-containing EGFP, a similar result was observed when the excitation was performed at the tryptophan-specific wavelength of 295 nm (Figure 7B). Strikingly, the fluorescence recovered to a much higher extend when the chromophore was excited at 488 nm (Figure 7E). It seems that S-Flp induces a much better yield of refolding compared to the other two variants. However, this beneficial effect was not seen when using 295 nm excitation due to unknown molecular interactions.
Subsequently, refolding velocity was monitored by recording fluorescence of both tryptophan, and the chromophore, separately, while the endpoint of the process was determined at 24 h after the start of renaturation. Only EGFP variants showed a relatively fast refolding kinetics that could be evaluated reliably, while none of the denatured NowGFP and KillerOrange variants could recover to a value that enabled further quantitative measurements. In EGFP, tryptophan emission recovery was twice as fast (completed in 750 s) compared to the recovery of chromophore emission (completed in 1,500 s), indicating the complexity of the underlying processes (Figure 8). At both excitation wavelengths, the refolding rate was elevated by the presence of S-Flp, in agreement with literature data25. At the same time, the Dhp-containing variant showed a refolding profile similar to wild-type.
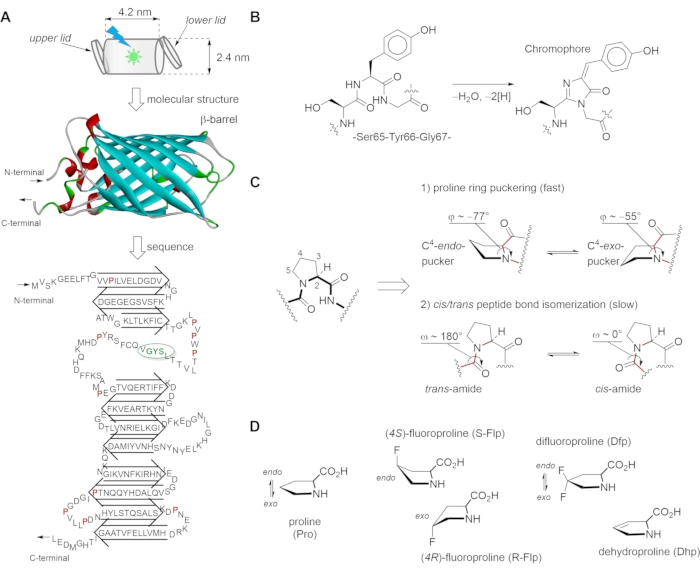
Figure 1: Green fluorescent protein (GFP) structural scaffold, chromophore building, proline conformational transitions and synthetic analogs used in this study. (A) The structure of GFP consists of the β-strands forming a nearly perfect barrel (i.e., a "can" with dimensions 4.2 nm x 2.4 nm) that is capped at both ends by α-helical lids. The 27 kDa GFP protein shows a tertiary structure consisting of eleven β-strands, two short α-helices, and the chromophore in the middle. The conformational states of adjacent prolines are linked to chromophore formation. (B) Autocatalytic maturation (condensation) of the chromophore occurs at residues Ser65, Tyr66, and Gly67, and proceeds in several steps: First, torsional adjustments in the polypeptide backbone to bring the carboxyl carbon of Thr65 into proximity to the amide nitrogen of Gly67. Then, the formation of a heterocyclic imidazoline-5-one ring system occurs upon nucleophilic attack on this carbon atom by the amide nitrogen of glycine and subsequent dehydration. Finally, the system gains visible fluorescence when oxidation of the tyrosine alpha-beta carbon bond by molecular oxygen leads to the extension of the conjugated system of the imidazoline ring system, at the end including the tyrosine phenyl ring and its para-oxygen substituent. The resulting para-hydroxybenzylidene imidazolinone chromophore in the center of the β-barrel is completely separated from the bulk solvent. (C) The skeletal structure formulas and geometries of 1) the proline ring (puckers) and 2) the preceding amide bond represents the main conformational transitions of the proline residue. (D) The proline analogs used in this work with the designated proline ring puckers. The figure was generated using ChemDraw and Discovery Studio Visualizer. The GFP structure is from PDB structure entry 2Q6P. Please click here to view a larger version of this figure.
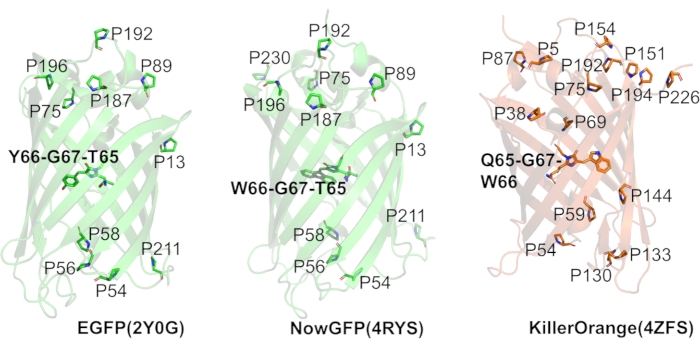
Figure 2: Fluorescent proteins used in this study. The panels show the ribbon representation of the typical β-barrel structures of three different variants of fluorescent proteins: EGFP, NowGFP, and KillerOrange, with ribbon color representing the color of fluorescence emission of each variant. Proline residues (one-letter code) are highlighted as sticks, and the appropriate positions are annotated. Chromophores are shown with initial amino acid composition in bold. All structure representations were produced with PyMol based on the following PDB structure entries: 2Q6P for EGFP, 4RYS for NowGFP, 4ZFS for KillerOrange. Please click here to view a larger version of this figure.
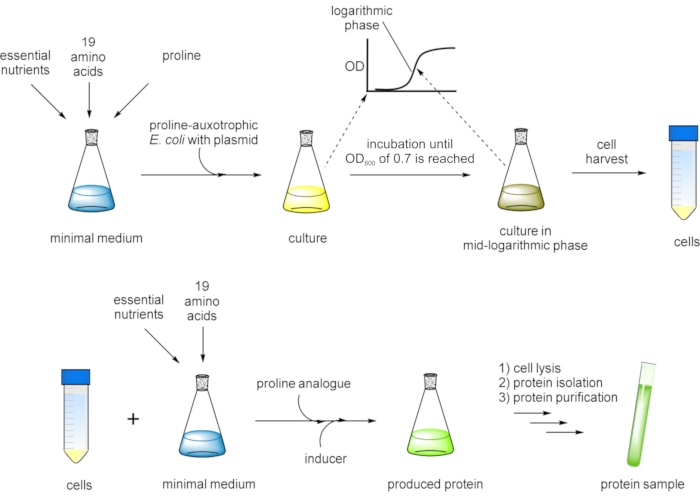
Figure 3: Flow chart presentation of the SPI method for residue-specific incorporation of non-canonical proline analogs. A proline-auxotrophic Escherichia coli (E. coli) host strain carrying the gene of interest on an expression plasmid is grown in a defined minimal medium with all 20 canonical amino acids until an OD600 of ~0.7 is reached at which the cell culture is in the mid-logarithmic growth phase. Cells are harvested and transferred into fresh minimal medium containing 19 canonical amino acids and a proline analog. After the addition of an inducer, protein expression is performed overnight. Finally, the target protein is isolated by cell lysis and purified prior to further analysis. In a variation of the protocol, the cells are grown in a defined minimal medium with 19 canonical amino acids, and proline is added in a limited amount (e.g., one-fifth of the concentration of the other amino acids). By this measure, the cells exhaust proline in the medium before they can exit the logarithmic growth phase, and then, subsequently, the analog is added, and the protein of interest production is induced. Please click here to view a larger version of this figure.
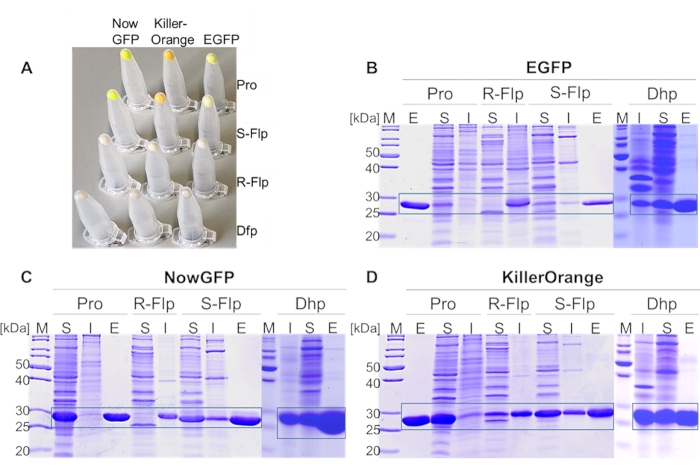
Figure 4: Expression analysis of EGFP, NowGFP, and KillerOrange variants. (A) Cell pellets from 1 mL of expression culture, normalized to OD600 = 2. SDS-PAGE analysis of (B) EGFP, (C) NowGFP, and (D) KillerOrange variants. Soluble (S) and insoluble fractions (I) of each fluorescent protein derivates were loaded on 15% acrylamide gel, as well as eluted fractions (E) from IMAC of soluble proteins. PageRuler Unstained Protein Ladder was used as a marker (M) in the lanes denoted by (M). The expected regions of the particular protein are framed. Incorporated amino acids at proline positions are Pro, R-Flp, S-Flp, and Dhp (in (A) cell pellets from fluorescent protein variants incorporating Dfp instead of Dhp are shown). Gels were stained by 1% (w/v) Coomassie Brillant Blue. Please click here to view a larger version of this figure.
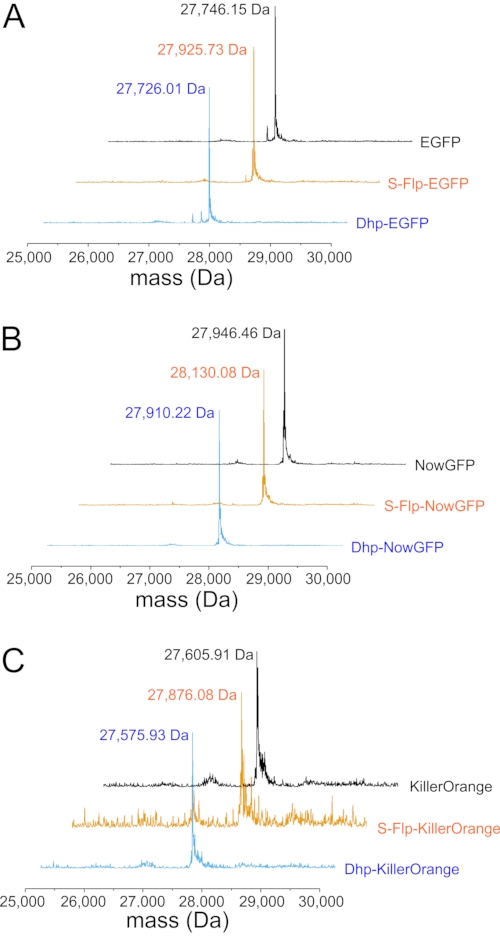
Figure 5: Mass spectrometric analysis of fluorescent protein variants. (A) Representative deconvoluted ESI-MS spectra of H6-tagged EGFP (black), S-Flp-EGFP (orange), and Dhp-EGFP (cyan) with the location of the main mass peaks provided as numbers (in Da). The calculated molecular masses [M+H]+ of the H6-tagged proteins are: For EGFP 27,745.33 Da (observed 27,746,15 Da); for S-Flp-EGFP 27,925.33 Da (observed 27,925.73 Da); for Dhp-EGFP 27,725.33 Da (observed 27,726.01 Da). (B) Representative deconvoluted ESI-MS spectra of H6-tagged NowGFP (black), S-Flp-NowGFP (orange), and Dhp-NowGFP (cyan) with the location of the main mass peaks provided as numbers (in Da). The calculated masses of the H6-tagged proteins are: For NowGFP 27,931.50 Da (observed 27,946.46 Da; the difference of ~16 Da is probably due to oxidation of a methionine in the protein); for S-Flp-NowGFP 28,129.50 Da (observed 28,130.08 Da); for Dhp-NowGFP 27,909.50 Da (observed 27,910.22 Da). (C) Representative deconvoluted ESI-MS spectra of H6-tagged KillerOrange (black), S-Flp-KillerOrange (orange), and Dhp-KillerOrange (cyan) with the location of the main mass peaks provided as numbers (in Da). The calculated masses of the H6-tagged proteins are: For KillerOrange 27,606.09 Da (observed 27,605.91 Da); for S-Flp-KillerOrange 27,876.09 Da (observed 27,876.08 Da); for Dhp-KillerOrange 27,576.09 Da (observed 27,575.93 Da). Deviations between the observed and calculated molecular masses of about 1 Da are within the error range of the ESI-MS equipment. Please click here to view a larger version of this figure.
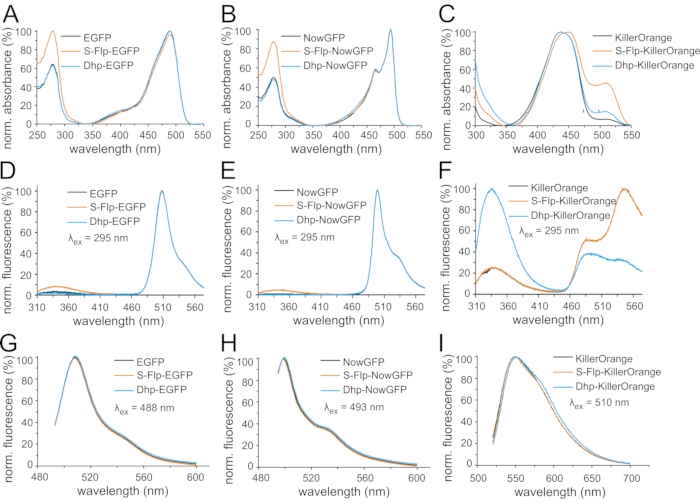
Figure 6: Light absorption and fluorescence emission spectra of fluorescent protein variants. Normalized UV-Vis absorption spectra are shown for the variants (A) of EGFP, (B) of NowGFP, and (C) of KillerOrange. Spectra were normalized to the maximum of chromophore absorbance (around 500 nm). Normalized fluorescence emission spectra are shown of the variants (D,G) of EGFP, (E,H) of NowGFP, and (F,I) of KillerOrange. Spectra in (D,E,F) were measured upon excitation with ultraviolet light (295 nm), for the spectra in (G,H,I) 488 nm, 493 nm, and 510 nm light were used for excitation, respectively, and the spectra were normalized to the respective maxima of chromophore emission (around 500 nm). In each panel, black curves correspond to the spectra of the fluorescent protein variant with native proline, orange curves indicate the spectra of S-Flp-substituted proteins, and blue curves correspond to Dhp-substituted proteins. Please click here to view a larger version of this figure.
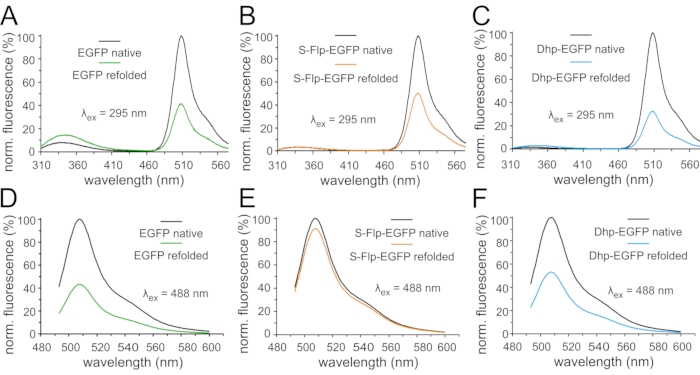
Figure 7: Fluorescence emission spectra of EGFP variants in refolding experiments. Normalized fluorescence emission spectra of 0.3 µM solutions of fluorescent protein variants in the native state and after denaturation and refolding: Spectra in (A,B,C) were measured upon excitation with ultraviolet light (295 nm) (A) for EGFP, (B) for S-Flp-EGFP, and (C) for Dhp-EGFP. Spectra in (D,E,F) were measured upon excitation with green light (488 nm) (D) for EFGP, (E) for S-Flp-EGFP, and (F) for Dhp-EGFP. The emission spectra of the native (black curves) and refolded samples (green corresponds to EGFP, orange to S-Flp-EGFP and blue to Dhp-EGFP, respectively) of each protein variant are normalized to the maximum fluorescence of the appropriate native state. Please click here to view a larger version of this figure.

Figure 8: Monitoring protein folding and chromophore maturation of EGFP variants with fluorescence. (A) Fluorescence emission in the region of Trp fluorescence (emission was set to 330 nm) recorded upon excitation with ultraviolet light (295 nm). (B) Development of the fluorescence amplitude in the region of chromophore emission upon excitation with green light (488 nm). The time-dependent fluorescence traces were normalized to unity (100%) according to the fluorescence amplitude reached at the end of the monitoring interval. In each panel, black curves correspond to the spectra of the fluorescent protein variant with native proline, orange curves indicate the spectra of S-Flp-substituted proteins and blue curves correspond to Dhp-substituted proteins. Please click here to view a larger version of this figure.
| Construct | Amino acid sequences (6xHis tag underlined): | |||
| EGFP-H6 | MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVP WPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTR AEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVN FKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDH MVLLEFVTAAGITLGMDELYKHHHHHH |
|||
| H6-NowGFP | MRGSHHQHHHGSVSKGEKLFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGK MSLKFICTTGKLPVPWPTLKTTLTWGMQCFARYPDHMKQHDFFKSAMPEGY VQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGVDFKEDGNILGHKLEYN AISGNANITADKQKNGIKAYFTIRHDVEDGSVLLADHYQQNTPIGDGPVLLPD NHYLSTQSKQSKDPNEKRDHMVLLEFVTAAGIPLGADELYK |
|||
| H6-KillerOrange | MRGSHHHHHHGSECGPALFQSDMTFKIFIDGEVNGQKFTIVADGSSKFPH GDFNVHAVCETGKLPMSWKPICHLIQWGEPFFARYPDGISHFAQECFPEG LSIDRTVRFENDGTMTSHHTYELSDTCVVSRITVNCDGFQPDGPIMRDQ LVDILPSETHMFPHGPNAVRQLAFIGFTTADGGLMMGHLDSKMTFNGSR AIEIPGPHFVTIITKQMRDTSDKRDHVCQREVAHAHSVPRITSAIGSDQD |
|||
Table 1: Primary structures of the target proteins. His-tags are underlined in each sequence.
| λ [nm] | ε [M-1·cm-1] (EGFP) | ε [M-1·cm-1] (S-Flp-EGFP) | ε [M-1·cm-1] (Dhp-EGFP) |
| 488 (≡ CRO) | 31,657 (± 1,341) | 22,950 (± 290) | 27,800 (± 542) |
| 280 (≡ Tyr+Trp) | 20,116 (± 172) | 23,800 (± 715) | 17,300 (± 554) |
| Values for extinction coefficient ε (in M-1·cm-1) are calculated from recorded UV-Vis absorption spectra of appropriate EGFP variants using known protein concentrations. Selected wavelength at 280 nm corresponds to the maximum absorbance of aromatic residues, tyrosine and tryptophan, and 488 nm represents the chromophore absorbance wavelength. | |||
Table 2: Extinction coefficients (ε) of EGFP variants at selected wavelengths. Values for the extinction coefficient ε (in M-1·cm-1) are calculated from recorded UV-Vis absorption spectra of appropriate EGFP variants using known protein concentrations. The selected wavelength of 280 nm corresponds to the maximum absorbance of aromatic residues, tyrosine, and tryptophan, whereas 488 nm represents the maximum chromophore absorbance wavelength.
Supplementary Material: Preparation of stock solutions and buffers Please click here to download this File.
Discussion
In nature, manipulations with protein structures and functions typically occur due to mutations, the phenomenon that leads to an exchange of an amino acid identity at certain positions in the protein sequence. This natural mechanism is widely applied as a biotechnological method for protein engineering in the form of mutagenesis, and it relies on the repertoire of the 20 canonical amino acids involved in the process. The exchange of proline residues is problematic, however. Due to its special backbone group architecture, it is hardly interchangeable with the remaining 19 residues for replacement72. For example, proline is typically known as a secondary structure breaker in polypeptide sequences because of its poor compatibility with the most common secondary structures, i.e., α-helix and β-strand. This proline feature is easily lost when the residue is mutated to another amino acid from the common repertoire. The replacement of proline with its chemical analogs offers an alternative approach, which enables to keep the basic backbone features of the parent proline residue while imposing bias on its specific conformational transitions or producing modulations of the molecular volume and polarity. For example, it is possible to supply bacterial cultures with analog structures such as hydroxy-, fluoro-, alkyl-, dehydroprolines, structures having variable ring sizes and more, thus facilitating the production of a protein containing specific proline residue alterations.
The selective pressure incorporation (SPI) method described in this study allows for a global, i.e., residue-specific replacement of all prolines in the target protein with related chemical analogs. The importance of the method is reflected by the fact that SPI allows creating sequence changes inaccessible to common mutagenesis techniques. For example, it allows the production of a target protein containing rather small structural changes that may typically not exceed one or two atom replacements/deletions/additions, as demonstrated in this study. Such protein modifications are dubbed "atomic mutations"73,74. In a fluorescent protein such as GFP, the result of this molecular intrusion can be seen in the velocity of folding, local polarities, protein packing, stability of the involved structural features. The changes in the absorbance and fluorescence properties are produced indirectly due to the impact on protein folding and residue microenvironments. The precision of the molecular changes performed by SPI is typically much higher, as compared to mutations of prolines to other canonical residues, the latter being typically detrimental for the protein folding, production, and isolation.
As a production method, the SPI approach uses the substrate tolerance of the aminoacyl-tRNA synthetase pocket towards chemical analogs of the native amino acid. The synthetase is responsible for the correct identification of the amino acid structure, while the incorporation into proteins occurs downstream in the translation process. Instrumentally, the protein production, isolation, and purification in SPI are performed in a way typical for any other recombinant protein expression techniques; however, with some additions to the protocol as follows: Proline, which is bound for replacement, is provided at the beginning of the fermentation process, such that the cells can grow and develop their intact cellular machinery. However, the cell culture is not allowed to reach the maximal optical density, to keep the cells in the logarithmic phase optimal for protein expression. There are two major variations of the SPI method at this point. In the first one, the concentration of proline is adjusted in the initial growth medium (a chemically defined medium) such that depletion of proline happens without any external intrusion. The cells exhaust proline in the medium before they can exit the logarithmic growth phase, and then, subsequently, the analog is added, and the protein of interest production is induced. In the second version of the method, the cells are grown in the medium containing proline until the middle of their logarithmic phase. At this point, the cells should be taken out and physically transferred into another medium, which no longer contains proline, only the analog, with subsequent induction of the protein of interest. In both versions, the analog and the protein induction reagent are provided to the pre-grown cells. The isolation and purification of the wild-type protein are performed in the same way as for the variants. In principle, every available Pro-auxotrophic strain can be used as an expression host. Nevertheless, expression tests to identify the most suitable host are advisable. Also, tests of different chemically defined media can be used to optimize protein yield.
There are certain requirements regarding the chemical analogs that need to be considered for SPI, such as solubility and concentration. The metabolic availability and uptake of amino acids are dependent on the number of dissolved molecules in the medium. To increase the solubility of a particular compound, slightly acidic or alkaline conditions can be chosen. Since the artificial molecules can cause growth inhibitory effects due to their cell toxicity, the concentration should be lowered to a minimum in order to avoid cell stress75.
A minor weakness of SPI is the decrease of incorporation efficiency with larger numbers of positions that need to be exchanged. In principle, a reduction of the amino acid frequency within the target biomolecule by site-directed mutagenesis can solve this problem. However, the structural and functional properties of a desired protein might be affected by changing the primary structure.
As mentioned before, SPI allows residue-specific replacement of the canonical amino acid. This implies that non-canonical amino acids are inserted into every position of the canonical amino acid within the target protein, including conserved residues that are indispensable for protein function or folding. Alternative methods for site-specific incorporation are the only possibility to overcome this issue3. In the past few decades, the orthogonal pair method has been developed that can produce proteins containing modified residues at predefined sites. The most common modification of this method is known as stop codon suppression. This method is based on an engineered orthogonal translation system dedicated for site-specific incorporation of synthetic amino acids76. More than 200 amino acids with different side-chain modifications have been incorporated into proteins to date using this approach77. However, these translation systems are still not suitable for insertions of proline analogs into target proteins. Furthermore, the method's performance is considered low in the case of minor amino acid modifications because some background promiscuity of the aminoacyl-tRNA synthetase typically remains in the engineered translation systems.
Using SPI, we produced a number of β-barrel fluorescent protein variants and studied outcomes of the exchange of proline with its unnatural analogs. In the case of proline replacement with R-Flp and Dfp, a dysfunctional protein was produced by the expression host. The effect is likely produced by protein misfolding. The latter may originate from the C4–exo conformation promoted by R-Flp, which is unfavored by the parent protein structures27. With Dfp, the misfolding is likely to be produced by the diminished velocity of the trans-to-cis peptide bond isomerization at the proline residue27. The latter is known to be among the limiting steps in the kinetic profile of the protein folding that affects β-barrel formation and subsequent chromophore maturation. Indeed, for both amino acids, R-Flp and Dfp, the protein production resulted in an aggregated and insoluble protein. Consequently, chromophore formation could not occur, and the fluorescence was lost entirely. With S-Flp and Dhp, however, proper protein maturation was observed, resulting in fluorescent protein samples for each analog/protein combination. Despite some modulations in the absorbance and fluorescence features of the protein, these largely remained similar to those of the wild-type proteins. The effect of the amino acid substitution was revealed in the refolding kinetics studies. The latter showed a faster refolding in the case of replacement with S-Flp. Model studies have shown that this residue may generate some improvement in the trans-to-cis amide rotation velocity and lead to the formation of the C4–endo conformation. Both these factors are likely to contribute to the beneficial kinetic effects of this residue in EGFP. In contrast, Dhp produced kinetic folding profiles maximally similar to the parent protein. The diversity of the outcomes produced by mere atomic mutations in the examined fluorescent proteins illustrates the potential of the SPI production method in altering target protein properties. The protein alterations induced by proline replacement with the analogs have further implications in the engineering of enzymes78,79,80 and ion channels81,82, as well as in general engineering of protein stability.
The basic limitation of the SPI method is its "all-or-none" mode in exchanging proline resides with related analogs. It would be of great advantage to be able to select precisely, which proline residues should be replaced with the analogs, and which ones should remain unmodified. However, at present, there is no technique that could perform such a sophisticated production using a microbial production host. Chemical synthesis of proteins83,84, as well as cell-free production85,86, are the two alternative methods that can produce position-specific proline modifications. Nonetheless, their operational complexity and low production yields make them inferior compared to the production in living cells. As of now, SPI remains the most operationally simple and robust approach for the production of complex proteins bearing atomic mutations. By introducing unnatural amino acid substitutes, the method allows modifying protein features in a targeted manner, as exemplified here by alterations in folding and light absorption/emission of fluorescent proteins generated by proline replacements.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the German Research Foundation (Cluster of Excellence "Unifying Systems in Catalysis) to T.F. and N.B. and by the Federal Ministry of Education and Science (BMBF Program "HSP 2020", TU-WIMIplus Project SynTUBio) to F.-J.S. and T.M.T.T.
Materials
| Acetonitrile | VWR | HiPerSolv CHROMANORM ULTRA for LC-MS, 83642 | LC-MS grade required |
| Acrylamide and bisacrylamide aqueous stock solution at a ratio of 37.5:1 (ROTIPHORESE Gel 30) | Carl Roth | 3029.1 | |
| Agar-agar | Carl Roth | 5210 | |
| Ammonium molybdate ((NH4)2MoO4) | Sigma-Aldrich | 277908 | |
| Ammonium peroxydisulphate (APS) | Carl Roth | 9592.2 | ≥98 %, p.a., ACS grade required |
| Ammonium sulfate ((NH4)2SO4) | Sigma-Aldrich | A4418 | |
| Ampicillin sodium salt | Carl Roth | K029 | |
| Biotin | Sigma-Aldrich | B4501 | |
| Bromophenol blue | Sigma-Aldrich | B0126 | |
| Calcium chloride (CaCl2) | Sigma-Aldrich | C5670 | |
| Coomassie Brillant Blue R 250 | Carl Roth | 3862 | |
| Copper sulfate (CuSO4) | Carl Roth | CP86.1 | |
| D-glucose | Carl Roth | 6780 | |
| 1,2-Bis-(dimethylamino)-ethane, N,N,N',N'-Tetramethylethylenediamine (TEMED) | Carl Roth | 2367.3 | ≥99 %, p.a., for electrophoresis |
| 1,4-dithiothreitol (DTT) | Carl Roth | 6908 | |
| Dichloromethane (DCM) | Sigma-Aldrich | 270997 | |
| di-potassium hydrogen phosphate (K2HPO4) | Carl Roth | P749.1 | |
| di-sodium hydrogen phosphate (Na2HPO4) | Carl Roth | X987 | |
| DNase I | Sigma-Aldrich | D5025 | |
| Dowex 50WX8-100 (hydrogen form) | Acros Organics / Thermo Fisher Scientific (Waltham, U.S.A.) | 10731181 | cation exchange resin |
| Ethanol | Carl Roth | 9065.1 | |
| Formic acid | VWR | HiPerSolv CHROMANORM for LC-MS, 84865 | LC-MS grade required |
| Glacial acetic acid | Carl Roth | 3738.5 | 100 %, p. a. |
| Glycerol | Carl Roth | 3783 | |
| Imidazole | Carl Roth | X998 | |
| Hydrogen chlroide (HCl) | Merck | 295426 | |
| Iron(II) chloride (FeCl2) | Sigma-Aldrich | 380024 | |
| Isopropanol | Carl Roth | AE73.1 | |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Sigma-Aldrich | I6758 | |
| Lysozyme | Sigma-Aldrich | L6876 | |
| Magnesium chloride (MgCl2) | Carl Roth | KK36.1 | |
| Magnesium sulfate (MgSO4) | Carl Roth | 8283.2 | |
| Manganese chloride (MnCl2) | Sigma-Aldrich | 63535 | |
| β-mercaptoethanol | Carl Roth | 4227.3 | |
| PageRuler Unstained Protein Ladder | Thermo Fisher Scientific | 26614 | |
| Potassium chloride (KCl) | Carl Roth | 6781.3 | |
| Potassium dihydrogen phosphate (KH2PO4) | Sigma-Aldrich | P5655 | |
| RNase A | Carl Roth | 7156 | |
| Sodium chloride (NaCl) | Carl Roth | P029 | |
| Sodium dihydrogen phosphate (NaH2PO4) | Carl Roth | T879 | |
| Sodium dodecyl sulphate (NaC12H25SO4) | Carl Roth | 0183 | |
| Thiamine | Sigma-Aldrich | T4625 | |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich | T6508 | |
| Tris hydrochloride (Tris-HCl) | Sigma-Aldrich | 857645 | |
| Tris(hydroxymethyl)-aminomethane (Tris) | Carl Roth | 5429 | |
| Tryptone | Carl Roth | 8952 | |
| Yeast extract | Carl Roth | 2363 | |
| Zinc chloride (ZnCl2) | Sigma-Aldrich | 229997 | |
| L-alanine | Sigma-Aldrich | A7627 | |
| L-arginine | Sigma-Aldrich | A5006 | |
| L-asparagine | Sigma-Aldrich | A8381 | |
| L-aspartic acid | Sigma-Aldrich | A0884 | |
| L-cysteine | Sigma-Aldrich | C7352 | |
| L-glutamic acid | Sigma-Aldrich | G2128 | |
| L-glutamine | Sigma-Aldrich | G3126 | |
| L-glycine | Sigma-Aldrich | G7126 | |
| L-histidine | Sigma-Aldrich | H8000 | |
| L-isoleucine | Sigma-Aldrich | I2752 | |
| L-leucine | Sigma-Aldrich | L8000 | |
| L-lysine | Sigma-Aldrich | L5501 | |
| L-methionine | Sigma-Aldrich | M9625 | |
| L-phenylalanine | Sigma-Aldrich | P2126 | |
| L-proline | Sigma-Aldrich | P0380 | |
| L-serine | Sigma-Aldrich | S4500 | |
| L-threonine | Sigma-Aldrich | T8625 | |
| L-tryptophan | Sigma-Aldrich | T0254 | |
| L-tyrosine | Sigma-Aldrich | T3754 | |
| L-valine | Sigma-Aldrich | V0500 | |
| (4S)-fluoroproline | Bachem | 4033274 | Make sure that all proline analogs are proline free, check content. Otherwise include a step to consume proline contaminations during expression. |
| (4R)-fluoroproline | Bachem | 4033275 | Make sure that all proline analogs are proline free, check content. Otherwise include a step to consume proline contaminations during expression |
| 3,4-dehydroproline | Bachem | 4003545 | Make sure that all proline analogs are proline free, check content. Otherwise include a step to consume proline contaminations during expression |
| 4,4-difluoroproline | Enamine | EN400-17448 | Make sure that all proline analogs are proline free, check content. Otherwise include a step to consume proline contaminations during expression |
| Conical polystyrene (Falcon) tubes, 15 mL | Fisher Scientific | 14-959-49B | |
| Conical polystyrene (Falcon) tubes, 50 mL | Fisher Scientific | 14-432-22 | |
| Dialysis membrane, Molecular Weight Cut-Off (MWCO) 5,000 | Spectrum Medical Industries | Spectra/Por MWCO 5000 dialysis membrane, 133198 | |
| Immobilized Metal ion Affinity Chromatography (IMAC) column 1 mL, Ni-NTA | GE Healthcare | HisTrap HP, 1 mL, 17-5247-01 | |
| Luer-Lock syringe, 5 mL | Carl Roth | EP96.1 | |
| Luer-Lock syringe, 20 mL | Carl Roth | T550.1 | |
| Luer-Lock syringe, 50 mL | Carl Roth | T552.1 | |
| Microcentrifuge tubes, 1.5 mL | Eppendorf | 30120086 | |
| Petri dishes (polystyrene, sterile) | Carl Roth | TA19 | |
| pQE-80L plasmid vector | Qiagen | no longer available | replaced by N-terminus pQE Vector set Cat No./ID: 32915 |
| Pro-auxotrophic E. coli strain JM83 | Addgene | 50348 | https://www.addgene.org/50348/ |
| Pro-auxotrophic E. coli strain JM83 | ATCC | 35607 | |
| Round-bottom polystyrene tubes, 14 mL | Fisher Scientific | Corning Falcon, 14-959-1B | |
| Syringe filter 0.45 µm with polyvinylidene difluoride (PVDF) membrane | Carl Roth | CCY1.1 | |
| High-Performance Liquid Chromatography (HPLC) column for LC-ESI-TOF-MS | Sigma-Aldrich | Supelco Discovery BIO Wide Pore C5 HPLC column, 3 µm particle size, 10 cm x 2.1 mm | with conical 0.1 mL glass inserts, screw caps and septa |
| HPLC autosampler vials 1.5 mL | Sigma-Aldrich | Supelco 854165 | |
| Mass spectrometer for LC-ESI-TOF-MS | Agilent | Agilent 6530 Accurate-Mass QTOF | |
| Mass spectrometry data analysis software | Agilent | MassHunter Qualitative Analysis software v. B.06.00 | |
| Benchtop centrifuge for 1.5 mL Eppendorf tubes | Eppendorf | 5427 R | |
| Cooling centrifuge for 50 mL Falcon tubes | Eppendorf | 5810 R | |
| Fast Protein Liquid Chromatography (FPLC) system | GE Healthcare | ÄKTA pure 25 L | |
| Fluorescence spectrometer | Perkin Elmer | LS 55 | |
| High pressure microfluidizer for bacterial cell disruption | Microfluidics | LM series with “Z” type chamber | |
| Orbital shaker for bacterial cultivation | Infors HT | Minitron | |
| Peristaltic pump for liquid chromatography (LC) | GE Healthcare | P-1 | |
| Ultrasonic homogenizer for bacterial cell disruption | Omnilab | Bandelin SONOPULS HD 3200, 5650182 | with MS72 sonifier tip |
| UV-Vis spectrophotometer | Biochrom | ULTROSPEC 2100 | |
| UV-Vis/NIR spectrophotometer | Perkin Elmer | LAMBDA 950 UV/Vis/NIR |
References
- Agostini, F., Völler, J. -. S., Koksch, B., Acevedo-Rocha, C. G., Kubyshkin, V., Budisa, N. Biocatalysis with unnatural amino acids: Enzymology meets xenobiology. Angewandte Chemie (International ed. In English). 56 (33), 9680-9703 (2017).
- Minks, C., Alefelder, S., Moroder, L., Huber, R., Budisa, N. Towards new protein engineering: In vivo building and folding of protein shuttles for drug delivery and targeting by the selective pressure incorporation (SPI) method. Tetrahedron. 56 (48), 9431-9442 (2000).
- Hoesl, M. G., Budisa, N. Expanding and engineering the genetic code in a single expression experiment. Chembiochem: A, European Journal of Chemical Biology. 12 (4), 552-555 (2011).
- Wiltschi, B., Budisa, N. Natural history and experimental evolution of the genetic code. Applied Microbiology and Biotechnology. 74 (4), 739-753 (2007).
- Hoesl, M. G., et al. Lipase congeners designed by genetic code engineering. ChemCatChem. 3 (1), 213-221 (2011).
- . FPbase avGFP Available from: https://www.fpbase.org/protein/avgfp/ (2011)
- Tsien, R. Y. The green fluorescent protein. Annual Review of Biochemistry. 67, 509-544 (1998).
- Zhang, Y., et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 418 (6895), 331-335 (2002).
- Misteli, T., Spector, D. L. Applications of the green fluorescent protein in cell biology and biotechnology. Nature Biotechnology. 15 (10), 961-964 (1997).
- Hanson, M. R., Köhler, R. H. GFP imaging: methodology and application to investigate cellular compartmentation in plants. Journal of Experimental Botany. 52 (356), 529-539 (2001).
- Sakamoto, S., Shoyama, Y., Tanaka, H., Morimoto, S. Application of green fluorescent protein in immunoassays. Advances in Bioscience and Biotechnology. 05 (6), 557-563 (2014).
- Akbar, M., Kim, H. Y. Green fluorescent protein tagging: A novel tool in biomedical research. Indian Journal of Biotechnology. 4, 466-470 (2005).
- Kobayashi, T., Morone, N., Kashiyama, T., Oyamada, H., Kurebayashi, N., Murayama, T. Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PloS One. 3 (12), 3822 (2008).
- Kent, K. P., Oltrogge, L. M., Boxer, S. G. Synthetic control of green fluorescent protein. Journal of the American Chemical Society. 131 (44), 15988-15989 (2009).
- Born, J., Pfeifer, F. Improved GFP variants to study gene expression in Haloarchaea. Frontiers in Microbiology. 10, 1200 (2019).
- Pakhomov, A. A., Martynov, V. I. GFP family: structural insights into spectral tuning. Chemistry & Biology. 15 (8), 755-764 (2008).
- Zimmer, M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chemical Reviews. 102 (3), 759-781 (2002).
- Valbuena, F. M., Fitzgerald, I., Strack, R. L., Andruska, N., Smith, L., Glick, B. S. A photostable monomeric superfolder green fluorescent protein. Traffic. 21 (8), 534-544 (2020).
- Craggs, T. D. Green fluorescent protein: structure, folding and chromophore maturation. Chemical Society Reviews. 38 (10), 2865-2875 (2009).
- Maddalo, S. L., Zimmer, M. The role of the protein matrix in green fluorescent protein fluorescence. Photochemistry and Photobiology. 82 (2), 367-372 (2006).
- Cui, G., Lan, Z., Thiel, W. Intramolecular hydrogen bonding plays a crucial role in the photophysics and photochemistry of the GFP chromophore. Journal of the American Chemical Society. 134 (3), 1662-1672 (2012).
- Yang, F., Moss, L. G., Phillips, G. N. The molecular structure of green fluorescent protein. Nature Biotechnology. 14 (10), 1246-1251 (1996).
- Andrews, B. T., Roy, M., Jennings, P. A. Chromophore packing leads to hysteresis in GFP. Journal of Molecular Biology. 392 (1), 218-227 (2009).
- Xie, J. -. B., Zhou, J. -. M. Trigger factor assisted folding of green fluorescent protein. 생화학. 47 (1), 348-357 (2008).
- Steiner, T., Hess, P., Bae, J. H., Wiltschi, B., Moroder, L., Budisa, N. Synthetic biology of proteins: tuning GFPs folding and stability with fluoroproline. PloS One. 3 (3), 1680 (2008).
- Andrews, B. T., Schoenfish, A. R., Roy, M., Waldo, G., Jennings, P. A. The rough energy landscape of superfolder GFP is linked to the chromophore. Journal of Molecular Biology. 373 (2), 476-490 (2007).
- Kubyshkin, V., Davis, R., Budisa, N. Biochemistry of fluoroprolines: the prospect of making fluorine a bioelement. Beilstein Journal of Organic Chemistry. 17, 439-460 (2021).
- Renner, C., Alefelder, S., Bae, J. H., Budisa, N., Huber, R., Moroder, L. Fluoroprolines as tools for protein design and engineering. Angewandte Chemie (International ed. In English). 40 (5), 923-925 (2001).
- Fukuda, H., Arai, M., Kuwajima, K. Folding of green fluorescent protein and the cycle3 mutant. 생화학. 39 (39), 12025-12032 (2000).
- Rosenman, D. J., et al. Green-lighting green fluorescent protein: faster and more efficient folding by eliminating a cis-trans peptide isomerization event. Protein Science: A Publication of the Protein Society. 23 (4), 400-410 (2014).
- Vitagliano, L., Berisio, R., Mastrangelo, A., Mazzarella, L., Zagari, A. Preferred proline puckerings in cis and trans peptide groups: implications for collagen stability. Protein Science: A Publication of the Protein Society. 10 (12), 2627-2632 (2001).
- Kubyshkin, V. Experimental lipophilicity scale for coded and noncoded amino acid residues. Organic and Biomolecular Chemistry. 19 (32), 7031-7040 (2021).
- Kubyshkin, V., Budisa, N. cis-trans-Amide isomerism of the 3,4-dehydroproline residue, the ‘unpuckered’ proline. Beilstein Journal of Organic Chemistry. 12, 589-593 (2016).
- Budisa, N. Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angewandte Chemie (International ed. In English). 43 (47), 6426-6463 (2004).
- Beatty, K. E., Tirrel, D. A. Noncanonical amino acids in protein science and engineering. Protein Engineering. Nucleic Acids and Molecular Biology. 22, (2009).
- Budisa, N. . Engineering the Genetic Code: Expanding the Amino Acid Repertoire for the Design of Novel Proteins. , (2006).
- Merkel, L., Budisa, N. Organic fluorine as a polypeptide building element: in vivo expression of fluorinated peptides, proteins and proteomes. Organic & Biomolecular Chemistry. 10 (36), 7241-7261 (2012).
- van Eldijk, M. B., van Hest, J. C. M. Residue-specific incorporation of non-canonical amino acids for protein engineering. Methods in Molecular Biology. 1728, 137-145 (2018).
- Hartman, M. C. T. Non-canonical amino acid substrates of E. coli aminoacyl-tRNA synthetases. Chembiochem: A European Journal of Chemical Biology. , (2021).
- Gomez, M. A. R., Ibba, M. Aminoacyl-tRNA synthetases. RNA. 26 (8), 910-936 (2020).
- Grant, M. M., Brown, A. S., Corwin, L. M., Troxler, R. F., Franzblau, C. Effect of l-azetidine 2-carboxylic acid on growth and proline metabolism in Escherichia coli. Biochimica et Biophysica Acta. 404 (2), 180-187 (1975).
- Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J., Huber, R. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. European Journal of Biochemistry. 230 (2), 788-796 (1995).
- Budisa, N., Pal, P. P. Designing novel spectral classes of proteins with a tryptophan-expanded genetic code. Biological Chemistry. 385 (10), 893-904 (2004).
- Hoesl, M. G., Budisa, N. Recent advances in genetic code engineering in Escherichia coli. Current Opinion in Biotechnology. 23 (5), 751-757 (2012).
- Nickling, J. H., et al. Antimicrobial peptides produced by selective pressure incorporation of non-canonical amino acids. Journal of Visualized Experiments: JoVE. (135), e57551 (2018).
- JoVE. Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: Electroporation. JoVE Science Education Database. , (2021).
- JoVE. Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: The Heat Shock Method. JoVE Science Education Database. , (2021).
- Sarkisyan, K. S., et al. fluorescent protein with anionic tryptophan-based chromophore and long fluorescence lifetime. Biophysical Journal. 109 (2), 380-389 (2015).
- Sarkisyan, K. S., et al. KillerOrange, a genetically encoded photosensitizer activated by blue and green light. PloS One. 10 (12), 0145287 (2015).
- Grigorenko, B. L., Krylov, A. I., Nemukhin, A. V. Molecular modeling clarifies the mechanism of chromophore maturation in the green fluorescent protein. Journal of the American Chemical Society. 139 (30), 10239-10249 (2017).
- Pletnev, V. Z., et al. Structure of the green fluorescent protein NowGFP with an anionic tryptophan-based chromophore. Acta Crystallographica. Section D, Biological Crystallography. 71, 1699-1707 (2015).
- Neidhardt, F. C., Bloch, P. L., Smith, D. F. Culture medium for enterobacteria. Journal of Bacteriology. 119 (3), 736-747 (1974).
- Hörnsten, E. G. On culturing Escherichia coli on a mineral salts medium during anaerobic conditions. Bioprocess Engineering. 12 (3), 157-162 (1995).
- Davis, B. D. The Isolation of biochemically deficient mutants of bacteria by means of penicillin. Proceedings of the National Academy of Sciences of the United States of America. 35 (1), 1-10 (1949).
- Sambrook, J., Russell, D. W. . Molecular Cloning: A Laboratory Manual. , (2001).
- Wang, Y. -. S., et al. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Molecular bioSystems. 7 (3), 714-717 (2011).
- JoVE. Basic Methods in Cellular and Molecular Biology. Separating Protein with SDS-PAGE. JoVE Science Education Database. , (2021).
- . ImageJ Available from: https://imagej.nih.gov/ij/ (2021)
- Baumann, T., et al. Engineering ‘Golden’ fluorescence by selective pressure incorporation of non-canonical amino acids and protein analysis by mass spectrometry and fluorescence. Journal of Visualized Experiments: JoVE. (134), e57017 (2018).
- . FPbase NowFFP Available from: https://www.fpbase.org/protein/nowgfp/ (2021)
- Markwardt, M. L., et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PloS One. 6 (3), 17896 (2011).
- Gotthard, G., von Stetten, D., Clavel, D., Noirclerc-Savoye, M., Royant, A. Chromophore isomer stabilization is critical to the efficient fluorescence of cyan fluorescent proteins. 생화학. 56 (49), 6418-6422 (2017).
- Lelimousin, M., et al. Intrinsic dynamics in ECFP and Cerulean control fluorescence quantum yield. 생화학. 48 (42), 10038-10046 (2009).
- . FPbase mKillerOrange Available from: https://www.fpbase.org/protein/mkillerorange/ (2021)
- Pletneva, N. V., et al. Crystal structure of phototoxic orange fluorescent proteins with a tryptophan-based chromophore. PloS One. 10 (12), 0145740 (2015).
- Royant, A., Noirclerc-Savoye, M. Stabilizing role of glutamic acid 222 in the structure of Enhanced Green Fluorescent Protein. Journal of Structural Biology. 174 (2), 385-390 (2011).
- Larregola, M., Moore, S., Budisa, N. Congeneric bio-adhesive mussel foot proteins designed by modified prolines revealed a chiral bias in unnatural translation. Biochemical and Biophysical Research Communications. 421 (4), 646-650 (2012).
- Yamaguchi, H., Miyazaki, M. Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules. 4 (1), 235-251 (2014).
- Ghisaidoobe, A. B. T., Chung, S. J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. International Journal of Molecular Sciences. 15 (12), 22518-22538 (2014).
- Pal, P. P., et al. Structural and spectral response of Aequorea victoria green fluorescent proteins to chromophore fluorination. 생화학. 44 (10), 3663-3672 (2005).
- Pletnev, S., et al. Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. The Journal of Biological Chemistry. 284 (46), 32028-32039 (2009).
- Kubyshkin, V., Budisa, N. Anticipating alien cells with alternative genetic codes: away from the alanine world. Current Opinion in Biotechnology. 60, 242-249 (2019).
- Minks, C., Huber, R., Moroder, L., Budisa, N. Atomic mutations at the single tryptophan residue of human recombinant annexin V: effects on structure, stability, and activity. 생화학. 38 (33), 10649-10659 (1999).
- Budisa, N., Huber, R., Golbik, R., Minks, C., Weyher, E., Moroder, L. Atomic mutations in annexin V thermodynamic studies of isomorphous protein variants. European Journal of Biochemistry. 253, 1-9 (1998).
- Lin, X., Yu, A. C. S., Chan, T. F. Efforts and challenges in engineering the genetic code. Life. 7, (2017).
- Völler, J. -. S., Budisa, N. Coupling genetic code expansion and metabolic engineering for synthetic cells. Current Opinion in Biotechnology. 48, 1-7 (2017).
- Liu, C. C., Schultz, P. G. Adding new chemistries to the genetic code. Annual Review of Biochemistry. 79, 413-444 (2010).
- Lukesch, M. S., Pavkov-Keller, T., Gruber, K., Zangger, K., Wiltschi, B. Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system. Scientific Reports. 9 (1), 2697 (2019).
- Acevedo-Rocha, C. G., et al. Non-canonical amino acids as a useful synthetic biological tool for lipase-catalysed reactions in hostile environments. Catalysis Science & Technology. 3 (5), 1198 (2013).
- Holzberger, B., Marx, A. Replacing 32 proline residues by a non-canonical amino acid results in a highly active DNA polymerase. Journal of the American Chemical Society. 132 (44), 15708-15713 (2010).
- Mosesso, R., Dougherty, D. A., Lummis, S. C. R. Proline residues in the transmembrane/extracellular domain interface loops have different behaviors in 5-HT3 and nACh receptors. ACS Chemical Neuroscience. 10 (7), 3327-3333 (2019).
- Mosesso, R., Dougherty, D. A., Lummis, S. C. R. Probing proline residues in the prokaryotic ligand-gated ion channel, ELIC. 생화학. 57 (27), 4036-4043 (2018).
- Torbeev, V. Deciphering protein folding using chemical protein synthesis. Total Chemical Synthesis of Proteins. 1st ed. , (2021).
- Torbeev, V. Y., Hilvert, D. Both the cis-trans equilibrium and isomerization dynamics of a single proline amide modulate β2-microglobulin amyloid assembly. Proceedings of the National Academy of Sciences of the United States of America. 110 (50), 20051-20056 (2013).
- Kawakami, T., Ishizawa, T., Murakami, H. Extensive reprogramming of the genetic code for genetically encoded synthesis of highly N-alkylated polycyclic peptidomimetics. Journal of the American Chemical Society. 135 (33), 12297-12304 (2013).
- Cui, Z., Johnston, W. A., Alexandrov, K. Cell-free approach for non-canonical amino acids incorporation into polypeptides. Frontiers in Bioengineering and Biotechnology. 8, 1031 (2020).

